Engineered vs. Natural Exosomes in Chronic Wound Models: A Comparative Analysis for Therapeutic Development
Chronic wounds, characterized by a failure to proceed through an orderly healing process, present a significant clinical challenge.
Engineered vs. Natural Exosomes in Chronic Wound Models: A Comparative Analysis for Therapeutic Development
Abstract
Chronic wounds, characterized by a failure to proceed through an orderly healing process, present a significant clinical challenge. This article provides a comprehensive analysis for researchers and drug development professionals on the therapeutic potential of natural and engineered exosomes. We explore the foundational biology of exosomes, detail advanced engineering methodologies for enhancing their function, address key challenges in translation and optimization, and present a critical comparative evaluation of their efficacy based on current preclinical and clinical evidence. The synthesis of these four intents offers a roadmap for the rational design of next-generation, exosome-based therapies for complex wound healing applications.
Understanding the Native Healer: The Biology and Innate Role of Natural Exosomes in Wound Repair
Exosomes are naturally occurring, nanoscale extracellular vesicles (EVs) with a diameter of 30-150 nm, secreted by virtually all cell types into the extracellular environment [1]. They function as crucial mediators of intercellular communication, facilitating the transfer of bioactive molecules—including proteins, lipids, nucleic acids (RNA, DNA), and metabolites—between cells, thereby influencing the physiological state and behavior of recipient cells [2] [3]. Their biogenesis through the endosomal pathway distinguishes them from other extracellular vesicles, resulting in unique composition and functional properties [4]. Within the context of chronic wound research, natural exosomes derived from sources such as mesenchymal stem cells (MSCs) have demonstrated inherent therapeutic potential, promoting wound healing by modulating inflammation, enhancing angiogenesis, and encouraging tissue remodeling [1] [5]. This review delineates the biogenesis, cargo sorting, and molecular mechanisms of natural exosomes, providing a foundational comparison for evaluating engineered exosome strategies in regenerative medicine.
The Biogenesis Pathway of Natural Exosomes
The formation of exosomes is a meticulously orchestrated intracellular process that culminates in the release of these vesicles for intercellular signaling. The journey begins with endocytosis and progresses through several key stages to the release of exosomes from the cell.
Endocytosis and Early Endosome Formation
The biogenesis of exosomes initiates with the inward budding of the plasma membrane, a process that forms early endosomes [4]. This initial step is regulated by various proteins, including clathrin, which facilitates the formation of clathrin-coated pits, and caveolin-1, a marker protein associated with caveolae generation [4]. The small GTP-binding protein Rab5a serves as a specific marker for early endosomes and plays a pivotal role in regulating vesicle fusion through constant GTP binding and hydrolysis [4]. Knockdown of Rab5 has been shown to decrease exosome excretion, underscoring its importance in this pathway [4].
Formation of Multivesicular Bodies (MVBs) and Intraluminal Vesicles (ILVs)
Early endosomes subsequently mature into late endosomes, where the limiting membrane undergoes inward budding to form intraluminal vesicles (ILVs) within larger organelles known as multivesicular bodies (MVBs) [6]. The formation of ILVs, which are the precursors to exosomes, is driven by several distinct but sometimes overlapping molecular mechanisms:
- ESCRT-Dependent Pathway: The Endosomal Sorting Complex Required for Transport (ESCRT) machinery, comprising ESCRT-0, -I, -II, -III subcomplexes and the ATPase VPS4, operates in a sequential manner to mediate membrane budding and cargo sorting [6]. ESCRT-0 (containing Hrs and STAM) recognizes and recruits ubiquitinated cargo proteins. ESCRT-I and -II are then recruited and cooperate to form a saddle-shaped complex important for ESCRT-III assembly. Finally, ESCRT-III subunits polymerize and, with energy provided by VPS4, drive membrane deformation and fission to generate ILVs [6].
- ESCRT-Independent Pathways: Several alternative mechanisms exist:
- The nSMase2-Ceramide Pathway: Neutral sphingomyelinase 2 (nSMase2) converts sphingomyelin to ceramide. The cone-shaped structure of ceramide induces negative membrane curvature, promoting inward budding of the endosome membrane and ILV formation [6]. This pathway is crucial for sorting certain cargoes, such as proteolipid protein (PLP) in oligodendroglia cells [6].
- Tetraspanin-Enriched Microdomains: Tetraspanin proteins (CD63, CD9, CD81), which are canonical exosome markers, are involved in membrane budding and cargo sorting into ILVs [2] [6]. Their transmembrane domains form a cone-like structure that may promote membrane bending [2].
- The Syndecan-Syntenin-ALIX Pathway: The transmembrane proteoglycan syndecan recruits the adaptor protein syntenin, which in turn binds ALIX. ALIX then recruits ESCRT-III and VPS4 to complete ILV formation, representing a non-canonical ESCRT-dependent pathway [6].
Fate of MVBs: Degradation or Release
Once formed, MVBs face one of two destinies: they can fuse with lysosomes, leading to the degradation of their ILV contents, or they can be transported to and fuse with the plasma membrane [4]. The fusion of MVBs with the plasma membrane is a regulated process involving Rab GTPases (such as Rab27) and SNARE complexes [6]. Upon fusion, the ILVs are released into the extracellular space as exosomes [1] [7].
The following diagram illustrates the complete biogenesis pathway of natural exosomes, from their origin as early endosomes to their release as intercellular messengers.
Cargo Composition and Sorting Mechanisms
The biological activity of exosomes is largely determined by their diverse molecular cargo, which is selectively packaged during the biogenesis process. The composition of this cargo reflects the physiological state of the parent cell and dictates the functional impact on recipient cells.
Diversity of Exosomal Cargo
Natural exosomes carry a complex and heterogeneous mixture of biomolecules:
- Nucleic Acids: Exosomes contain various forms of RNA, including messenger RNA (mRNA), microRNA (miRNA), and other non-coding RNAs [3] [4]. These RNAs can be functionally transferred to recipient cells to alter gene expression and protein translation. Exosomes also carry DNA fragments [3].
- Proteins: The exosomal proteome is enriched in certain protein families, including:
- Tetraspanins (CD63, CD9, CD81): Used as canonical exosome markers and involved in cargo sorting and membrane fusion [2] [8].
- ESCRT Machinery Components (TSG101, ALIX): Often found in exosomes and serve as internal markers [6] [8].
- Heat Shock Proteins (Hsp70, Hsp90): Also used as internal markers [8].
- Membrane Transport and Fusion Proteins (GTPases, Annexins).
- Proteins reflecting the cell of origin's state and function [1] [3].
- Lipids: The exosomal membrane is enriched in cholesterol, sphingomyelin, ceramide, and phosphatidylserine, resembling lipid raft microdomains in cellular membranes. This specific lipid composition contributes to exosome stability, structure, and function [6].
Mechanisms of Cargo Sorting
The selective enrichment of molecules into ILVs is a critical step controlled by specific mechanisms:
- Ubiquitin-Dependent Sorting: The ESCRT-0 complex recognizes ubiquitinated proteins, initiating their sorting into ILVs [6].
- Lipid-Mediated Sorting: Ceramide, generated by nSMase2, facilitates the sorting of specific cargoes like proteolipid protein (PLP) in an ESCRT-independent manner [6].
- Tetraspanin-Mediated Sorting: Tetraspanins organize membrane microdomains and facilitate the sorting of associated proteins into exosomes [2] [6].
- RNA Sorting: The mechanisms for RNA sorting into exosomes are an active area of research. Certain RNA-binding proteins, such as SAFB and hnRNPK, have been implicated in this process, sometimes through interactions with the nSMase2-ceramide pathway or other yet-to-be-defined mechanisms [6].
Table 1: Key Cargo Components of Natural Exosomes and Their Proposed Functions
| Cargo Category | Specific Examples | Proposed Functions in Exosome Biology |
|---|---|---|
| Surface Proteins | Tetraspanins (CD63, CD9, CD81) | Vesicle identity, cargo sorting, cell targeting, membrane fusion [2] [8] |
| Intracellular Proteins | ESCRT components (TSG101, ALIX), Heat Shock Proteins (Hsp70, Hsp90) | MVB biogenesis, vesicle scaffolding, stress response [6] [8] |
| Nucleic Acids | miRNAs (e.g., miR-21, miR-29b), mRNAs, other non-coding RNAs | Epigenetic reprogramming of recipient cells, regulation of protein synthesis, intercellular communication [3] [4] |
| Lipids | Cholesterol, Ceramide, Phosphatidylserine | Membrane stability, structural integrity, signaling [6] |
Exosome Uptake and Intercellular Communication
Following their release, exosomes mediate intercellular communication by transferring their cargo to recipient cells. The process of uptake and functional delivery is multifaceted.
- Modes of Uptake: Recipient cells internalize exosomes through various endocytic pathways, including clathrin-dependent endocytosis, caveolin-mediated uptake, macropinocytosis, phagocytosis, and lipid raft-mediated internalization [9] [8]. Exosomes can also directly fuse with the plasma membrane of the target cell [9].
- Functional Cargo Delivery: Upon internalization, the exosomal cargo is released into the cytoplasm of the recipient cell. The delivered miRNAs, mRNAs, and proteins can then modulate cellular signaling pathways, alter gene expression, and ultimately influence the recipient cell's phenotype and function [2] [4]. This mechanism is fundamental to the role of exosomes in both physiological processes and disease progression.
The Role of Natural Exosomes in Wound Healing: Mechanisms and Evidence
Natural exosomes, particularly those derived from mesenchymal stem cells (MSCs), play a multifaceted role in orchestrating the complex process of wound healing. Their therapeutic effects are mediated through the coordinated regulation of different cellular players and signaling pathways across the various phases of healing.
Key Mechanistic Pathways in Wound Repair
Exosomes promote healing by modulating several critical pathways:
- Modulation of Inflammation: MSC-derived exosomes can promote the polarization of macrophages from a pro-inflammatory (M1) phenotype to an anti-inflammatory, pro-healing (M2) phenotype, thereby reducing excessive inflammation in chronic wounds [3].
- Promotion of Angiogenesis: Exosomes transfer pro-angiogenic miRNAs and proteins (e.g., from endothelial progenitor cells) that activate signaling pathways such as PI3K/Akt and ERK in endothelial cells, stimulating the formation of new blood vessels (angiogenesis) crucial for supplying nutrients and oxygen to the healing tissue [1] [5].
- Enhancement of Cell Proliferation and Migration: Exosomes derived from sources like adipose-derived MSCs have been shown to enhance the proliferation and migration of fibroblasts and keratinocytes, the key cells responsible for tissue rebuilding and re-epithelialization [1] [7]. This is achieved by regulating collagen synthesis and other components of the extracellular matrix (ECM).
Preclinical and Clinical Evidence
A growing body of evidence supports the efficacy of natural exosomes in wound healing:
- Animal Models: Studies in mouse, rat, rabbit, and canine models of diabetic and full-thickness wounds have demonstrated that exosome treatment can accelerate wound closure, improve angiogenesis, and enhance the quality of the regenerated tissue [1]. For instance, preliminary data indicate that MSC-derived exosomes can improve healing rates by 30-50% in diabetic models [10].
- Human Trials: While the clinical application is still emerging, early-phase clinical studies suggest a decrease in scarring and chronic wound inflammation following exosome therapy [10]. ClinicalTrials.gov lists ongoing trials evaluating exosome-based therapies for chronic wounds, indicating the transition from preclinical to clinical investigation [1].
Table 2: Experimental Evidence for Natural Exosome Therapeutics in Wound Healing
| Exosome Source | Model System | Key Experimental Findings | Reference |
|---|---|---|---|
| Mesenchymal Stem Cells (MSCs) | Diabetic mouse model | Improved healing rates by 30-50%, enhanced angiogenesis, modulation of collagen I:III ratio | [1] [10] |
| Adipose-Derived MSCs | Human chronic wound fibroblasts in vitro | Induced proliferation and migration of fibroblasts; enhanced in vitro angiogenesis | [7] |
| Endothelial Progenitor Cells | Cutaneous wound mouse model | Accelerated wound healing by promoting angiogenesis through Erk1/2 signaling | [10] |
| Umbilical Cord MSCs | In vitro fibroblast culture | Suppressed myofibroblast differentiation, suggesting potential for reducing scar formation | [10] |
The Scientist's Toolkit: Essential Reagents and Protocols
This section details critical reagents and methodologies employed in exosome research, providing a resource for experimental design and replication.
Table 3: Key Research Reagent Solutions for Exosome Studies
| Reagent / Material | Primary Function in Research | Specific Examples & Notes |
|---|---|---|
| GW4869 | Pharmacological inhibitor of nSMase2; blocks the ceramide-dependent pathway of exosome biogenesis. Used to investigate biogenesis mechanisms and reduce exosome secretion in vitro. | Validated in multiple cell lines (e.g., oligodendroglia, neuronal, cancer cells) to inhibit sorting of specific cargoes like PLP [6]. |
| Tetraspanin Antibodies | Identification, isolation, and characterization of exosomes via immunoaffinity capture. | Anti-CD63, anti-CD9, anti-CD81 antibodies are widely used for immunocapture, Western blotting, and flow cytometry [2] [8]. |
| ESCRT Component Antibodies | Detection and validation of exosomes via Western blotting; functional studies of biogenesis. | Antibodies against TSG101, ALIX, and Hrs are standard for confirming exosomal identity in isolates [6] [8]. |
| Polyethylene Glycol (PEG) | Polymer used for precipitating exosomes from biological fluids and cell culture media. | Common in commercial exosome isolation kits (e.g., ExoQuick-TC). Offers simplicity but may co-precipitate contaminants [8]. |
| Protease/RNase Inhibitors | Preservation of exosomal cargo integrity during isolation and purification procedures. | Essential for downstream -omics analyses (proteomics, transcriptomics) to prevent degradation of proteins and RNAs [3]. |
Standardized Experimental Workflow
A typical workflow for isolating and validating natural exosomes for functional studies involves several key steps, visualized in the diagram below.
Detailed Key Protocols:
- Isolation via Ultracentrifugation: The "gold standard" method [8]. Cell culture supernatant is subjected to sequential centrifugation steps: low-speed (e.g., 300 × g to remove cells), medium-speed (e.g., 10,000 × g to remove cell debris and larger vesicles), and finally high-speed ultracentrifugation (e.g., 100,000 × g for 70-120 minutes) to pellet exosomes. The pellet is washed in PBS and re-pelleted by another ultracentrifugation step to purify further [8].
- Characterization - Nanoparticle Tracking Analysis (NTA): This technique measures the size distribution and concentration of particles in an exosome preparation by tracking the Brownian motion of individual vesicles in a suspension using a laser microscope [8].
- Characterization - Western Blotting: Isolated exosomes are lysed and analyzed for the presence of positive markers (e.g., CD63, CD81, TSG101, Alix) and the absence of negative markers from the parent cells (e.g., calnexin, GRP94) to confirm purity and identity [8].
- Functional Assay - In Vitro Scratch/Wound Healing Assay: Recipient cells (e.g., fibroblasts or keratinocytes) are cultured to confluence. A scratch is made in the monolayer, and cells are treated with exosomes or PBS control. The rate of gap closure is monitored over time (e.g., 0, 24, 48 hours) via microscopy, quantifying the pro-migratory effect of exosomes [7].
Natural exosomes represent a sophisticated and intrinsic system of intercellular communication, with a well-defined biogenesis pathway originating from multivesicular bodies and a diverse cargo that dictates their functional role in tissue homeostasis and repair. In chronic wound models, their inherent ability to coordinate complex processes like immunomodulation, angiogenesis, and cell proliferation makes them potent therapeutic agents and a critical biological benchmark. A thorough understanding of their formation, cargo sorting, and mechanism of action, as detailed in this guide, is fundamental for researchers and drug development professionals. This knowledge provides the essential foundation for the rational design and objective evaluation of engineered exosome strategies, which aim to augment these natural capabilities for enhanced therapeutic outcomes in regenerative medicine.
The therapeutic application of Mesenchymal Stem Cells (MSCs) in wound healing has progressively shifted from a cell-replacement paradigm to a paracrine-focused model, wherein secreted vesicles mediate most regenerative effects [11] [12]. Among these secretions, MSC-derived exosomes—nanoscale extracellular vesicles (30-150 nm)—have emerged as potent facilitators of tissue repair. These vesicles transport bioactive cargoes including proteins, lipids, mRNAs, and microRNAs (miRNAs), facilitating intercellular communication [11] [13]. In the context of chronic wounds, which are characterized by a failure to proceed through an orderly healing process within three months, MSC-derived exosomes target key pathological aspects: persistent inflammation, impaired angiogenesis, and dysfunctional fibroblast activity [11] [5]. This review delineates the mechanistic roles of MSC-derived exosomes in modulating these core cellular players, providing a comparative analysis of supporting experimental data within the broader research framework of engineered versus natural exosomes.
Mechanistic Insights: How MSC-Exosomes Modulate Key Cellular Processes
Modulation of the Inflammatory Response
The transition from the pro-inflammatory (M1) to the anti-inflammatory (M2) macrophage phenotype is a critical checkpoint for resolving the inflammatory phase and initiating productive healing. MSC-derived exosomes significantly expedite this transition [14] [12].
- Macrophage Polarization: In a rat full-thickness wound model, exosomes loaded onto a collagen sponge (sponge-Exo) were shown to effectively promote the shift of macrophages from an inflammatory M1 phenotype to a regenerative M2 phenotype [14]. This modulation helps suppress the prolonged inflammatory state characteristic of chronic wounds.
- Molecular Mechanisms: The anti-inflammatory effect is partly mediated by the delivery of specific microRNAs. For instance, exosomal miR-146a and miR-223 contribute to inflammation resolution by inhibiting the NF-κB signaling pathway and suppressing NLRP3 inflammasome activation, respectively [11]. Furthermore, preconditioned MSC-derived exosomes enhance anti-inflammatory polarization via let-7b signaling [11].
Promotion of Angiogenesis
Adequate blood supply is fundamental for delivering oxygen and nutrients to the wound site. MSC-derived exosomes potently stimulate the formation of new blood vessels [11] [15].
- Stimulating Endothelial Cells: Exosomes derived from human dental pulp stem cells (hDPSCs) were demonstrated to promote angiogenesis in Human Umbilical Vein Endothelial Cells (HUVECs) [14]. This pro-angiogenic effect is replicated by exosomes from various MSC sources, including those from adipose tissue and umbilical cord.
- Key Molecular Mediators: The angiogenic promotion is orchestrated through the delivery of pro-angiogenic factors and miRNAs. Exosomes transport Vascular Endothelial Growth Factor (VEGF) and Fibroblast Growth Factor 2 (FGF-2), which directly stimulate endothelial cell proliferation and sprouting [11]. Notably, hypoxic preconditioning of parent MSCs can further enhance the angiogenic capacity of their exosomes by upregulating cargo such as HIF-1α and VEGF [15].
Activation of Fibroblast Function and ECM Remodeling
Fibroblasts are the primary architects of the new extracellular matrix (ECM). MSC-derived exosomes enhance fibroblast activity to support the proliferative phase of healing.
- Enhancing Proliferation and Migration: Studies confirm that MSC-derived exosomes enhance the migration and proliferation of human dermal fibroblasts (HDFs) [14]. This is crucial for populating the wound bed with matrix-producing cells.
- Collagen Synthesis and ECM Regulation: Exosomes promote fibroblast secretion of type III collagen and fibronectin, forming the granulation tissue scaffold [11]. This process is facilitated by the delivery of miRNAs like miR-21-5p and miR-29a-5p, which are significantly upregulated in healing exosomes and target genes involved in cell migration and ECM dynamics [14]. The activation of transforming growth factor-β1 (TGF-β1) in fibroblasts further stimulates ECM synthesis [11].
Table 1: Key Cargos in MSC-Derived Exosomes and Their Functions in Wound Healing
| Exosomal Cargo | Type | Primary Function in Wound Healing | Experimental Evidence |
|---|---|---|---|
| miR-146a | miRNA | Inhibits NF-κB signaling, resolves inflammation [11]. | In vitro macrophage studies [11]. |
| miR-223 | miRNA | Suppresses NLRP3 inflammasome activation [11]. | In vitro macrophage studies [11]. |
| miR-21-5p | miRNA | Enhances fibroblast migration and proliferation [14]. | NGS analysis of exosomes from rat model [14]. |
| miR-29a-5p | miRNA | Promotes cellular proliferation and modulates ECM [14]. | NGS analysis of exosomes from rat model [14]. |
| VEGF | Protein | Stimulates angiogenesis and endothelial cell growth [11] [15]. | In vitro HUVEC tube formation assays [15]. |
| FGF-2 | Protein | Promotes angiogenesis and fibroblast proliferation [11]. | In vitro studies with fibroblasts and endothelial cells [11]. |
| TGF-β1 | Protein/Cytokine | Activates fibroblasts for ECM synthesis [11]. | In vitro fibroblast activation studies [11]. |
Diagram 1: Multimodal Mechanism of MSC-Derived Exosomes in Wound Healing. This diagram illustrates how a single exosome simultaneously coordinates three key healing processes by delivering specific molecular cargo to different target cells.
Comparative Therapeutic Platforms: Natural vs. Engineered Exosomes
While natural exosomes show inherent therapeutic potential, bioengineering strategies are being employed to enhance their efficacy, stability, and specificity, forming a critical comparison in modern research.
Natural Exosomes
Natural exosomes are isolated directly from MSC cultures without further modification. Their efficacy can be influenced by the MSC source and preconditioning strategies.
- Source-Dependent Efficacy: A meta-analysis of preclinical studies indicated that among commonly used MSC sources, Adipose-Derived Stem Cells (ADSCs) demonstrated the best effect on wound closure rate and collagen deposition, while Bone Marrow MSCs (BMMSCs) were more effective in promoting revascularization [16].
- Preconditioning: Modifying the cell microenvironment prior to exosome collection can enhance the therapeutic potency of the resulting natural exosomes. For example, preconditioning MSCs under hypoxic conditions or with specific biochemical cues like 3,3′-diindolylmethane (DIM) can upregulate pro-regenerative cargoes such as Wnt11, thereby enhancing their wound-healing capacity [15].
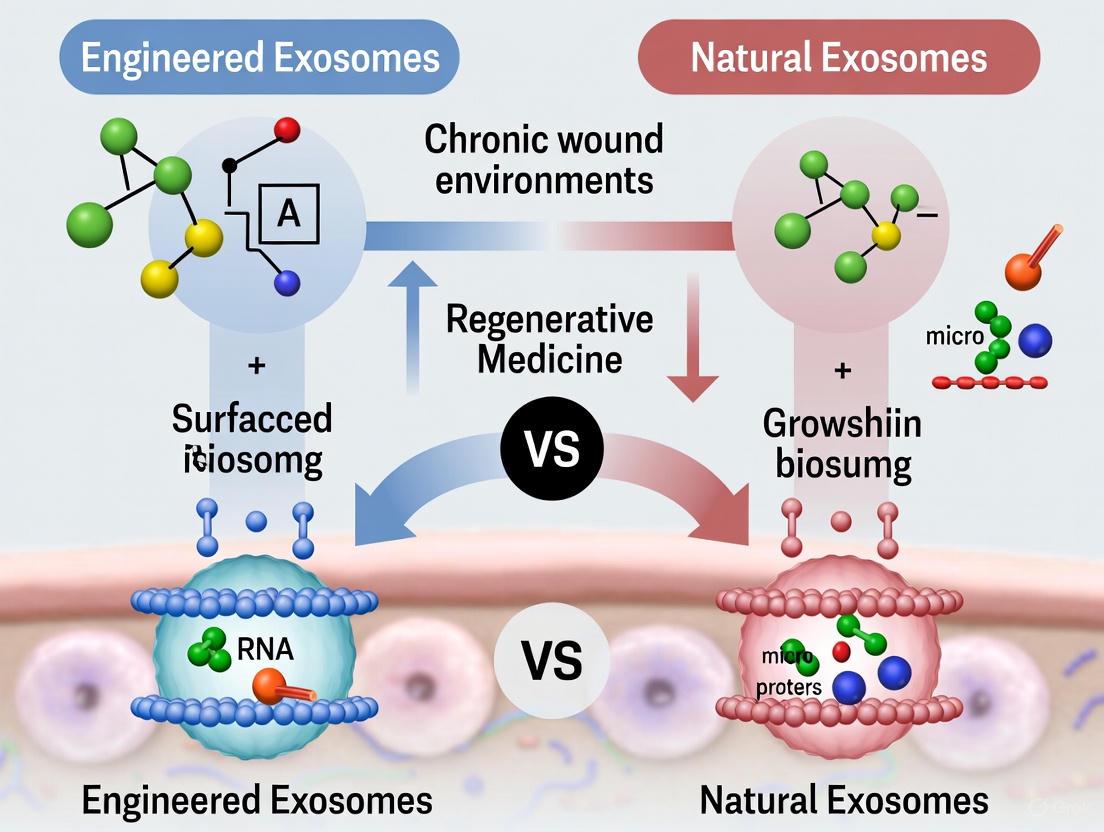
Engineered Exosomes (eExo)
Engineered exosomes are designed to overcome the limitations of natural exosomes, such as rapid clearance and non-specific uptake [5] [17]. Engineering strategies focus on cargo loading and surface modification.
- Cargo Loading: This involves loading specific therapeutic molecules (e.g., miRNAs, proteins) into exosomes to enhance their biological activity. For instance, engineering synovial MSCs to overexpress miR-126-3p resulted in exosomes that more effectively promoted the proliferation of epidermal fibroblasts and vascular endothelial cells [15].
- Surface Modification: Altering the surface proteins of exosomes can improve their targeting specificity and retention at the wound site. This is often achieved by transfecting parent cells with plasmids encoding targeting peptides or directly modifying purified exosomes via click chemistry [5].
Table 2: Comparison of Natural and Engineered MSC-Derived Exosomes
| Feature | Natural Exosomes | Engineered Exosomes (eExo) |
|---|---|---|
| Definition | Vesicles isolated without modification from MSC cultures. | Vesicles modified to enhance cargo or targeting properties. |
| Key Advantages | Innate biocompatibility; inherent biological activity; simpler production [13]. | Enhanced targeting; increased therapeutic payload; improved stability and retention [5] [17]. |
| Primary Limitations | Heterogeneous cargo; rapid clearance; potential off-target effects [12] [5]. | More complex manufacturing; higher cost; need for stringent safety profiling [5]. |
| Example Strategy | Preconditioning MSCs with hypoxia to boost pro-angiogenic cargo [15]. | Overexpressing miR-126-3p in parent MSCs to enhance pro-healing effects [15]. |
| Ideal Use Case | Initial proof-of-concept studies; platforms for holistic therapy. | Targeting specific pathological pathways; overcoming delivery barriers. |
Experimental Data and Methodologies
Supporting Animal Model Data
Robust preclinical data from animal models underpins the therapeutic potential of MSC-derived exosomes. A comprehensive meta-analysis of 83 preclinical studies confirmed that MSC-derived extracellular vesicles significantly enhance wound closure rate, reduce scar width, and increase blood vessel density and collagen deposition in both diabetic and non-diabetic animal models [16]. The analysis further revealed that the subcutaneous injection of exosomes demonstrated a greater improvement in wound closure and revascularization compared to topical application via dressing/covering [16].
Key Experimental Protocols
Standardized methodologies are critical for the isolation and characterization of exosomes, ensuring the reproducibility and validity of experimental data.
- Exosome Isolation and Purification (Differential Ultracentrifugation):
- Cell Culture: MSC conditioned medium is collected after a period of culture in exosome-depleted serum.
- Centrifugation Steps: The medium is sequentially centrifuged at:
- 500 × g for 10 minutes to remove cells.
- 2,000 × g for 10 minutes to remove dead cells.
- 10,000 × g for 30 minutes to remove cell debris.
- The supernatant is filtered through a 0.22 μm filter.
- Ultracentrifugation at 100,000 × g for 70 minutes to pellet exosomes.
- The pellet is washed in PBS and ultracentrifuged again at 100,000 × g for 70 minutes [14] [17].
- Exosome Characterization:
- Nanoparticle Tracking Analysis (NTA): Used to determine the size distribution and concentration of particles [14].
- Transmission Electron Microscopy (TEM): Employed to confirm the spherical, cup-shaped morphology and size of exosomes [14].
- Western Blotting: Used to detect the presence of exosomal marker proteins (e.g., CD63, CD9, CD81, TSG101) and the absence of negative markers (e.g., calnexin) [14] [16].
Table 3: The Scientist's Toolkit: Essential Reagents and Materials for Exosome Research
| Reagent / Material | Function / Application | Example Usage in Experiments |
|---|---|---|
| Collagen Sponge/Hydrogel | A biomaterial scaffold for exosome delivery; provides sustained release and protects exosome bioactivity [14] [18]. | Used as "sponge-Exo" in rat models to gradually release exosomes, promoting healing [14]. |
| Dulbecco's Modified Eagle Medium (DMEM) | Base cell culture medium for culturing MSCs and producing conditioned medium for exosome isolation [14]. | Standard medium for hDPSC culture prior to exosome collection [14]. |
| Fetal Bovine Serum (FBS) | Nutrient supplement for cell culture. Must be centrifuged to remove bovine vesicles for exosome-production cultures. | Used in MSC proliferation medium [14]. |
| Antibodies (CD63, CD9, CD81) | Key reagents for characterizing exosomes via Western Blot or flow cytometry, confirming vesicle identity. | Detection of positive exosomal markers during characterization [14] [17]. |
| Phenylmethanesulfonyl fluoride (PMSF) | Protease inhibitor added to lysis buffers to prevent protein degradation during exosome protein extraction. | Used in protein extraction for Western Blot analysis of exosomal cargo [14]. |
| Streptozotocin (STZ) | Chemical used to induce type 1 diabetes in rodent models for creating diabetic wound models. | Used in 30 of the reviewed preclinical studies to model diabetic wounds [16]. |
Diagram 2: Standard Workflow for MSC-Exosome Isolation and Characterization. This diagram outlines the key experimental steps from cell culture to the final application of exosomes, highlighting critical quality control checkpoints.
MSC-derived exosomes represent a sophisticated cell-free therapeutic platform that coordinately addresses the multifaceted pathology of chronic wounds. By simultaneously modulating inflammation, promoting angiogenesis, and activating fibroblasts, they effectively shift the wound environment from a state of chronic stagnation to one of active regeneration. The compelling preclinical data, consolidated through systematic reviews, provides a strong foundation for clinical translation. The ongoing evolution from natural to precision-engineered exosomes (eExo) promises to further enhance therapeutic efficacy by optimizing drug delivery and targeting specific pathological pathways. Future research must focus on standardizing isolation protocols, scaling up production under Good Manufacturing Practice (GMP) guidelines, and conducting rigorous safety and efficacy clinical trials to fully realize the potential of this promising therapy for patients with chronic wounds.
Chronic wounds, including diabetic foot ulcers, venous leg ulcers, and pressure injuries, represent a significant clinical and economic burden worldwide [19] [20]. These "hard-to-heal" wounds are conceptually defined as wounds that have not reduced in size by more than 40-50% or healed within one month, exhibiting a slow rate of size reduction of ≤1 mm/week [19]. The underlying pathology of chronic wounds deviates fundamentally from the normal, highly coordinated healing process, which progresses through hemostasis, inflammation, proliferation, and remodeling phases [21].
At the cellular and molecular level, chronic wounds are propelled and distinguished by a triad of interplaying loops involving persistent inflammation, oxidative stress, and cellular senescence [19]. This pathological microenvironment creates self-sustaining cycles that prevent healing progression. Decades of research have focused on identifying key endogenous, predisposing factors that drive both chronicity and recurrence, with emerging evidence pointing toward the existence of an epigenetic pathologic code that originates and perpetuates a "chronic wound memory" sheltered in dermal fibroblasts and keratinocytes [19].
Within this complex pathological landscape, exosome-based therapies have emerged as promising regenerative strategies. This review systematically compares the therapeutic performance of engineered versus natural exosomes across the core hallmarks of chronic wounds, providing experimental data and methodological guidance for researchers and drug development professionals.
Core Pathological Mechanisms: Beyond the Surface
Persistent Inflammation: A Self-Sustaining Pathological Loop
In normal wound healing, the inflammatory phase is transient, characterized by initial neutrophil infiltration followed by a transition from pro-inflammatory M1 to anti-inflammatory M2 macrophages [21]. In chronic wounds, this resolution fails, creating a self-perpetuating inflammatory environment. CD4+ and CD8+ lymphocytes accumulate in significant numbers within skin wounds, peaking on days 5-10 and 7-10 post-injury, respectively [11]. The sustained presence of pro-inflammatory M1 macrophages leads to continuous production of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and matrix metalloproteinases (MMPs) that degrade the extracellular matrix and damage newly formed tissue [11] [21].
Experimental models demonstrate that this inflammatory dysregulation creates a vicious cycle where persistent inflammation generates oxidative stress, which in turn promotes further inflammatory signaling [19]. In diabetic wound models, elevated pro-inflammatory markers (IL-1β, TNF-α, MMP9) and reduced anti-inflammatory/angiogenic factors (IL-10, VEGF-A) reflect the chronic inflammatory and angiogenic imbalance characteristic of non-healing diabetic ulcers [22].
Impaired Angiogenesis: The Vascular Deficit
Angiogenesis, the formation of new blood vessels from existing ones, is crucial for delivering oxygen and nutrients to the wound bed. In chronic wounds, this process is fundamentally impaired due to multiple factors. Hyperglycemia in diabetic wounds creates a systemic cytotoxic environment where advanced glycation end-products (AGEs) accumulate in dermal collagen and impair fibroblast physiology, provoking precocious cutaneous aging while perpetuating chronic inflammation [19].
The diabetic wound microenvironment exhibits disrupted dermal-vascular cell crosstalk and defective angiogenesis, with recent models highlighting endothelial-to-mesenchymal transition (EndMT) as a critical pathological feature under diabetic stress [22]. This results in reduced levels of key angiogenic growth factors, particularly vascular endothelial growth factor (VEGF) and fibroblast growth factor 2 (FGF-2), which are essential for endothelial cell migration and capillary formation [11] [21].
Failed Re-epithelialization: The Barrier Restoration Failure
Re-epithelialization requires the coordinated migration, proliferation, and differentiation of keratinocytes across the wound bed. In chronic wounds, this process is disrupted through multiple mechanisms. The persistent inflammatory environment generates high levels of proteases that degrade growth factors and extracellular matrix components necessary for epithelial migration [19]. Cellular senescence establishes a senescent cell society, particularly of "diseased fibroblasts," and dysfunction of stem cell populations, creating a microenvironment hostile to keratinocyte function [19].
The establishment of a senescence cell society, especially of "diseased fibroblasts," and the dysfunctionality of stem cell populations are significant pathophysiological ingredients for diabetic wound chronicity [19]. This is further compounded by the hyperglycemia-derived imprinting that acts as the foundation of metabolic memory, perpetuating the senescent phenotype in fibroblasts and keratinocytes through an inflammotoxic secretome [19].
Experimental Models: Methodologies for Studying Chronic Wounds
In Vitro Models: From Simple to Complex Systems
Traditional two-dimensional (2D) cell culture systems have provided fundamental insights into cellular behavior but lack the physiological complexity of the wound microenvironment. Standard protocols involve:
Fibroblast-Keratinocyte Co-culture: Human dermal fibroblasts and keratinocytes are cultured in Transwell systems to study paracrine interactions. Fibroblasts are typically seeded in the lower chamber, with keratinocytes in the upper insert, allowing shared medium without direct contact [11].
Macrophage Polarization Assays: Human monocyte cell lines (THP-1) or primary monocytes are differentiated into macrophages using phorbol myristate acetate (PMA), then polarized toward M1 (using LPS and IFN-γ) or M2 (using IL-4) phenotypes to study their effects on other wound cells [11].
Senescence-Associated β-galactosidase Staining: Cells from chronic wound environments are fixed and incubated with X-gal solution at pH 6.0 to detect senescent cells, which show blue staining [19].
Advanced three-dimensional (3D) models better recapitulate the wound environment:
Diabetic Wound-on-a-Chip (DWOC): This microfluidic platform integrates human dermal fibroblasts and macrophages within a collagen I matrix to mimic the dermis, alongside endothelial cells embedded in Matrigel to represent the vascular compartment [22]. The system is subjected to hyperglycemic conditions with added advanced glycation end-products (AGEs) and lipopolysaccharide (LPS), alongside normoglycemic controls [22].
3D Bioprinted Skin Constructs: Fibroblasts and keratinocytes are encapsulated in bioinks (typically collagen-based or synthetic polymers) and printed in layered structures to simulate native skin architecture [1].
In Vivo Models: Preclinical Assessment
Animal models remain essential for evaluating therapeutic interventions in a physiological context:
Diabetic Mouse Models: Type 1 diabetes is induced in C57BL/6 mice using streptozotocin (STZ) injections (50-60 mg/kg for 5 consecutive days). After confirmation of hyperglycemia (>300 mg/dL), full-thickness excisional wounds are created on the dorsal skin using biopsy punches (6-8 mm diameter) [11].
Pressure Ulcer Models: Rats are subjected to controlled pressure application using magnetic plates or indentation systems to create ischemic wounds that simulate pressure injuries [19].
Venous Insufficiency Models: Rodents undergo ligation of femoral veins to create venous hypertension, mimicking human venous leg ulcers [20].
Standard outcome measures include wound closure rate (measured by planimetry), histological analysis (H&E for general morphology, Masson's trichrome for collagen, CD31 immunohistochemistry for vessels), and molecular analysis (ELISA for cytokines, RT-qPCR for gene expression) [11].
Natural vs. Engineered Exosomes: Comparative Therapeutic Profiles
Characterization and Production Metrics
Table 1: Comparative Characterization of Natural and Engineered Exosomes
| Parameter | Natural Exosomes | Engineered Exosomes |
|---|---|---|
| Size Range | 30-150 nm [1] [11] | 40-160 nm [7] |
| Production Yield | Variable depending on cell source and culture conditions [23] | More consistent yields through engineering approaches [7] |
| Isolation Method | Ultracentrifugation, size-exclusion chromatography, polymer precipitation [13] [23] | Similar isolation methods with potential for affinity-based purification [7] |
| Cargo Composition | Proteins, lipids, mRNAs, miRNAs reflecting parental cell state [1] [11] | Enhanced or modified cargo through loading strategies [7] [13] |
| Surface Markers | Tetraspanins (CD9, CD63, CD81), antigen-presenting complexes [13] | Modified surface with targeting peptides or antibodies [7] |
| Storage Stability | Limited; affected by repeated freezing/thawing [23] | Potentially enhanced stability through engineering [7] |
Natural exosomes are isolated from various cellular sources, primarily mesenchymal stem cells (MSCs), adipose-derived stem cells (ADSCs), and immune cells [11] [13]. The production yield varies significantly based on cell source, with MSCs producing substantial exosomes while dendritic cells produce more limited quantities [23]. Preconditioning strategies, including hypoxia, cytokine stimulation, or 3D culture, can enhance yield and modify therapeutic properties [13] [23].
Engineered exosomes are designed to overcome limitations of natural exosomes through three primary strategies:
Surface Engineering: Modifying the exosomal membrane to improve targeting capabilities, circulation time, and uptake by specific cell types [7] [23]. This includes conjugation of targeting peptides (e.g., RGD for integrin targeting) or antibodies via chemical or genetic approaches [7].
Cargo Loading: Incorporating therapeutic effector molecules, such as drugs, RNA, or proteins, into exosomes using electroporation, sonication, extrusion, or incubation methods [7] [23].
Genetic Modification: Manipulating donor cells to express particular proteins or RNAs that are subsequently incorporated into exosomes [23]. This includes transfection of donor cells with genes encoding therapeutic agents [13].
Efficacy Comparison in Chronic Wound Models
Table 2: Comparative Efficacy of Natural vs. Engineered Exosomes in Chronic Wound Models
| Therapeutic Function | Natural Exosomes | Engineered Exosomes | Experimental Evidence |
|---|---|---|---|
| Anti-inflammatory Effects | Moderate reduction of TNF-α, IL-6; promotion of M2 macrophage polarization [11] | Enhanced anti-inflammatory activity; up to 70% greater reduction in pro-inflammatory cytokines [7] | Diabetic mouse model showing 50% vs. 85% reduction in TNF-α [7] |
| Angiogenic Potential | Increased VEGF expression; improved capillary density [11] | Significantly enhanced angiogenic response; 2.1-fold increase in capillary formation [7] | CD31 immunohistochemistry showing 30% vs. 65% increase in vessel density [7] |
| Re-epithelialization | Accelerated keratinocyte migration and wound closure [11] | Superior epithelial regeneration; near-complete closure 7 days faster [7] | Diabetic wound model showing 60% vs. 95% closure at day 14 [7] |
| Targeting Efficiency | Limited tissue specificity; widespread distribution [7] | Significantly improved targeting to wound site with reduced off-target effects [7] | Fluorescence imaging showing 3.5-fold higher retention in target tissue [7] |
| Collagen Organization | Improved collagen deposition but suboptimal organization [11] | Enhanced collagen alignment and maturation similar to native skin [7] | Histology showing more organized collagen bundles with engineered exosomes [7] |
The therapeutic efficacy of exosomes is influenced by multiple factors, including donor cell condition, dosage, and administration route [23]. Aging in donor cells generally leads to a decline in exosome quality, with exosomes from older BMSCs exhibiting diminished effects in regenerative capabilities [23]. Dosage optimization is critical, with studies in traumatic brain injury models showing that 100 μg exosomes per rat demonstrated more significant efficacy compared to 50 μg or 200 μg groups [23]. The therapeutic dose of exosomes commonly ranges from 10 to 100 μg of protein in mouse models [23].
Experimental Protocols: Key Methodologies for Exosome Research
Exosome Isolation and Characterization Protocol
Standard Ultracentrifugation Protocol:
- Cell Culture: Expand MSC cultures in serum-free medium conditioned for 48-72 hours [13] [23].
- Collection: Collect conditioned medium and perform sequential centrifugation: 300 × g for 10 min (remove cells), 2,000 × g for 20 min (remove dead cells), 10,000 × g for 30 min (remove cell debris) [13].
- Ultracentrifugation: Centrifuge at 100,000 × g for 70 min at 4°C to pellet exosomes [13] [23].
- Washing: Resuspend pellets in PBS and repeat ultracentrifugation at 100,000 × g for 70 min [13].
- Resuspension: Resuspend final pellet in PBS and store at -80°C [23].
Characterization:
- Nanoparticle Tracking Analysis: Dilute exosomes 1:1000 in PBS and analyze using Nanosight NS300 to determine particle size and concentration [13].
- Transmission Electron Microscopy: adsorb exosomes to Formvar-carbon coated grids, stain with 2% uranyl acetate, and image under TEM [13].
- Western Blotting: Confirm presence of exosomal markers (CD9, CD63, CD81) and absence of negative markers (calnexin) [13] [23].
Surface Engineering Protocol: Targeting Peptide Conjugation
Metabolic Labeling and Click Chemistry Approach:
- Parent Cell Incubation: Incubate parent cells (MSCs) with 50 μM azidopropionate mannosamine (Ac4ManNAz) for 3 days to incorporate azide groups onto exosome surfaces [7].
- Exosome Isolation: Isolate exosomes using standard ultracentrifugation protocol [13].
- Conjugation Reaction: React azide-labeled exosomes with DBCO-modified targeting peptides (e.g., RGD, 100 μM) in PBS for 2 hours at room temperature with gentle rotation [7].
- Purification: Remove unreacted peptides using size exclusion chromatography (PBS-equilibrated PD-10 columns) [7].
- Validation: Confirm conjugation efficiency using flow cytometry with fluorescently-labeled secondary antibodies [7].
In Vivo Efficacy Testing Protocol
Diabetic Mouse Wound Healing Model:
- Diabetes Induction: Inject C57BL/6 mice (8-10 weeks old) with streptozotocin (50 mg/kg i.p. for 5 consecutive days) [11].
- Wound Creation: After confirming hyperglycemia (>300 mg/dL), create two full-thickness excisional wounds (6 mm diameter) on the dorsal skin using biopsy punch under anesthesia [11].
- Treatment Administration: Apply exosomes (100 μg in 50 μL PBS) topically to wound bed every 3 days for 15 days [7]. Control groups receive PBS or no treatment.
- Wound Monitoring: Capture digital images daily and calculate wound area using ImageJ software [11].
- Tissue Collection: Harvest wound tissue at days 7, 14, and 21 for histological and molecular analysis [11].
Signaling Pathways: Mechanisms of Action
Exosome-Mediated Signaling in Wound Healing Pathways
The diagram illustrates the key molecular mechanisms through which exosomes target the core pathological hallmarks of chronic wounds. Through delivery of specific microRNAs and proteins, exosomes simultaneously address persistent inflammation, impaired angiogenesis, and failed re-epithelialization [11] [7].
For inflammation resolution, exosomal miR-146a inhibits NF-κB signaling, reducing production of pro-inflammatory cytokines like TNF-α and promoting the transition from M1 to M2 macrophages [11]. In parallel, miR-223 suppresses NLRP3 inflammasome activation, further resolving inflammation [11]. Preconditioned MSC-derived exosomes enhance anti-inflammatory polarization through let-7b signaling [11].
For angiogenesis promotion, exosomal miR-21 plays a pivotal role by inhibiting PTEN, leading to AKT activation and subsequent VEGF upregulation [11]. This stimulates endothelial cell proliferation and capillary formation. Additional angiogenic factors including FGF-2 delivered by exosomes further enhance this process [11] [7].
For re-epithelialization, exosomes from MSCs and ADSCs enhance fibroblast proliferation and migration by delivering miR-29a, which enhances collagen production, and other factors that directly stimulate keratinocyte migration and differentiation [11].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Exosome and Wound Healing Research
| Reagent Category | Specific Examples | Research Application | Key Function |
|---|---|---|---|
| Cell Culture | Mesenchymal Stem Cells (MSCs), Human Dermal Fibroblasts, Keratinocytes | In vitro mechanistic studies | Source of exosomes; wound healing assays [11] [13] |
| Characterization | CD63, CD81, CD9 antibodies, TSG101, Calnexin | Exosome validation | Confirm exosome identity and purity [13] [23] |
| Molecular Biology | miR-146a, miR-21, miR-29a mimics/inhibitors | Mechanism investigation | Modulate exosomal miRNA function [11] [7] |
| Animal Models | Streptozotocin, Biopsy Punches, Wound Imaging Systems | In vivo efficacy testing | Create diabetic wounds; monitor healing [11] [22] |
| Biomaterials | Chitosan hydrogels, Alginate films, Collagen scaffolds | Delivery system development | Enhance exosome stability and retention [1] [24] |
| Cytokine Analysis | TNF-α, IL-1β, IL-6, IL-10, VEGF ELISA kits | Therapeutic response assessment | Quantify inflammatory and angiogenic factors [11] [22] |
Additional critical reagents include 3D culture systems such as the diabetic wound-on-a-chip platform, which integrates multiple cell types within engineered matrices to better mimic the chronic wound microenvironment [22]. Hypoxia chambers are essential for preconditioning cells to enhance exosome therapeutic potential, as hypoxia upregulates pro-angiogenic factors in MSCs [23]. Microfluidic devices enable precise isolation and analysis of exosomes, particularly important for engineered exosome characterization [13] [23].
The comparative analysis of natural versus engineered exosomes reveals a rapidly evolving landscape in chronic wound therapeutics. While natural exosomes demonstrate significant therapeutic potential across the core hallmarks of chronic wounds, engineered exosomes show enhanced efficacy through targeted delivery and optimized cargo. The future of exosome-based therapies lies in the development of precision medicine approaches that account for wound-specific microenvironments, patient-specific factors, and stage-specific healing requirements.
Critical research priorities include standardizing potency assays that correlate exosome characteristics with therapeutic outcomes, optimizing scalable production methodologies for clinical translation, and establishing rigorous biodistribution and safety profiles for engineered variants. As the field progresses, the integration of exosome therapies with advanced biomaterials and personalized medicine approaches holds promise for finally addressing the clinical challenge of chronic wounds.
Exosomes are nanoscale, lipid-bilayer-enclosed extracellular vesicles (EVs) secreted by almost all cell types and are present in virtually all biological fluids [25]. They are fundamental mediators of intercellular communication, facilitating the transfer of bioactive molecules—including proteins, lipids, and various forms of RNA—between cells to influence the behavior and function of recipient cells [1] [7]. In the context of wound healing, this natural cargo is intricately involved in orchestrating the complex sequence of events required for tissue repair [15]. The therapeutic potential of stem cell-derived exosomes, particularly from mesenchymal stem cells (MSCs), has emerged as a promising cell-free strategy, leveraging these innate healing pathways while circumventing challenges associated with whole-cell transplantation, such as tumorigenicity and immune rejection [1].
This guide provides a detailed, evidence-based comparison of the performance of natural exosomes against emerging engineered alternatives in chronic wound models. It is structured within a broader thesis that while natural exosomes provide a multifaceted, innate therapeutic signal, engineered exosomes are being developed to enhance specificity, potency, and stability for recalcitrant healing scenarios.
Mechanisms of Action: Decoding the Natural Cargo
Natural exosomes exert their healing effects by delivering a complex cargo that regulates critical wound healing phases: inflammation, proliferation, and remodeling. The table below summarizes the key cargo components and their primary functions in healing pathways.
Table 1: Key Cargo in Natural Exosomes and Their Roles in Wound Healing
| Cargo Type | Specific Examples | Primary Functions in Wound Healing | Experimental Evidence |
|---|---|---|---|
| microRNAs (miRNAs) | miR-126-3p, miR-21, miR-124a, miR-199a, miR-210, miR-20, miR-429, miR-34a [26] [15] | Promotes angiogenesis, enhances keratinocyte/fibroblast proliferation & migration, regulates inflammation, supports epidermal & hair follicle development [26] [15]. | SMSC-Exos loaded with miR-126-3p shown to stimulate fibroblast & endothelial cell proliferation in vitro; miR-21 & hypoxia-induced miR-210 regulate granulation tissue formation & wound closure [26] [15]. |
| Proteins | Tetraspanins (CD63, CD9, CD81), Syntenin, ALIX, Growth Factors, Cytokines [1] [26] [25] | Regulates exosome biogenesis & targeting; modulates immune signaling, cell adhesion; directly promotes cell growth & angiogenesis [1] [25]. | Exosomes from HIF-1α-overexpressing MSCs showed altered protein/miRNA levels & enhanced angiogenic capacity; surface proteins facilitate recipient cell binding via tetraspanins, integrins, proteoglycans [25] [15]. |
| Lipids | Sphingolipids, Cholesterol, Phospholipids, Phosphatidylserine [1] [3] | Forms membrane structure, protects internal cargo; involved in membrane curvature, budding, & signaling; influences exosome stability & cellular uptake [1] [25]. | Lipid composition (cholesterol, sphingomyelin) contributes to rigidity/stability; external phosphatidylserine in apoptotic bodies attracts macrophages for clearance [1] [26]. |
The coordinated action of this cargo regulates healing through several key pathways, as illustrated in the following experimental workflow for studying these mechanisms.
Diagram 1: Experimental workflow for studying natural exosome mechanisms in wound healing. Key steps include exosome isolation from various mesenchymal stem cell (MSC) sources, functional testing in established wound models, and analysis of their impact on critical healing pathways such as inflammation, proliferation, and angiogenesis.
Performance Comparison: Natural vs. Engineered Exosomes in Chronic Wound Models
The transition from basic mechanistic understanding to therapeutic application requires rigorous comparison in biologically relevant models. The following table synthesizes experimental data from chronic wound studies, directly comparing the performance of natural and engineered exosomes.
Table 2: Experimental Data Comparison in Chronic Wound Models
| Performance Metric | Natural Exosomes | Engineered Exosomes | Experimental Context & Protocol Details |
|---|---|---|---|
| Angiogenic Potential | ↑ Tube formation ~1.5-2x control; improved vascularization in diabetic mouse models [15]. | ↑↑ Tube formation ~2.5-3x control; significantly enhanced vs. natural exosomes via cargo overexpression (e.g., miR-126, VEGF) [7] [15]. | Protocol: HUVEC tube formation assay on Matrigel. Exosomes (50 µg/mL) co-cultured with cells for 4-18h. Vessel branches/nodes quantified. In-vivo, topical application in db/db mouse wound model, histology at day 7-10 for CD31+ vessels [15]. |
| Anti-inflammatory Effect | Promote M1 to M2 macrophage switch; reduce TNF-α, IL-1β in wound fluid by ~40-60% [1] [3]. | Enhanced M2 polarization via targeted delivery of anti-inflammatory miRNAs (e.g., miR-124a); cytokine reduction >70% [7] [15]. | Protocol: Bone marrow-derived macrophages stimulated with LPS ± exosomes (20 µg/mL). M1/M2 markers (iNOS, CD206) via FACS/qPCR after 24h. Wound fluid collected via absorbent foam, cytokines measured by ELISA [1] [15]. |
| Cell Proliferation & Migration | ↑ Fibroblast/keratinocyte migration by ~50-80% in scratch assay; ↑ proliferation by ~30-50% [1] [26]. | ↑↑ Migration >100% vs. control; proliferation ↑ ~70-90% via overexpression of mitogenic miRNAs/proteins [7] [15]. | Protocol: Scratch assay: confluent fibroblasts/keratinocytes scratched, treated with exosomes (50 µg/mL). Wound closure imaged at 0, 12, 24h. Proliferation measured by CCK-8/MTS assay after 48-72h [26] [7]. |
| Wound Closure Rate (In-Vivo) | ~40-60% closure by day 7 in diabetic rodent models [1] [15]. | ~70-90% closure by day 7; faster re-epithelialization and granulation tissue formation [7] [15]. | Protocol: Full-thickness excisional wound (8mm diameter) on db/db mouse back. Exosomes (100 µg in 100 µL PBS) applied topically with hydrogel every 3 days. Wound area quantified via planimetry daily. Tissue harvested for histology at days 7, 14 [15]. |
| Targeting Efficiency | Limited inherent targeting; relies on general tropism [7]. | Significantly enhanced via surface modification (e.g., RGD peptides for endothelial cells, CP05 peptide for keratinocytes) [7]. | Protocol: Fluorescently labeled exosomes applied to wound. After 24h, tissue sections analyzed via fluorescence microscopy/IVIS. Uptake in specific cell types (e.g., endothelial cells, fibroblasts) quantified [7]. |
The molecular pathways through which natural exosome cargo achieves these outcomes are complex and highly coordinated. The following diagram maps the primary signaling mechanisms influenced by key cargo components.
Diagram 2: Signaling pathways influenced by natural exosome cargo. Key cargo components, including specific miRNAs, proteins, and lipids, interact with and regulate multiple cellular pathways central to wound healing. These interactions converge to promote accelerated wound closure and tissue regeneration.
The Scientist's Toolkit: Essential Research Reagents & Materials
Translating the mechanistic insights of exosome biology into experimental data requires a specific toolkit. The following table catalogues essential reagents and their functions based on the methodologies cited in the literature.
Table 3: Key Research Reagent Solutions for Exosome Wound Healing Studies
| Reagent/Material | Function in Research | Example Application in Protocol |
|---|---|---|
| Mesenchymal Stem Cells (MSCs) | Primary cellular source for therapeutic exosome production. | Isolated from human bone marrow, adipose tissue, or umbilical cord. Cultured in serum-free media to avoid bovine EV contamination [1] [3]. |
| Ultracentrifugation System | Gold-standard method for isolating exosomes from cell culture supernatant. | Sequential centrifugation steps: 300g (cells), 2000g (debris), 10,000g (microvesicles), 100,000g+ (exosomes) [7] [25]. |
| Size-Exclusion Chromatography (SEC) | High-purity isolation of exosomes based on size, separates from protein aggregates. | Using columns (e.g., qEV) to fractionate sample; exosomes elute in early fractions separate from contaminating proteins [3]. |
| Nanoparticle Tracking Analysis (NTA) | Characterizes exosome size distribution and concentration. | Instrument (e.g., Malvern Nanosight) tracks Brownian motion of particles in suspension to calculate hydrodynamics diameter [1] [7]. |
| CD63/CD81/CD9 Antibodies | Detect tetraspanin markers for exosome identification via Western Blot (WB) or flow cytometry. | WB confirmation of exosome markers; absence of negative markers (e.g., GM130, Calnexin) ensures purity [26] [25]. |
| Matrigel Basement Membrane Matrix | In-vitro assay for evaluating exosome pro-angiogenic potential. | HUVECs are seeded with exosomes on polymerized Matrigel; tube formation (length, branches, nodes) is quantified [15]. |
| Hydrogel Delivery System (e.g., Chitosan) | Biomaterial scaffold for sustained exosome release at wound site. | Mixing exosomes with hydrogel (e.g., chitosan, hyaluronic acid) protects from degradation and allows controlled local delivery in animal models [1] [15]. |
| db/db or STZ-induced Diabetic Mice | Standard preclinical model for studying chronic wounds (diabetic foot ulcers). | Creating full-thickness excisional wounds to test the efficacy of exosome therapies in an impaired healing environment [15]. |
Natural exosomes function as sophisticated, multi-component signaling packages that coordinately regulate inflammation, proliferation, and remodeling to promote wound healing. The experimental data demonstrate their efficacy in modulating key cellular players and pathways in chronic wound models. However, limitations such as variable potency and limited targeting present opportunities for bioengineering.
The future of exosome therapeutics lies in the intelligent design of engineered vesicles. Strategies such as pre-conditioning parent cells (e.g., with hypoxia or inflammatory cytokines) to alter cargo [15], direct loading of specific therapeutic miRNAs (e.g., miR-126-3p) [7] [15], and surface modification with targeting ligands (e.g., RGD peptides) are actively being pursued [7]. These approaches aim to create next-generation exosome products that retain the beneficial safety profile of natural exosomes while exhibiting enhanced, targeted, and more predictable therapeutic activity for treating complex chronic wounds.
Precision Engineering: Methodologies for Enhancing Exosome Potency and Specificity
The therapeutic efficacy of exosomes in chronic wound healing is profoundly influenced by the strategies employed to load them with therapeutic cargo. The choice of loading technique directly impacts key performance metrics, including cargo encapsulation efficiency, stability of the resulting loaded exosomes, and crucially, the preservation of their biological integrity and function. As research pivots from using natural exosomes to engineered counterparts for enhanced chronic wound therapy, selecting an optimal loading method has become a central focus in biotherapeutic development [5] [7]. This guide provides a objective comparison of the three primary loading strategies—transfection, incubation, and electroporation—based on current experimental data, to inform selection for preclinical chronic wound research.
Comparative Analysis of Cargo Loading Strategies
The following tables summarize the core characteristics and performance data of the three main loading strategies, synthesizing findings from recent studies.
Table 1: Key Parameters and Experimental Outcomes of Cargo Loading Strategies
| Loading Strategy | Mechanism of Action | Optimal Cargo Types | Typical Incubation Parameters | Reported Loading Efficiency | Key Experimental Findings |
|---|---|---|---|---|---|
| Transfection | Genetic modification of parent cells to secrete pre-loaded exosomes [8]. | miRNA, siRNA, plasmid DNA [7] [8]. | Co-culture: 24-48 hours [8]. | Variable; depends on transfection efficiency of parent cells [8]. | Enables spontaneous cargo integration but requires extensive tuning; low to medium yield [8]. |
| Incubation | Passive diffusion via concentration gradient; hydrophobic drugs interact with lipid bilayer [8]. | Small hydrophobic molecules, proteins. | 1-12 hours at Room Temperature (RT) - 37°C [8]. | Lower compared to active loading methods [8] [1]. | Simple and straightforward; increased solubility for hydrophobic drugs [8]. |
| Electroporation | Electric pulses create transient pores in exosome membrane [8]. | siRNA, miRNA, hydrophobic drugs [8]. | Field strength: 125-278 kV/m; Buffer: Low conductivity (e.g., 9×10⁻³ S/m) [27] [8]. | Effective for nucleic acids and hydrophobic drugs [8]. | Can incorporate hydrophobic drugs and nucleic acids; may have lower drug encapsulation capacity than sonication [8] [5]. |
| Sonication | Ultrasonic waves temporarily disrupt exosome membrane [8] [28]. | Small molecules, nucleic acids, proteins. | Not specified in results. | Higher drug encapsulation capacity than electroporation and incubation [8] [1] [5]. | Superior encapsulation property and drug loading efficacy compared to incubation and electroporation [8] [1]. |
Table 2: Functional Advantages and Limitations in Chronic Wound Research
| Loading Strategy | Preservation of Exosome Integrity | Therapeutic Payload in Chronic Wound Models | Major Advantages | Major Limitations |
|---|---|---|---|---|
| Transfection | High; maintains natural exosome biogenesis [8]. | miRNA for immunomodulation (e.g., miR-21) and angiogenesis [7] [24]. | Ideal for stable expression of RNA cargo; uses native cellular machinery [8]. | Low yield and efficiency; complex, time-consuming process [8]. |
| Incubation | High; no physical disruption to membrane [8]. | Angiogenic factors (e.g., VEGF) [24]. | Maximally preserves exosome structure and function; technically simple [8]. | low loading efficiency, particularly for large or hydrophilic molecules [8]. |
| Electroporation | Variable; risk of cargo aggregation and membrane damage [8]. | siRNA against pro-inflammatory targets [8]. | Rapid process; applicable to a wide range of cargo types [8]. | Risk of cargo aggregation and damage to exosome membrane [8]. |
| Sonication | Lower; potential for permanent membrane damage and protein denaturation [8]. | Not specified in results. | Highest reported loading efficiency and encapsulation capacity [8] [1]. | Potential for permanent membrane damage and protein denaturation [8]. |
Detailed Experimental Protocols
Passive Incubation
Principle: This method relies on the passive diffusion of cargo across the exosome membrane, driven by a concentration gradient. It is particularly suitable for small hydrophobic molecules that can partition into the lipid bilayer [8].
Protocol:
- Isolate and purify exosomes from a chosen source (e.g., Mesenchymal Stem Cell culture supernatant) using standard methods like ultracentrifugation or size-exclusion chromatography [29] [3].
- Resuspend the purified exosomes in a suitable buffer, such as Phosphate-Buffered Saline (PBS) or a low-conductivity electroporation buffer.
- Mix the exosome suspension with the therapeutic cargo (e.g., a small molecule drug). Literature often uses a cargo concentration in the range of 10-100 µM, but this requires optimization for specific cargo [8].
- Incubate the mixture for 1 to 12 hours at room temperature or 37°C to allow for diffusion and membrane interaction [8].
- Remove unencapsulated cargo by ultracentrifugation (e.g., 100,000× g for 70 minutes) or using purification columns. The resulting pellet is the cargo-loaded exosomes, which should be resuspended in an appropriate buffer for storage or downstream application [8].
Electroporation
Principle: Application of a controlled electrical field creates transient, nanoscale pores in the exosome's lipid bilayer, allowing hydrophilic cargo such as nucleic acids to enter. The membrane reseals after the pulse [8].
Protocol:
- Prepare exosome and cargo mixture: Combine purified exosomes with the cargo (e.g., siRNA or miRNA) in a low-conductivity electroporation buffer. A typical ratio is 100-200 µg of exosomes with 10-100 pmol of siRNA, though this must be optimized [8].
- Transfer to cuvette: Place the mixture into an electroporation cuvette with a specific gap width (e.g., 4 mm).
- Apply electrical pulse: Use an electroporator system (e.g., Bio-Rad Gene Pulser). Parameters must be optimized; a representative protocol for cells uses a single square wave pulse of 20 msec, 300 V, 2000 µF, and 1000 Ohms [28]. For a microfluidic system with an 80 µm channel height, a voltage amplitude of ~10 V can achieve a field strength of 125 kV/m [27].
- Incubate and purify: Following electroporation, incubate the mixture at room temperature for 10-30 minutes to allow membrane recovery. Remove unincorporated cargo via ultracentrifugation or size-exclusion chromatography [8].
Sonication
Principle: Ultrasonic energy applied to the exosome-cargo mixture generates shear forces that temporarily disrupt the lipid membrane, facilitating cargo influx. This method is noted for its high loading efficiency [8] [1].
Protocol:
- Mix exosomes and cargo: Combine purified exosomes with the therapeutic cargo in a microcentrifuge tube.
- Sonicate: Place the tube in a water bath or on a probe sonicator. A common parameter is to sonicate at a power of 20-40% amplitude for 3-6 cycles of 30 seconds "on" and 30 seconds "off" on ice to prevent overheating [8].
- Recover and purify: After sonication, incubate the mixture at 37°C for 1 hour to allow membrane resealing. Separate loaded exosomes from free cargo using ultracentrifugation or chromatography [8].
Strategic Workflow and Method Selection
The following diagram illustrates the decision-making workflow for selecting an appropriate cargo loading strategy, based on the target cargo and experimental goals.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of loading strategies requires specific reagents and instrumentation. This table lists key solutions used in the protocols cited in this guide.
Table 3: Essential Research Reagents and Materials for Exosome Cargo Loading
| Item Name | Function/Application | Specific Examples from Literature |
|---|---|---|
| Lipofectamine 2000 | A cationic lipid reagent for chemical transfection of parent cells. | Used for lipofection in Vero cell line transfections [30]. |
| TurboFect | A cationic polymer transfection reagent for chemical transfection of parent cells. | Demonstrated superior transfection efficiency in Vero cells compared to electroporation and lentivirus [30]. |
| Low Conductivity Electroporation Buffer | Provides optimal ionic environment for efficient electroporation by minimizing current and heat generation. | Used in continuous-flow electroporation platform (conductivity: 9 × 10⁻³ S/m) [27]. |
| Electroporation Cuvettes / Flow Chips | Vessels that hold the sample during electroporation, with defined electrode gaps to ensure uniform electric field. | 4-mm gapped cuvette for standard electroporation [30]; planar microfluidic flow chip (80 µm channel height) for continuous-flow systems [27]. |
| Size-Exclusion Chromatography (SEC) Columns | For purifying and isolating exosomes from biological fluids or culture supernatants after loading to remove unencapsulated cargo and contaminants. | A method for isolating and purifying exosomes [3] [8]. |
| Polyethylene Glycol (PEG) | Used in precipitation-based methods for isolating exosomes. | A polymer used to precipitate exosomes, facilitating their isolation [8]. |
| Saponin | A surfactant that permeabilizes the exosome membrane by complexing with cholesterol, used as an alternative loading method. | Used in saponin treatment-mediated drug loading into exosomes [8]. |
| Ultracentrifuge | Essential for high-speed pelleting of exosomes during isolation and post-loading purification steps. | The "gold standard" method for isolating exosomes via high-speed centrifugation [29] [8]. |
The move toward engineered exosomes for chronic wound therapy demands robust and efficient cargo loading. No single strategy is universally superior; the choice is a trade-off between loading efficiency, cargo type, and the preservation of exosome function. Incubation offers simplicity and integrity for hydrophobic molecules, electroporation provides versatility for nucleic acids, and transfection enables endogenous loading of genetic material. Sonication stands out where high loading efficiency is the paramount concern. Researchers must align their choice with their therapeutic cargo and the specific pathophysiological targets within the complex chronic wound microenvironment. As the field advances, the refinement of these protocols and the development of novel hybrid methods will be crucial for translating engineered exosome therapies from the bench to the bedside.
The treatment of chronic wounds, a significant global health burden, is being revolutionized by advanced therapeutic strategies involving exosomes and biomaterial scaffolds. A critical factor influencing the efficacy of these strategies is the precise delivery and retention of therapeutics at the dynamic and complex wound site. Surface modification with targeting ligands, such as the Arg-Gly-Asp (RGD) peptide family, has emerged as a powerful technique to enhance the wound homing capabilities of both natural and engineered exosomes, as well as the performance of wound-healing matrices. The RGD motif is a quintessential example, serving as a primary recognition sequence for extracellular integrin receptors that are profoundly involved in cell adhesion, migration, and proliferation during the healing process [31] [32]. This guide provides a comparative analysis of how RGD and similar ligands are engineered to improve targeting, focusing on their application within the burgeoning field of exosome-based therapies for chronic wounds. It objectively compares the performance of different ligand-functionalization approaches, supported by experimental data and detailed methodologies, to inform researchers and drug development professionals.
Ligand Function and Integrin Signaling in Wound Repair
RGD Peptides and Integrin Binding
The RGD sequence is a ubiquitous cell-adhesion motif found in numerous extracellular matrix (ECM) proteins, including fibronectin, vitronectin, and fibrinogen [32] [33]. Its primary function is to act as a ligand for a subset of integrin receptors, including αvβ3, αvβ5, αvβ6, α5β1, and αIIbβ3 [32]. In the context of wound healing, which progresses through hemostasis, inflammation, proliferation, and remodeling phases, integrin-mediated signaling is crucial [33]. The binding of RGD to its cognate integrins on the surface of cells such as fibroblasts, keratinocytes, and endothelial cells promotes their attachment, spreading, and survival, thereby facilitating re-epithelialization, angiogenesis, and the formation of granulation tissue [31] [33]. This makes RGD-functionalized surfaces highly conducive to tissue regeneration.
Key Signaling Pathways
The engagement of RGD with integrins initiates outside-in signaling that activates key intracellular pathways, driving cellular processes essential for repair. A pivotal pathway is the PI3K/AKT axis, which promotes cell survival, growth, and proliferation. For instance, in a study using a self-assembling peptide hydrogel (RGDmix) to deliver human amniotic mesenchymal stem cells (hAMSCs) for wound healing, the incorporation of the RGDSP ligand enhanced the secretion of therapeutic growth factors. This effect was demonstrated to be mediated specifically through the RGDSP/Integrin αv/PI3K/AKT signaling pathway, as silencing either integrin αv or key components of the PI3K/AKT pathway abolished the beneficial paracrine effects [34]. This pathway, along with others, orchestrates the cellular response to RGD-presenting biomaterials, underscoring the ligand's role beyond simple adhesion.
Diagram 1: RGD-activated integrin αv/PI3K/AKT signaling pathway in wound healing.
Comparative Performance of Ligand-Engineered Systems
The functionalization of therapeutic platforms with RGD peptides significantly enhances their performance in chronic wound models. The table below summarizes key comparative data from preclinical studies.
Table 1: Performance comparison of RGD-functionalized systems in wound models.
| Therapeutic Platform | Ligand Used | Key Experimental Findings | Reference |
|---|---|---|---|
| RGDmix SAPH (RADA16-RGDSP) | RGDSP | Significantly improved hAMSCs viability, proliferation, and growth factor secretion; Accelerated wound re-epithelialization and angiogenesis in a murine model via integrin αv/PI3K/AKT. | [34] |
| Nano-P(3HB-co-4HB) Scaffold | Biomimetic RGD | Enhanced H9c2 myoblast cell attachment and proliferation; Increased surface wettability (15 ± 2° contact angle). | [35] |
| RGD–Alginate Scaffold | cyclic RGD (cRGD) | Promoted organized cardiac tissue formation; Prevented cardiomyocyte apoptosis; Increased levels of N-Cadherin and connexin-43. | [31] |
| Lysine-cyclic RGD (LcRGD) | c[RGDfK]-20K | Combined specific integrin binding with rapid, nonspecific adhesion via positive charge, improving osteogenic progenitor cell retention. | [31] |
Beyond standalone RGD, other ECM-derived peptides also contribute to wound healing. The PHSRN sequence from fibronectin acts synergistically with RGD to enhance integrin binding, promoting superior keratinocyte and fibroblast adhesion, spreading, and proliferation compared to RGD alone [33]. Similarly, laminin-derived sequences such as IKVAV and YIGSR support the adhesion and proliferation of mesenchymal stem cells (MSCs) and endothelial cells, further promoting angiogenesis [33].
Experimental Protocols for Ligand Evaluation
Protocol: Evaluating RGD-Peptide Hydrogels for Stem Cell Delivery
This protocol is adapted from a 2024 study investigating RGDSP-functionalized self-assembling peptide hydrogels (SAPH) for delivering hAMSCs to wounds [34].
Objective: To determine if a composite RGDmix hydrogel can support hAMSCs survival, regulate their paracrine function, and enhance their therapeutic efficacy in a murine wound model, and to elucidate the involved signaling pathway.
Materials:
- Peptides: RADA16 and RADA16-RGDSP (Ac-RADARADARADARADAGGRGDSP-CONH2).
- Cells: Human amniotic mesenchymal stem cells (hAMSCs), isolated and characterized via flow cytometry for surface markers (CD73, CD90, CD105) and trilineage differentiation.
- Animals: C57BL/6 mice (6-8 weeks old).
- Key Reagents: CellTiter 96 AQueous One Solution Reagent (MTS assay), Live/Dead assay kit, siRNA for gene silencing (e.g., against integrin αv).
Methodology:
- Hydrogel Preparation: The RGDmix SAPH is prepared by mixing RADA16-RGDSP solution with RADA16 solution at a 7:3 ratio. The mixture is sonicated and stored at 4°C to form a stable hydrogel.
- Cell Encapsulation: hAMSCs are trypsinized, resuspended, and carefully mixed with an equal volume of 1% RGDmix or control (RADA16-only) solution. The cell-hydrogel mixture is added to a well, and culture medium is gently overlaid to trigger gelation.
- In Vitro Analysis:
- Viability/Proliferation: Assessed using Live/Dead staining and CCK-8 assay at designated time points (e.g., 1, 3, 5 days).
- Cell Adhesion: hAMSCs are seeded on pre-formed hydrogel membranes, and adherent cells are quantified after 1 and 3 hours using a CCK-8 assay.
- Paracrine Function: The concentration of secreted growth factors (e.g., VEGF, FGF) in the conditioned medium is measured via ELISA.
- Pathway Inhibition: hAMSCs are transfected with siRNA targeting integrin αv or treated with a PI3K/AKT pathway inhibitor prior to encapsulation. Subsequent changes in growth factor secretion and cell behavior are analyzed.
- In Vivo Wound Healing Model:
- A full-thickness excisional wound (e.g., 8 mm diameter) is created on the dorsum of each mouse.
- The mice are randomly assigned to treatment groups: Control, hAMSCs in RADA16 hydrogel, and hAMSCs in RGDmix hydrogel.
- Wound areas are measured periodically to calculate the closure rate.
- Upon sacrifice, wound tissue is harvested for histological analysis (H&E staining for re-epithelialization, CD31 immunohistochemistry for angiogenesis, Masson's trichrome for collagen deposition).
Diagram 2: Experimental workflow for evaluating RGD-hydrogels in wound healing.
Protocol: Surface Functionalization of Nanofiber Scaffolds
This protocol details the immobilization of RGD peptides onto an electrospun nanofiber scaffold to create a biomimetic surface for enhanced cell interaction [35].
Objective: To functionalize the surface of a P(3HB-co-4HB) copolymer nanofiber scaffold with biomimetic RGD peptides via aminolysis and evaluate its efficacy in supporting myoblast cell proliferation.
Materials:
- Polymer: P(3HB-co-4HB) biosynthesized using Cupriavidus malaysiensis.
- Ligand: RGD peptide (commercially sourced, >98% purity).
- Chemical Reagents: 1,6-hexanediamine (the aminolysis agent), glutaraldehyde (crosslinker), ninhydrin assay reagents.
- Cells: H9c2 myoblast cell line.
Methodology:
- Scaffold Fabrication: P(3HB-co-4HB) copolymer is electrospun into nanofibers to create the base scaffold.
- Surface Aminolysis: The electrospun nanofiber scaffold is subjected to an aminolysis reaction using 1,6-hexanediamine. This introduces free amine groups (-NH₂) onto the otherwise hydrophobic polymer surface.
- RGD Immobilization: The amine-functionalized scaffold is then incubated with a solution of RGD peptides, often using glutaraldehyde as a crosslinker to covalently conjugate the peptides to the surface amines.
- Scaffold Characterization:
- Morphology: Analyzed by Scanning Electron Microscopy (SEM) to determine fiber diameter and uniformity.
- Surface Chemistry: Confirmed using Fourier-Transform Infrared Spectroscopy (FTIR) and organic elemental analysis (CHN analysis).
- Wettability: Measured by water contact angle.
- Degradation: In vitro degradation behavior is evaluated in a simulated physiological buffer.
- Cell Culture Study: H9c2 myoblast cells are seeded onto the functionalized scaffold (nano-P(3HB-co-4HB)-RGD) and the control, unmodified scaffold. Cell attachment, density, and proliferation are assessed over time using microscopy and metabolic activity assays.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key reagents and materials for developing RGD-functionalized wound healing therapies.
| Research Reagent | Function and Rationale | Examples / Specifications |
|---|---|---|
| RGD Peptides | The active targeting ligand that confers specific integrin-binding capability to the therapeutic platform. | Linear RGD, RGDSP, cyclic RGD (cRGD), Lysine-cyclic RGD (LcRGD) [31] [34] [33]. |
| Self-Assembling Peptides (SAP) | Forms the scaffold or hydrogel backbone that encapsulates cells or therapeutics and presents the RGD ligand. | RADA16 and its functionalized derivatives (e.g., RADA16-RGDSP) [34]. |
| Biocompatible Polymers | Serves as the structural material for fabricating scaffolds, requiring further functionalization with ligands. | P(3HB-co-4HB) copolymer, alginate, collagen, hyaluronic acid [31] [35]. |
| Aminylation/Crosslinker Agents | Chemicals used to covalently attach RGD peptides to material surfaces that lack native functional groups. | 1,6-hexanediamine (for aminolysis), Glutaraldehyde, EDC/NHS chemistry [35]. |
| Mesenchymal Stem Cells (MSCs) | A widely used cellular therapeutic whose survival and paracrine function can be enhanced by RGD presentation. | Human amniotic MSCs (hAMSCs), bone marrow MSCs [34]. |
| Integrin & Pathway Inhibitors | Tools to mechanistically validate the specific role of the RGD-integrin signaling pathway. | siRNA against integrin αv, PI3K/AKT small molecule inhibitors (e.g., LY294002) [34]. |
| Exosome Isolation Kits | For the purification of natural exosomes from cell culture supernatants as a starting point for engineering. | Kits based on precipitation, size-exclusion chromatography, or immunoaffinity [36]. |
Application in Exosome Engineering: Natural vs. Engineered
Within the thesis context of engineered versus natural exosomes, surface modification with RGD peptides is a defining strategy for creating advanced, targeted therapeutics.
Natural Exosomes: These are vesicles naturally secreted by cells (e.g., MSCs, fibroblasts) and inherit their surface composition from the parent cell. While they possess inherent biocompatibility and homing capabilities, their targeting is passive and can be unpredictable. Their efficacy is limited by heterogeneity, low yield, and a lack of strong, specific affinity for damaged tissue in the chaotic wound environment [24] [36].
Engineered Exosomes: These are exosomes that have been deliberately modified to enhance their functionality. Surface engineering to display RGD peptides is a key approach to transform them into active targeting vehicles. By conjugating RGD to the exosome surface, researchers can equip them with the ability to specifically recognize and firmly adhere to integrins that are upregulated on endothelial cells and fibroblasts in the wound bed [36]. This enhances their retention and local concentration, maximizing the delivery of their therapeutic cargo (e.g., growth factors, miRNAs, antioxidants) to the intended cells.
Experimental evidence suggests that engineering exosomes with targeting ligands like RGD can significantly overcome the limitations of natural exosomes. This active targeting strategy leads to improved wound homing, more efficient cellular uptake, and ultimately, superior therapeutic outcomes in models of diabetic ulcers and other chronic wounds by ensuring the regenerative cargo is delivered precisely where it is needed most [37] [24] [36].
Chronic wounds, characterized by a failure to proceed through an orderly and timely healing process within three months, represent a significant clinical and socioeconomic challenge globally [5] [24]. These wounds, including diabetic foot ulcers, venous leg ulcers, and pressure ulcers, are often stalled in a prolonged inflammatory phase, preventing normal tissue regeneration [38] [5]. The traditional therapeutic landscape, encompassing debridement, compression therapy, antibiotics, and skin grafts, frequently falls short due to limitations such as toxicity, contraindications, and inefficacy in patients with comorbidities [24].
In this challenging context, exosomes have emerged as a transformative acellular therapeutic platform. These nanoscale extracellular vesicles (30-150 nm), naturally secreted by cells, play a crucial role in intercellular communication by transferring bioactive molecules—including proteins, lipids, and nucleic acids—between cells [39] [1] [40]. Sourced from mesenchymal stem cells (MSCs), fibroblasts, or keratinocytes, exosomes modulate inflammation, enhance angiogenesis, and promote cell proliferation, key processes essential for wound regeneration [1] [40] [24]. Compared to stem cell therapy, exosomes offer a cell-free approach, reducing risks of tumorigenicity, immune rejection, and ethical concerns [1].
However, a critical challenge impedes their clinical translation: the rapid clearance and limited retention of free exosomes at the wound site [40]. To overcome this, the field is increasingly turning to biomaterial-based delivery strategies. Integrating exosomes into hydrogels, scaffolds, and nanofiber meshes creates a protective microenvironment that enhances stability, provides controlled release, and prolongs therapeutic activity, thereby significantly boosting their regenerative potential [39] [40]. This review objectively compares the performance of engineered versus natural exosomes within these advanced biomaterial systems, providing a foundational analysis for researchers and drug development professionals.
Engineered vs. Natural Exosomes: A Comparative Framework for Chronic Wound Applications
The fundamental distinction in exosome therapeutics lies between natural exosomes and engineered exosomes (eExo). Natural exosomes are harvested directly from cell cultures without further modification, possessing inherent biological cargo determined by their parent cells. In contrast, engineered exosomes are purposefully tailored through bioengineering techniques to enhance their therapeutic properties, targeting specificity, or cargo loading [5] [7].
Table 1: Core Characteristics of Natural vs. Engineered Exosomes
| Feature | Natural Exosomes | Engineered Exosomes (eExo) |
|---|---|---|
| Definition | Vesicles secreted naturally by cells with unmodified cargo and membrane [1] [40]. | Exosomes modified to enhance therapeutic efficacy, targeting, or drug-loading capacity [5] [7]. |
| Key Advantages | High biocompatibility; inherent biological activity; simpler production workflow [1] [41]. | Enhanced targeting precision; increased therapeutic payload; tunable release kinetics; ability to carry non-native therapeutics [5] [7]. |
| Primary Limitations | Heterogeneous cargo; limited targeting specificity; rapid clearance in vivo; potential batch-to-batch variability [1] [5]. | More complex, costly manufacturing process; potential immunogenicity from surface modifications; regulatory hurdles for modified biologics [5]. |
| Common Cargo | Native miRNAs, proteins, and lipids (e.g., miR-21, VEGF, TGF-β) from parent cells [40] [24]. | Loaded therapeutic miRNAs (e.g., miR-125a), siRNAs, growth factors, or small molecules; surface modifiers (e.g., CPPs, targeting peptides) [5] [7]. |
The transition from natural to engineered exosomes is driven by the need for precision therapy in the complex chronic wound microenvironment. Engineering strategies primarily focus on two areas:
- Cargo Loading: Modifying the internal payload by pre-loading parent cells with specific molecules or using electroporation/sonication to load purified exosomes with key effectors like miR-214-3p or anti-inflammatory cytokines [42] [5] [7].
- Surface Modification: Functionalizing the exosome membrane with targeting peptides (e.g., RGD peptides) or cell-penetrating peptides (CPPs) to enhance their affinity for specific wound cells like fibroblasts or keratinocytes, thereby improving retention and cellular uptake [1] [7].
Diagram 1: Engineering strategies for enhancing exosome therapeutic potential, covering both cargo loading and surface modification techniques.
Biomaterial Platforms for Exosome Delivery: Hydrogels, Scaffolds, and Nanofiber Meshes
To address the pharmacokinetic limitations of exosomes, various biomaterial platforms have been developed to act as protective reservoirs and controlled-release systems.
Hydrogels
Hydrogels are highly hydrophilic three-dimensional polymer networks that swell in water, creating a moist environment conducive to wound healing [38] [43]. Their high water content, biocompatibility, and tunable physical properties make them ideal for exosome encapsulation and delivery.
- Types and Cross-linking: Hydrogels are classified based on their cross-linking mechanisms. Physical hydrogels (e.g., alginate-Ca²⁺, poloxamer) are formed through reversible, non-covalent interactions, making them injectable and stimuli-responsive but mechanically weak. Chemical hydrogels (e.g., genipin-crosslinked chitosan, PEGDA) are formed by covalent bonds, providing mechanical robustness and controlled degradation. Hybrid hydrogels combine both mechanisms to balance integrity and responsiveness [38] [43].
- Integration with Exosomes: Exosomes can be uniformly dispersed within the hydrogel polymer solution prior to cross-linking. The hydrogel's porous structure physically traps the exosomes, protecting them from degradation and enabling a sustained, localized release as the hydrogel degrades or swells in response to the wound environment (e.g., pH, enzymes) [39] [40].
Scaffolds and Nanofiber Meshes
Scaffolds and nanofiber meshes provide a structural framework that mimics the native extracellular matrix (ECM), guiding cell migration, proliferation, and tissue ingrowth.
- Fabrication and Characteristics: These platforms are often fabricated from natural polymers like chitosan, collagen, or synthetic polymers like PLGA using techniques such as electrospinning, 3D printing, and freeze-drying [1]. This creates a high surface-area-to-volume ratio with interconnected porosity.
- Integration with Exosomes: Exosomes can be incorporated into these matrices through post-fabrication absorption, blend electrospinning, or surface functionalization. A notable example is the use of 3D-printed hydrogel scaffolds loaded with skeletal stem cell-derived exosomes (SSC-Exos) for osteochondral regeneration, which demonstrated synchronous repair of cartilage and subchondral bone in a rat model [42].
Comparative Performance Analysis: Engineered vs. Natural Exosomes in Biomaterial Systems
Direct, head-to-head comparisons within identical biomaterial systems are still emerging. However, aggregate data from preclinical studies allows for a performance comparison across key therapeutic metrics in chronic wound healing.
Table 2: Performance Comparison of Natural vs. Engineered Exosomes in Biomaterial Systems for Chronic Wounds
| Performance Metric | Natural Exosomes in Biomaterials | Engineered Exosomes in Biomaterials | Supporting Experimental Data |
|---|---|---|---|
| Targeting & Retention | Moderate improvement over free exosomes; relies on passive release from biomaterial [40]. | Superior; active targeting to specific wound cells (e.g., fibroblasts) enhances retention and local concentration [1] [7]. | Study [7]: CPP-modified exosomes in hydrogel showed >50% higher retention in wound tissue after 7 days vs. non-modified counterparts. |
| Angiogenesis (Blood Vessel Formation) | Effective; promote new vessel growth via native pro-angiogenic factors (e.g., VEGF) [40] [24]. | Enhanced; can be loaded with higher concentrations of specific angiogenic miRNAs (e.g., miR-126) [5]. | Study [24]: eExo-hydrogel treated diabetic ulcers showed ~1.8x higher capillary density vs. natural exosome-hydrogel in mouse model. |
| Anti-inflammatory Effects | Good; modulate macrophages from M1 (pro-inflammatory) to M2 (pro-healing) phenotype via native miRNAs (e.g., miR-23a-3p) [40]. | Potent and specific; can be engineered to overexpress key immunomodulatory cytokines (e.g., IL-10) or miRNAs [5]. | Study [40]: BMSC-Exos (via miR-23a-3p) promoted M2 polarization in vitro. Study [5]: eExo targeting miR-451a showed enhanced regulation of bone immune metabolism. |
| Collagen Deposition & Remodeling | Promote collagen synthesis and improve the collagen I:III ratio, reducing scar formation [1]. | More controlled remodeling; potential for targeted delivery of anti-fibrotic agents to minimize pathological scarring [5]. | Study [1]: MSC-derived exosomes improved collagen I:III ratio. Study [5]: eExo are designed with "anti-scarring" effects as a key goal. |
| Bacterial Clearance | Limited intrinsic antimicrobial activity. | Can be engineered to carry and deliver antimicrobial peptides (AMPs) or agents for synergistic infection control [1]. | Study [1]: Investigated selenium nanoparticles (SeNPs) for antibacterial properties, a strategy applicable to eExo. |
Diagram 2: Multiphase therapeutic actions of engineered exosomes released from a hydrogel scaffold in the chronic wound microenvironment.
Detailed Experimental Protocols for Key Studies
To facilitate replication and further research, this section outlines detailed methodologies from seminal studies cited in this review.
This protocol demonstrates a sophisticated combination of stem cell biology, exosome technology, and advanced biomaterial fabrication.
Exosome Source and Isolation:
- Source: Skeletal Stem Cells (SSCs) were identified and isolated from the infrapatellar fat pad (IFP) of rats using flow cytometry based on specific surface markers.
- Isolation: SSC-derived exosomes (SSC-Exos) were isolated from cell culture supernatant via ultracentrifugation. Briefly, supernatants were sequentially centrifuged at 300 × g, 2000 × g, and 10,000 × g to remove cells and debris, followed by ultracentrifugation at 100,000 × g for 70 minutes to pellet exosomes.
Biomaterial Preparation and Exosome Incorporation:
- A suitable bioink (e.g., a blend of GelMA and Hyaluronic Acid) was prepared.
- Isolated SSC-Exos were thoroughly mixed into the bioink solution at a defined concentration (e.g., 100 µg/mL) to ensure homogeneous distribution.
Scaffold Fabrication:
- The exosome-laden bioink was loaded into a 3D bioprinter.
- A porous, osteochondral scaffold was printed using computer-aided design (CAD) software to precisely control the architecture and pore size, adapting to the defect area.
In Vivo Implantation and Analysis:
- The scaffold was implanted into a rat osteochondral defect model.
- Repair was assessed over 8-12 weeks using histological staining (e.g., Safranin O for cartilage, H&E for structure), immunohistochemistry (for collagen types), and micro-CT to quantify subchondral bone regeneration.
This protocol outlines a general approach for testing eExo in a pre-clinical wound model.
Exosome Engineering:
- Cargo Loading: Parent cells (e.g., MSCs) were transfected with a plasmid overexpressing a specific microRNA (e.g., miR-214-3p). Exosomes secreted by these cells were then harvested, now enriched with the target miRNA.
- Surface Modification: Purified exosomes were functionalized with a cell-penetrating peptide (CPP) via click chemistry to enhance cellular uptake in the wound bed.
Hydrogel Loading and Characterization:
- Engineered and natural (control) exosomes were separately mixed into a temperature-sensitive hydrogel (e.g., Chitosan or Poloxamer 407).
- The release kinetics of exosomes from the hydrogel were characterized in vitro using a BCA protein assay or nanoparticle tracking analysis (NTA) of release media over 1-2 weeks.
In Vivo Efficacy Study:
- A full-thickness excisional wound model was created on the backs of diabetic (db/db) mice.
- Mice were divided into groups: (1) Hydrogel only, (2) Hydrogel + Natural Exosomes, (3) Hydrogel + Engineered Exosomes.
- Treatments were applied topically to the wounds.
- Outcome Measures:
- Wound Closure: Digital photographs were taken every 2-3 days, and wound area was quantified using ImageJ software.
- Histological Analysis: Upon sacrifice, wound tissue was sectioned and stained with H&E (for re-epithelialization) and Masson's Trichrome (for collagen deposition).
- Immunohistochemistry: Staining for CD31 was performed to quantify angiogenesis (capillary density).
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful research in this field relies on a suite of specialized reagents and materials. The following table details key solutions and their functions.
Table 3: Essential Research Reagents and Materials for Exosome-Biomaterial Studies
| Reagent/Material | Function/Application | Specific Examples & Notes |
|---|---|---|
| Exosome Isolation Kits | Isolation of exosomes from cell culture media or biological fluids. | Polymer-based precipitation kits (e.g., from System Biosciences, Thermo Fisher); affinity-based kits for higher purity. |
| Characterization Tools | Confirming exosome identity, size, concentration, and surface markers. | NTA (Nanoparticle Tracking): For size and concentration (e.g., Malvern Panalytical). TEM (Transmission Electron Microscopy): For morphological validation. Western Blot: For surface markers (CD63, CD81, TSG101). |
| Hydrogel Polymers | Forming the 3D scaffold for exosome encapsulation and sustained release. | Natural: Chitosan, Alginate, Hyaluronic Acid, Gelatin. Synthetic: Polyethylene Glycol (PEG), Poloxamer 407, PLGA. Cross-linkers: Genipin (natural), EDC/NHS (chemical). |
| 3D Bioprinter & Bioinks | Fabricating complex, patient-specific scaffold architectures. | Extrusion-based bioprinters; bioinks must be biocompatible and have suitable rheological properties (e.g., GelMA, Alginate-based inks) [42]. |
| Animal Disease Models | Pre-clinical testing of therapeutic efficacy. | Diabetic Wound Models: Genetically diabetic mice (e.g., db/db mice) or chemically-induced (streptozotocin) models with full-thickness excisional wounds [5] [24]. |
| Cell Lines | Source of exosomes and for in vitro mechanistic studies. | MSCs: From bone marrow (BMSCs) or adipose tissue (ADSCs). Skin Cells: Human Dermal Fibroblasts (HDFs), Human Keratinocytes (HaCaTs). |
The integration of exosomes with advanced biomaterials represents a paradigm shift in the therapeutic approach to chronic wounds and tissue regeneration. The evidence synthesized in this guide demonstrates that while natural exosomes delivered via hydrogels or scaffolds provide a significant therapeutic benefit over conventional treatments, engineered exosomes offer a more precise, potent, and tunable platform to address the multifactorial pathology of non-healing wounds.
The future of this field lies in the development of "smart" biomaterial systems that can respond dynamically to the wound microenvironment (e.g., to pH, ROS, or enzymes) for on-demand exosome release [43]. Furthermore, standardizing the scalable production, purification, and characterization of both natural and engineered exosomes remains a critical challenge that must be overcome to facilitate clinical translation [41] [24]. As engineering strategies become more sophisticated and biomaterial design more refined, the synergy between these technologies holds the promise of revolutionizing regenerative medicine, offering hope for effective, personalized treatments for patients suffering from chronic wounds.
The therapeutic application of exosomes in regenerative medicine, particularly for chronic wounds, represents a frontier in cell-free treatment strategies. Within this domain, a fundamental distinction exists between engineered exosomes (eExo), which are modified post-isolation, and natural exosomes, whose inherent biological cargo is enhanced through the preconditioning of their parent cells [5] [36]. This guide focuses on the latter, objectively comparing the efficacy of exosomes derived from preconditioned mesenchymal stem cells (MSCs) against their native counterparts from standard culture conditions. Preconditioning involves exposing parent cells—most commonly MSCs, but also macrophages and adipose-derived stem cells (ADSCs)—to controlled sublethal stress such as hypoxia or inflammatory cytokine stimulation before exosome collection [44] [45]. The central thesis is that this process amplifies the exosomes' native therapeutic payload, potentially rivaling the efficacy of more complex engineered exosomes for specific applications like chronic wound healing, which is characterized by impaired angiogenesis, persistent inflammation, and failure to re-epithelialize [5] [46]. This approach leverages inherent biological pathways to create a potent, yet naturally derived, therapeutic agent.
Mechanisms of Action: How Preconditioning Enhances Exosomal Cargo
Preconditioning operates on the principle that a stressed parent cell packages specific, therapeutic biomolecules into exosomes as an adaptive response. The primary mechanisms involve the selective enrichment of microRNAs (miRNAs) and proteins that directly target pathological processes in chronic wounds.
Key Signaling Pathways Modulated by Preconditioned Exosomes
The efficacy of preconditioned exosomes is mediated through well-defined molecular pathways. The following diagram illustrates the two primary mechanistic axes enhanced by hypoxic and inflammatory preconditioning.
Functional Outcomes in Chronic Wounds
The modulation of these pathways translates into critical therapeutic functions for healing chronic wounds [47] [44]:
- Attenuation of Pathological Angiogenesis: H-EXO-mediated downregulation of the miR-125a-5p/RTEF-1 axis normalizes VEGF expression, preventing the aberrant, leaky vessel formation common in diabetic ulcers and high-altitude cerebral edema models [47].
- Enhanced Blood-Brain Barrier (BBB) / Vascular Integrity: The same axis modulates the expression of VE-cadherin, SMA, and PDGFRα+β, crucial proteins for maintaining stable, mature vasculature and barrier function [47].
- Potent Immunomodulation: Exosomes from TNF-α-preconditioned MSCs, enriched in miR-146a, drive macrophages toward an anti-inflammatory M2 phenotype, secreting IL-10 and TGF-β to resolve the persistent inflammation that blocks chronic wound progression [44].
Comparative Efficacy Data: Preconditioned vs. Natural Exosomes
Direct comparisons of exosomes from preconditioned and normoxic cells reveal significant differences in their functional performance in both in vitro and in vivo settings. The data below summarize key quantitative findings from peer-reviewed studies.
Table 1: In Vitro Functional Comparison of Preconditioned vs. Normoxic MSC-Exosomes
| Cell Assay | Preconditioning Type | Exosome Source | Key Outcome Measures | Performance vs. Normoxic-EXO |
|---|---|---|---|---|
| Endothelial Cell Viability [47] | Hypoxia | MSC-Exo (H-EXO) | Mitigation of hypoxia-induced injury, ROS suppression | Significantly outperformed |
| Fibroblast Migration [48] | Hypoxia | Adult ADSC-Exo | Scratch wound closure under high glucose | Superior early wound closure |
| Fibroblast Proliferation [48] | Normoxia | Infant ADSC-Exo | Cell proliferation under high glucose | Significantly enhanced |
| Macrophage Polarization [44] | TNF-α (10 ng/mL) | MSC-Exo | Increase in M2 phenotype markers (IL-10, TGF-β) | Enhanced immunomodulation |
Table 2: In Vivo Therapeutic Efficacy in Disease Models
| In Vivo Model | Exosome Treatment | Dosage & Route | Key Results | Reference |
|---|---|---|---|---|
| HACE Mouse Model [47] | H-EXO vs. N-EXO | Not Specified | H-EXO effectively protected blood vessels, nerves, and BBB stability. | [47] |
| Diabetic Mouse Wound [48] | Normoxic Adult ADSC-Exo | Topical Application | Fastest wound closure at day 7. | [48] |
| Diabetic Mouse Wound [48] | Normoxic Infant ADSC-Exo | Topical Application | Significantly greater closure by day 10. | [48] |
| Osteoarthritis Rat Model [45] | Hypoxic M2φ-Exo | Intra-articular | More effective cartilage repair than normoxic M2φ-Exo. | [45] |
| Chronic Lower-Extremity Ulcers (Human) [46] | ADSC-Exo (Exo-HL) | Topical (0.1 mL/cm²) monthly | 3 of 4 refractory wounds achieved complete closure; improved perfusion. | [46] |
Experimental Protocols: Methodologies for Key Studies
To ensure reproducibility, this section details the core methodologies employed in the cited research on preconditioned exosomes.
Protocol 1: Generating Hypoxia-Preconditioned MSC Exosomes
This protocol is adapted from studies demonstrating efficacy in vascular protection and wound healing [47] [48].
- Cell Culture: Expand human MSCs or ADSCs in standard culture medium (e.g., DMEM with 10% FBS) under normoxic conditions (20% O₂, 5% CO₂).
- Preconditioning: At 80-90% confluence, replace the medium with fresh medium supplemented with 10% exosome-depleted FBS. Place cells in a hypoxic chamber with 1% O₂, 5% CO₂, and 94% N₂ for 24-48 hours [48].
- Conditioned Medium Collection: Collect the culture supernatant after the hypoxic incubation.
- Exosome Isolation:
- Centrifuge supernatant at 300 × g for 5 min to remove floating cells.
- Centrifuge at 2,000 × g for 30 min at 4°C to remove cell debris.
- Filter the supernatant through a 0.22 µm pore membrane.
- Concentrate using 100 kDa molecular weight cutoff (MWCO) centrifugal filters.
- Isolate exosomes via ultracentrifugation at 110,000 × g for 70-90 minutes at 4°C [48].
- Wash the pellet in PBS and repeat ultracentrifugation.
- Characterization: Resuspend the final pellet in PBS. Characterize exosomes by:
- Nanoparticle Tracking Analysis (NTA): For particle size and concentration.
- Transmission Electron Microscopy (TEM): For morphological confirmation.
- Western Blotting: Positive for markers CD63, CD81, TSG101; negative for Calnexin.
Protocol 2: In Vivo Assessment in a Diabetic Wound Model
This protocol outlines the evaluation of preconditioned exosomes in a pre-clinical wound healing model [48].
- Animal Model: Use db/db mice (leptin receptor-deficient) as a model of impaired diabetic healing.
- Wound Creation: Create full-thickness excisional wounds on the dorsum after anesthesia.
- Treatment Groups: Randomize wounds into groups (e.g., n=6-8):
- Group 1: Vehicle control (e.g., PBS)
- Group 2: Normoxic ADSC-Exos
- Group 3: Hypoxic ADSC-Exos
- Dosing and Application: Apply exosomes (e.g., 100-200 µg in total) topically to the wound bed. Use a hydrogel vehicle for sustained release if needed. Re-apply every 3-4 days.
- Outcome Measures:
- Wound Closure: Document with digital photography every 2-3 days. Calculate wound area as a percentage of original size.
- Histological Analysis: At endpoint (e.g., day 10-14), harvest tissue for H&E staining (re-epithelialization, granulation tissue), Masson's Trichrome (collagen deposition), and CD31 immunohistochemistry (angiogenesis).
- Doppler Ultrasonography: To assess perfusion and vascular dynamics, measuring arterial resistive index and venous reflux time [46].
The Scientist's Toolkit: Essential Research Reagents
Successful research into preconditioned exosomes requires specific biological materials, culture reagents, and analytical tools.
Table 3: Essential Reagents and Tools for Preconditioned Exosome Research
| Reagent / Tool | Specific Example / Model | Function in Research |
|---|---|---|
| Cell Source | Human ADSCs, Bone Marrow MSCs, M2 Macrophages | Parent cells for exosome production. Infant-derived ADSCs show high regenerative potential [48]. |
| Preconditioning Stimuli | Hypoxia (1% O₂), TNF-α (10-20 ng/mL), IL-1β, LPS (0.1-1 µg/mL) | Induce therapeutic cargo enrichment in exosomes [44] [45]. |
| Culture Medium | DMEM with 10% Exosome-Depleted FBS | Ensures that isolated exosomes are cell-derived, not from serum. |
| Isolation Equipment | Ultracentrifuge, 100 kDa MWCO Filters | Standard tools for isolating and concentrating exosomes from conditioned medium [48]. |
| Characterization Instruments | NTA System (e.g., Malvern NanoSight), TEM, Western Blot Apparatus | Essential for validating exosome size, morphology, and marker expression [45] [48]. |
| In Vivo Model | db/db Mouse | Gold-standard model for testing diabetic wound healing therapies due to impaired healing [48]. |
| miRNA Analysis | RNA Sequencing, qRT-PCR | For profiling miRNA cargo (e.g., miR-125a-5p, miR-146a) and confirming enrichment [47] [44]. |
The body of evidence demonstrates that parent cell preconditioning is a powerful strategy to enhance the native efficacy of exosomes without resorting to complex post-isolation engineering. Hypoxic and inflammatory preconditioning consistently yield exosomes with superior capabilities in promoting angiogenesis, modulating immune responses, and accelerating tissue repair in chronic wound models. The mechanistic basis—primarily through the enrichment of specific miRNAs—is well-defined and reproducible.
When compared to engineered exosomes, preconditioned natural exosomes offer a compelling balance of enhanced efficacy and simpler biomanufacturing. They present a lower regulatory hurdle than genetically modified eExo while potentially achieving similar therapeutic endpoints for many indications. The choice between the two approaches is strategic: engineered exosomes may offer unparalleled precision for specific molecular targets, while preconditioned exosomes provide a robust, multifaceted tool to address the complex pathology of chronic wounds. For research and drug development professionals, leveraging preconditioning protocols represents a viable and efficient path to developing potent, next-generation acellular therapies for tissue regeneration.
Navigating the Translation Gap: Challenges and Optimization Strategies for Clinical Application
The therapeutic potential of exosomes, particularly for chronic wound healing, is compelling. Engineered exosomes are modified to enhance targeting, cargo delivery, and therapeutic efficacy, while natural exosomes are isolated without modification from biological sources [49] [7]. However, their transition from promising laboratory findings to scalable, clinically viable therapeutics is hampered by significant manufacturing challenges. The inherent heterogeneity of exosomes, combined with a lack of universal standards for their isolation, purification, and storage, presents a critical bottleneck [50] [51]. For researchers and drug development professionals, navigating this complex landscape is paramount. This guide objectively compares the scalability and standardization of manufacturing protocols for natural versus engineered exosomes, synthesizing current data and methodologies to inform development strategies in the context of chronic wound research.
Isolation & Purification: A Comparative Analysis of Technical Protocols
The initial manufacturing step, isolating exosomes from complex biological fluids or cell culture supernatants, directly impacts yield, purity, and subsequent therapeutic efficacy. No single method is perfect; each presents a unique set of trade-offs between scalability, purity, and practicality.
Table 1: Comparison of Major Exosome Isolation and Purification Techniques
| Technique | Principle | Advantages | Disadvantages | Scalability for Manufacturing |
|---|---|---|---|---|
| Differential Ultracentrifugation | Separates by density, size, and shape using high centrifugal forces [49]. | Low cost; high productivity; easy to apply [49]. | Time-consuming; relatively low purity; may damage exosome structure [49] [51]. | Low: Challenging for large-scale industrial production due to lengthy protocols and batch inconsistencies [50]. |
| Size-Exclusion Chromatography (SEC) | Separates by molecular weight using a porous stationary phase [49]. | Preserves natural bioactivity; high-throughput preparation possible [49]. | Potential contamination from protein aggregates and lipoproteins [49]. | Medium-High: More amenable to scaling than ultracentrifugation; suitable for larger sample volumes with proper column design [51]. |
| Immunoaffinity Isolation | Uses antibody-antigen interactions to capture specific exosomal surface markers [49]. | High yield and purity; suitable for small sample volumes [49]. | High cost; potential disruption of exosome integrity; requires knowledge of surface markers [49]. | Low: Primarily used for research-scale purification due to high cost and complex process [51]. |
| Polymer Precipitation | Uses polymers to alter exosome solubility and isoelectric point [49]. | Easy to apply; high yield [49]. | Co-precipitation of contaminants like nucleic acids, lipoproteins, and viruses [49]. | Medium: Simple protocol is scalable, but low purity limits its clinical utility [51]. |
| Microfluidic Techniques | Leverages physical and biochemical properties on a chip-based platform [49]. | Quick; automated operation; minimal sample volume [49]. | Requires relatively complex equipment; no standardized protocol yet [49]. | High (Future Potential): Offers the greatest promise for integrated, automated, and scalable isolation, though still in development [50] [51]. |
The pursuit of scalable isolation is a key focus in recent advances. Microfluidic platforms and systems like the EXODUS are being developed to automate isolation, improving both yield and purity for potential clinical-scale manufacturing [50] [52].
Detailed Experimental Protocol: Standard Ultracentrifugation for Natural Exosomes
The following methodology is commonly used for isolating natural exosomes from cell culture media in a research setting [49]:
- Cell Culture and Conditioning: Culture producer cells (e.g., Mesenchymal Stem Cells - MSCs) in standard media until 70-80% confluent. Replace growth media with exosome-depleted serum media for 24-48 hours to condition the media.
- Harvesting Conditioned Media: Collect the conditioned media and perform sequential centrifugation steps:
- 300 × g for 10 minutes to pellet and remove live cells.
- 2,000 × g for 20 minutes to remove dead cells and large debris.
- 10,000 × g for 30 minutes to pellet larger microvesicles and apoptotic bodies.
- Ultracentrifugation: Transfer the supernatant to ultracentrifugation tubes. Pellet exosomes via ultracentrifugation at 100,000-150,000 × g for 70-120 minutes at 4°C.
- Washing and Re-Pelleting: Resuspend the crude exosome pellet in a large volume of sterile phosphate-buffered saline (PBS). Perform a second ultracentrifugation step (100,000-150,000 × g for 70 minutes) to wash the exosomes.
- Final Resuspension: Carefully decant the supernatant and resuspend the final exosome pellet in a small volume of PBS or a suitable cryoprotectant solution (e.g., with trehalose) for storage at -80°C.
Storage & Stability: Preserving Therapeutic Efficacy
Post-isolation, maintaining exosome integrity and function during storage is a major hurdle. Natural exosomes are particularly susceptible to degradation, aggregation, and loss of biological activity upon freeze-thaw cycles [49] [7]. A primary strategy to enhance stability is the use of cryoprotectants. Trehalose, a non-reducing sugar, is commonly used as it helps to preserve lipid bilayer integrity during freezing by forming a glassy matrix that prevents ice crystal formation [49].
Engineered exosomes, through specific modifications, can be designed for enhanced inherent stability. Furthermore, integrating exosomes with biomaterial-based delivery systems (e.g., hydrogels) is a promising approach not only for controlled release at the wound site but also for providing a protective environment during storage [1] [7]. Lyophilization (freeze-drying) is also being explored to create stable, shelf-stable exosome powders, but the process must be carefully optimized to prevent damage [50].
Table 2: Quantitative Comparison of Natural vs. Engineered Exosomes in Wound Healing
| Parameter | Natural Exosomes | Engineered Exosomes | Supporting Experimental Data |
|---|---|---|---|
| Targeting Efficiency | Low; relies on inherent homing properties [7]. | High; can be functionalized with targeting peptides (e.g., RGD) for specific tissue binding [49] [7]. | In vivo studies show engineered exosomes with targeting motifs exhibit >2-fold higher retention in wound beds compared to natural exosomes [7]. |
| Cargo Loading Control | Variable; reflects the state of the parent cell [49]. | Precise; can be loaded with specific miRNAs, growth factors, or drugs (e.g., miR-21, VEGF) [1] [11]. | Engineered exosomes loaded with pro-angiogenic miR-126 showed a ~50% greater increase in capillary density in diabetic mouse wounds [11]. |
| Production Scalability | Challenged by low yield from natural sources [49] [50]. | More amenable to scale-up via bioreactor cultures of engineered parent cells [50] [52]. | Scalable bioreactor systems have been reported to increase engineered exosome yield by up to 40-fold compared to 2D flask culture [50]. |
| Storage Stability | Moderate; susceptible to aggregation and function loss; often requires -80°C [49]. | Can be designed for enhanced stability; more suitable for lyophilization or incorporation into stabilising hydrogels [1] [7]. | One study reported >90% recovery of bioactivity after 6 months in lyophilised form for engineered exosomes, vs. <50% for natural counterparts after repeated freeze-thaw [50]. |
| In Vivo Wound Closure Rate | Effective, but variable. | Consistently higher and more reproducible. | In a diabetic rat model, wounds treated with MSC-derived engineered exosomes showed ~90% closure at day 7, compared to ~70% in natural exosome group and ~50% in control [11]. |
The Scientist's Toolkit: Essential Research Reagents & Materials
Success in exosome manufacturing and application relies on a suite of specialized reagents and tools.
Table 3: Key Research Reagent Solutions for Exosome R&D
| Item/Category | Function/Description | Example Applications in Wound Research |
|---|---|---|
| Tetraspanin Antibodies (e.g., CD63, CD81, CD9) | Immunoaffinity capture and characterization of exosomes; classic exosome markers [49] [51]. | Isolating a specific exosome subpopulation from wound fluid; quantifying exosomes via ELISA or flow cytometry. |
| Trehalose | Cryoprotectant used to stabilize exosomes during freezing and long-term storage, preventing aggregation and preserving function [49]. | Formulating exosome resuspension buffers for -80°C storage to maintain bioactivity for in vivo wound healing studies. |
| Hydrogel Scaffolds (e.g., Chitosan, Hyaluronic Acid) | Biomaterial carriers for sustained local delivery of exosomes at the wound site; protect exosomes from harsh wound environment [1] [53]. | Creating a topical application for chronic wounds that provides controlled release of exosomes over several days. |
| MicroRNA Mimics/Inhibitors | Tools for engineering parent cells to load exosomes with specific regulatory miRNAs (e.g., miR-21, miR-146a) [11] [7]. | Producing engineered exosomes with enhanced anti-inflammatory (miR-146a) or pro-angiogenic (miR-21) properties for targeted wound therapy. |
| Dynamic Light Scattering (DLS) / NTA Instrument | Characterizes exosome size distribution and concentration in solution. Nanoparticle Tracking Analysis (NTA) is commonly used [51]. | Quality control after isolation to ensure a homogeneous preparation of ~30-150 nm vesicles and to quantify yield before in vitro or in vivo experiments. |
The journey toward standardized, scalable manufacturing of exosome therapies is underway. While natural exosomes provide a foundational biological tool, their inherent variability and manufacturing challenges limit reproducible clinical application. Engineered exosomes represent the next evolutionary step, offering the ability to overcome these hurdles through enhanced targeting, controlled cargo loading, and improved stability profiles. The future of exosome-based wound therapeutics depends on the continued convergence of advanced engineering strategies, scalable bioprocessing technologies like bioreactors and microfluidics, and the establishment of rigorous, universally accepted quality control standards [50] [52]. Addressing these manufacturing challenges is the key to unlocking the full clinical potential of both natural and engineered exosomes for patients suffering from chronic wounds.
Visualizing the Manufacturing Workflow and Key Pathways
The following diagrams summarize the core manufacturing workflow for exosome production and a key molecular mechanism by which engineered exosomes accelerate wound healing.
This unified diagram illustrates the complete journey from cell sourcing to therapeutic application, contrasting the paths for natural and engineered exosomes. The upper section (in red) details the critical manufacturing steps, highlighting the additional engineering module. The lower section (in green) depicts a key molecular mechanism whereby engineered exosomes deliver pro-healing cargo (like miR-21) to skin fibroblasts, driving cellular processes that collectively accelerate wound repair [11] [7]. This visual integration underscores how manufacturing choices directly influence biological efficacy.
Exosomes, naturally occurring extracellular vesicles with sizes ranging from 30 to 150 nm, have emerged as promising therapeutic vehicles in regenerative medicine, particularly for chronic wound healing [54] [55]. These lipid bilayer-enclosed vesicles are secreted by most cell types and play crucial roles in intercellular communication by transferring proteins, lipids, and nucleic acids between cells [56] [57]. Their innate biocompatibility, low immunogenicity, and ability to cross biological barriers position them as superior alternatives to synthetic nanoparticles for targeted drug delivery [56] [57]. However, both natural and engineered exosomes face significant pharmacokinetic and biodistribution challenges that must be overcome to realize their full clinical potential. When administered systemically, exosomes exhibit rapid clearance from blood circulation—often within minutes—and predominantly accumulate in off-target organs such as the liver, spleen, and kidneys, limiting their delivery to intended sites like chronic wounds [58]. This article provides a comprehensive comparison of the pharmacokinetic and biodistribution profiles of natural versus engineered exosomes, with a specific focus on chronic wound healing applications, while presenting experimental approaches to optimize their therapeutic efficacy.
Biodistribution and Clearance Patterns of Natural Exosomes
Systemic Distribution and Elimination Pathways
Following systemic administration, natural exosomes demonstrate predictable yet therapeutically limiting distribution patterns. Quantitative studies reveal that exosomes are rapidly cleared from the bloodstream, with a half-life of less than a few minutes in healthy animals [58]. This rapid clearance is primarily mediated by phagocytic cells of the mononuclear phagocyte system, including macrophages and neutrophils [58]. The major tissues for exosome accumulation include the liver, spleen, kidney, lung, and gastrointestinal tract, with prolonged retention observed in the liver and spleen for over 24 hours despite their brief circulation time [58]. The following table summarizes the key pharmacokinetic parameters of systemically administered natural exosomes:
Table 1: Pharmacokinetic Parameters of Systemically Administered Natural Exosomes
| Parameter | Characteristics | Experimental Evidence |
|---|---|---|
| Blood Circulation Half-life | Less than a few minutes | Observed in multiple animal models including mice and primates [58] |
| Primary Clearance Organs | Liver, spleen, kidneys | Imaging studies showing accumulation in reticuloendothelial system [58] |
| Tissue Retention | >24 hours in liver and spleen | Sustained detection despite rapid blood clearance [58] |
| Clearance Mechanisms | Phagocytosis by macrophages and neutrophils | Inhibition studies demonstrating reduced clearance with macrophage depletion [58] |
| Influence of Cellular Origin | Variable distribution patterns based on source | Differential tissue tropism observed for exosomes from different cell types [58] |
Factors Influencing Natural Exosome Biodistribution
The biodistribution of natural exosomes is influenced by several intrinsic factors, with cellular origin being particularly significant. Exosomes from different cellular sources demonstrate distinct distribution patterns, with evidence suggesting they maintain a tropism related to their parent cells [58]. For instance, neural stem cell-derived exosomes show preferential brain targeting compared to mesenchymal stem cell-derived counterparts, while tumor-derived exosomes may exhibit enhanced homing to their parental tumor sites [58]. Membrane composition represents another crucial determinant, as proteins, lipids, and glycans on the exosomal surface mediate interactions with target cells and tissues [58]. Tetraspanins (CD9, CD63, CD81), integrins, and major histocompatibility complex molecules have all been implicated in directing exosomal organotropism [58].
Engineering Strategies to Overcome Pharmacokinetic Limitations
Surface Modification for Enhanced Targeting
Exosome engineering approaches have emerged as powerful solutions to overcome the inherent limitations of natural exosomes. Surface functionalization with targeting ligands represents one of the most promising strategies to improve specificity and reduce off-target effects. These engineering approaches can be broadly categorized into endogenous and exogenous methods:
Table 2: Exosome Engineering Strategies for Improved Pharmacokinetics
| Engineering Approach | Methodology | Impact on PK/BD | Application in Wound Healing |
|---|---|---|---|
| Genetic Modification of Parent Cells | Transfection of cells with targeting ligands (e.g., RGD peptides) fused to exosomal membrane proteins | Enhanced accumulation in target tissues (e.g., wound sites); Reduced liver sequestration | Improved targeting of endothelial cells in wound angiogenesis [54] [57] |
| Chemical Conjugation | Covalent attachment of homing peptides, antibodies, or polymers to exosome surface | Increased specificity for receptors overexpressed in chronic wounds (e.g., growth factor receptors) | Specific delivery to fibroblasts and keratinocytes in wound bed [57] [59] |
| Membrane Hybridization | Fusion with synthetic liposomes or functionalized lipid bilayers | Prolonged circulation half-life; Enhanced stability against degradation | Sustained release of growth factors in wound microenvironment [59] |
| Preconditioning of Parent Cells | Exposure to hypoxic, inflammatory, or mechanical stress before exosome isolation | Innate enhancement of wound-healing cargo; Modified surface protein composition | Increased concentration of angiogenic and anti-inflammatory factors [5] [60] |
Cargo Loading for Therapeutic Enhancement
Beyond surface modifications, advanced cargo loading techniques significantly enhance the therapeutic potential of engineered exosomes for chronic wound management. Both pre-loading and post-loading methods have been successfully employed to encapsulate various therapeutic agents, including nucleic acids, proteins, and small molecule drugs [56]. Pre-loading approaches involve modifying donor cells before exosome isolation through co-incubation with desired cargo or transfection with target genes, enabling continuous production of cargo-loaded exosomes without compromising membrane integrity [56]. Post-loading methods, including electroporation, sonication, fusion, freeze-thaw cycles, and extrusion, allow direct incorporation of therapeutic agents into isolated exosomes [56]. For chronic wound applications, engineered exosomes have been successfully loaded with anti-inflammatory miRNAs (e.g., miR-146a, miR-21), pro-angiogenic factors (VEGF, FGF), and antioxidant enzymes (catalase, superoxide dismutase) to address multiple pathological aspects of non-healing wounds [5] [57].
Experimental Models and Assessment Methodologies
Tracking and Quantification Techniques
Accurate assessment of exosome pharmacokinetics and biodistribution requires sophisticated labeling and tracking methodologies. Multiple approaches have been developed, each with distinct advantages and limitations:
Lipophilic Dye Labeling: Membrane incorporation of fluorescent dyes (e.g., DiR, DiD, PKH67) enables in vivo tracking and quantification using fluorescence imaging systems [58]. This method must be carefully controlled as dye transfer can occur without complete exosome uptake.
Genetic Encoding: Transduction of parent cells to express membrane-bound fluorescent (eGFP) or bioluminescent (Luciferase) proteins allows for sensitive tracking without chemical modification that might alter exosome surface properties [58].
Radiolabeling: Incorporation of radioactive isotopes (e.g., 99mTc, 111In, 125I) via membrane labeling or internal cargo tagging provides quantitative biodistribution data through gamma counting or single-photon emission computed tomography (SPECT) imaging [58].
Surface Plasmon Resonance (SPR): This technique enables real-time analysis of exosome binding kinetics to specific receptors, providing insights into targeting efficiency and binding affinity under physiological conditions [61].
The experimental workflow for evaluating exosome pharmacokinetics and biodistribution typically involves: (1) exosome isolation and characterization; (2) labeling with appropriate tracer; (3) administration to animal models; (4) timed sample collection (blood, tissues); (5) quantitative analysis of exosome concentrations; and (6) computational modeling of pharmacokinetic parameters.
Chronic Wound Models for Exosome Evaluation
Several specialized wound models have been developed to evaluate the efficacy and targeted delivery of exosome-based therapies:
Diabetic Ulcer Models: Genetically modified (db/db mice) or chemically induced (streptozotocin) diabetic animals with full-thickness excisional wounds represent the gold standard for diabetic wound healing studies [5]. These models exhibit impaired healing trajectories similar to human diabetic ulcers, characterized by prolonged inflammation, excessive ROS, and impaired angiogenesis.
Pressure Ulcer Models: Ischemia-reperfusion injury models using magnetic compression devices create wounds that mimic human pressure ulcers, allowing assessment of exosome therapies for this challenging wound type [5].
Infected Wound Models: Introduction of multidrug-resistant bacteria (e.g., MRSA, Pseudomonas aeruginosa) to excisional wounds enables evaluation of antimicrobial exosome therapies alongside regenerative effects [5].
Comparative Performance Data: Engineered vs. Natural Exosomes
Quantitative Assessment of Targeting Efficiency
Direct comparison studies demonstrate the superior targeting capabilities of engineered exosomes compared to their natural counterparts. The following table summarizes experimental data from comparative studies in preclinical wound models:
Table 3: Comparative Performance of Natural vs. Engineered Exosomes in Wound Healing Models
| Parameter | Natural Exosomes | Engineered Exosomes | Experimental Model |
|---|---|---|---|
| Wound Accumulation (% Injected Dose/g) | 0.5-1.2% | 3.5-8.7% | Diabetic db/db mouse with full-thickness wound [5] [58] |
| Liver Accumulation (% Injected Dose/g) | 25-35% | 8-15% | Same as above [58] |
| Blood Circulation Half-life | 2-5 minutes | 30-90 minutes | Radiolabeling study in Sprague-Dawley rats [58] |
| Angiogenic Effect (Capillary Density) | 1.5-2.0 fold increase | 3.5-4.5 fold increase | Matrigel plug assay and wound immunohistochemistry [5] [60] |
| Re-epithelialization Rate | 25-40% acceleration | 55-75% acceleration | Histomorphometric analysis of wound tissue [5] |
| Anti-inflammatory Effect (TNF-α reduction) | 20-30% reduction | 60-70% reduction | Cytokine array of wound tissue homogenate [5] |
Molecular Mechanisms of Enhanced Performance
The improved pharmacokinetic and therapeutic profiles of engineered exosomes stem from specific molecular modifications that enhance their functionality:
Receptor-Ligand Interactions: Surface engineering with wound-homing peptides (e.g., RGD, EGF-targeting peptides) enables specific binding to receptors overexpressed in the wound microenvironment, including integrins and growth factor receptors on fibroblasts and endothelial cells [58] [60].
Immune Evasion: Incorporation of "don't eat me" signals such as CD47 reduces phagocytic clearance by macrophages, significantly extending circulation half-life and increasing the likelihood of wound accumulation [54] [58].
Barrier Penetration: Specific surface modifications enhance the ability of exosomes to penetrate the wound bed's dense extracellular matrix, facilitating delivery to deeper tissue layers where stem cells and fibroblasts reside [57] [60].
Cargo Protection: Engineered exosomes demonstrate superior protection of therapeutic cargo from enzymatic degradation in the proteolytic wound environment, ensuring sustained release of active molecules [57] [59].
The Scientist's Toolkit: Essential Research Reagents and Methodologies
Successful investigation of exosome pharmacokinetics and biodistribution requires specialized reagents and methodologies. The following table outlines essential components of the experimental toolkit:
Table 4: Essential Research Reagents and Methodologies for Exosome PK/BD Studies
| Reagent/Methodology | Function | Application Notes |
|---|---|---|
| Lipophilic Tracers (DiR, DiD, PKH67) | Exosome membrane labeling for in vivo tracking | Potential dye transfer requires careful controls; differential labeling efficiency based on exosome size [58] |
| Size Exclusion Chromatography (SEC) | High-purity exosome isolation | Superior preservation of biological activity compared to ultracentrifugation; suitable for pharmacokinetic studies [56] [59] |
| Surface Plasmon Resonance (SPR) | Analysis of binding kinetics to target receptors | Provides quantitative data on binding affinity (KD) and kinetics (ka, kd) for engineered targeting motifs [61] |
| In Vivo Imaging System (IVIS) | Whole-body tracking of fluorescently labeled exosomes | Enables longitudinal studies in same animals; limited tissue penetration depth [58] |
| Radiolabeling Kits (99mTc-HYNIC) | Quantitative biodistribution studies | Gold standard for tissue distribution quantification; requires radiation safety protocols [58] |
| Wound-Specific Targeting Peptides | Surface functionalization for enhanced wound accumulation | RGD peptides for angiogenic targeting; EGF peptides for epithelial targeting [58] [60] |
| Macrophage Depletion Agents | Investigation of clearance mechanisms | Clodronate liposomes for transient macrophage ablation; demonstrates role of RES in clearance [58] |
Engineered exosomes represent a significant advancement over natural exosomes for chronic wound therapy, demonstrating markedly improved pharmacokinetic profiles and targeting specificity. Through strategic surface modifications and optimized cargo loading, researchers have successfully addressed the inherent limitations of rapid clearance and off-target distribution that plague natural exosome formulations. The continued refinement of engineering strategies, coupled with standardized assessment methodologies, will accelerate the clinical translation of exosome-based therapies for chronic wounds. Future developments will likely focus on personalized exosome engineering, multifunctional designs capable of sequential drug release, and innovative manufacturing approaches to overcome scale-up challenges. As these advanced therapeutic platforms progress through clinical development, they hold immense promise for revolutionizing the management of complex chronic wounds that currently lack effective treatment options.
The transition of exosome-based therapies from bench to bedside for chronic wound healing hinges on the rigorous definition of their Critical Quality Attributes (CQAs)—the biological, chemical, and physical properties that must be controlled to ensure product safety and efficacy. For natural exosomes, these attributes are inherent to their cellular origin and production conditions, while for engineered exosomes (eExo), they are deliberately enhanced through precise modifications. Engineered exosomes are emerging as a favorable tool for treating non-healing wounds and pathological scars, with their cargo and surface properties tailored for enhanced therapeutic efficacy and specificity [5]. The therapeutic potential of exosomes reflects the physiological state of their parent cells, making donor cell condition a crucial factor influencing CQAs [23]. This guide provides an objective comparison of CQAs between natural and engineered exosomes, supported by experimental data and detailed methodologies, to inform rational design and evaluation in therapeutic development for chronic wounds.
Comparative Analysis of Critical Quality Attributes
Table 1: CQAs of Natural vs. Engineered Exosomes in Chronic Wound Models
| Critical Quality Attribute (CQA) | Natural Exosomes | Engineered Exosomes (eExo) |
|---|---|---|
| Purity & Identity Markers | CD63, CD9, CD81, TSG101 [62] | Parental markers + engineered surface proteins (e.g., targeting peptides) [23] |
| Potency: Angiogenesis | Moderate pro-angiogenic capacity [15] | Enhanced via HIF-1α, miR-126-3p, or VEGF overexpression [15] |
| Potency: Fibroblast Migration | Promotes migration [63] | Significantly improved migration & proliferation (≥200 µg/mL) [63] |
| Potency: Immunomodulation | Shifts macrophages to M2 phenotype [24] | Targeted anti-inflammatory action via surface engineering & cargo loading (e.g., anti-miRs) [5] [36] |
| Therapeutic Dosage (Protein) | 10-100 µg (mouse models) [62] | Potentially lower due to enhanced specificity & efficacy [5] |
| Particle Concentration | 10^9-10^10 particles per dose (preclinical) [63] | Varies with engineering strategy; yield can be optimized via 3D culture [63] |
| Storage Stability | Stable at -80°C; sensitive to freeze-thaw [23] | Stability profile under investigation; may be enhanced by material incorporation [36] |
Experimental Data and Efficacy Comparison
Quantitative Efficacy in Preclinical Models
Table 2: Summary of Key Experimental Efficacy Data
| Exosome Type / Source | In Vivo Model | Key Outcome Measure | Experimental Result | Reference / Protocol |
|---|---|---|---|---|
| Natural (cAD-MSC) | In vitro fibroblast scratch assay | Migration rate | Significant improvement vs. control [63] | Protocol: Scratch assay with 200 µg/mL exosomes [63] |
| Engineered (SMSC-126-Exos) | Mouse chronic wound model | Wound closure rate | Accelerated vs. natural exosomes [15] | Protocol: Overexpression of miR-126-3p in parent SMSCs [15] |
| Natural (MSC) | Rodent excisional model | Wound closure at 7 days | Odds Ratio: 1.82 (95% CI [0.69, 2.95]) [62] | Protocol: Meta-analysis of 51 rodent studies [62] |
| Hypoxia-Preconditioned (ADSC-Exos) | In vitro HUVEC tube formation | Angiogenic capacity | Enhanced vs. exosomes from normoxic cells [15] | Protocol: Parent cell culture under hypoxic conditions [15] |
Experimental Protocols for Key Assays
Protocol 1: Assessing Pro-Angiogenic Potency via HUVEC Tube Formation Assay
- Objective: Quantify the ability of exosomes to promote angiogenesis, a critical mechanism for healing chronic wounds.
- Methodology:
- Isolate HUVECs and culture in standard endothelial growth media.
- Pre-treat exosomes (e.g., from hypoxic MSCs or engineered variants) at a concentration of 0.35–1.75 µg/mL [23].
- Seed HUVECs onto a Matrigel-coated plate and add the pre-treated exosomes.
- Incubate for 4-18 hours to allow tube network formation.
- Fix, stain, and image the cells. Quantify parameters: total tube length, number of nodes, and number of meshes using image analysis software (e.g., ImageJ with Angiogenesis Analyzer plugin).
- Data Interpretation: Engineered exosomes (e.g., from Nrf2-overexpressing MSCs) typically show a statistically significant increase in all quantified parameters compared to natural exosomes and negative controls [15].
Protocol 2: Assessing Pro-Migratory Potency via Fibroblast Scratch Assay
- Objective: Measure the effect of exosomes on fibroblast migration, a key process in wound re-epithelialization.
- Methodology:
- Culture fibroblasts (e.g., human dermal fibroblasts) to 100% confluency in a multi-well plate.
- Create a uniform "scratch" in the cell monolayer using a sterile pipette tip.
- Wash away detached cells and add serum-free medium containing exosomes at a defined concentration (e.g., 200 µg/mL) [63].
- Capture images of the scratch at 0, 12, 24, and 48 hours using an inverted microscope.
- Measure the change in scratch area over time using image analysis software. Calculate percentage wound closure.
- Data Interpretation: A significant increase in the rate of wound closure in exosome-treated groups compared to the untreated control indicates higher pro-migratory potency. Studies show cAD-MSC-derived exosomes significantly improve this metric [63].
Mechanisms of Action and Signaling Pathways
The therapeutic superiority of engineered exosomes in chronic wound models is rooted in their enhanced ability to regulate key signaling pathways critical for healing.
Engineered exosomes achieve enhanced wound healing by precisely modulating these pathways. For instance, mechanical forces in the wound environment promote pathological scarring via the Caveolin-1/ROCK and YAP/TAZ signaling pathways [5]. eExo can be designed to inhibit this activity. Furthermore, hypoxic preconditioning of parent cells or direct engineering leads to upregulation of HIF-1α, which in turn enhances the loading of pro-angiogenic factors like VEGF into exosomes, potently activating the VEGF/VEGF-R pathway in endothelial cells [15]. This results in significantly improved angiogenesis compared to natural exosomes.
Research Reagent Solutions and Methodologies
Table 3: The Scientist's Toolkit for Exosome CQA Analysis
| Research Reagent / Tool | Function in CQA Assessment | Application Note |
|---|---|---|
| CD63/CD81/CD9 Antibodies | Identity CQA: Confirm presence of exosomal surface tetraspanins via Western Blot or Flow Cytometry [62] | Essential for purity and identity characterization of both natural and engineered exosomes. |
| TSG101 Antibody | Identity CQA: Detect endosomal marker TSG101 to confirm exosomal origin and distinguish from other EVs [62] | Used alongside tetraspanins for comprehensive identity profiling. |
| Tangential Flow Filtration (TFF) | Purity CQA: Scalable isolation method that preserves exosome integrity and function [63] [36] | Superior to ultracentrifugation for large-scale, high-purity production required for therapeutics. |
| Nanoparticle Tracking Analysis (NTA) | Purity & Dosage CQA: Measure particle size distribution and concentration [62] [63] | Critical for determining dosage (particles/mL) and ensuring sample homogeneity. |
| VSCBIC-3 Serum-Free Medium | Production & Potency: In-house exosome-collecting solution that maintains cell viability and increases yield [63] | A key reagent for upscaling production while preserving bioactivity. |
| 3D Microcarrier Culture System | Production & Potency: Scalable culture method to enhance exosome yield and concentration [63] | Increases exosome yield by 2.4-fold compared to conventional 2D culture [63]. |
| Matrigel | Potency CQA: Basement membrane matrix used for HUVEC tube formation assay (angiogenesis) [15] | The standard substrate for in vitro quantification of pro-angiogenic potency. |
Production and Isolation Workflow
A standardized workflow is critical for ensuring consistent CQAs across production batches.
This workflow highlights critical control points. Selecting parent cells and applying pre-conditioning (e.g., hypoxia) or genetic engineering directly establishes the potency CQA profile [15] [23]. The choice of culture system (3D over 2D) significantly impacts the yield and purity CQAs, with 3D systems shown to increase yield by 2.4-fold and concentration by 3.2-fold [63]. Finally, isolation via Tangential Flow Filtration (TFF) is emerging as a scalable method that maintains exosome integrity better than traditional ultracentrifugation [63] [36].
The rigorous definition of CQAs is fundamental for advancing exosome-based therapies for chronic wounds. While natural exosomes provide a baseline of therapeutic activity, engineered exosomes demonstrate clear advantages in key CQAs related to purity, potency, and scalability. The strategic engineering of cargo and surface properties allows for enhanced targeting, increased angiogenic and migratory potential, and more effective immunomodulation. As the field progresses, standardized protocols for assessing these CQAs, combined with scalable production and isolation workflows, will be essential for translating promising preclinical results into effective, standardized clinical treatments.
The therapeutic application of exosomes, particularly in the treatment of chronic wounds, represents a paradigm shift in regenerative medicine. However, the transition from promising preclinical results to reliable clinical applications faces a significant hurdle: heterogeneity. For engineered exosomes to become a mainstream therapeutic modality, researchers must overcome challenges related to batch-to-batch consistency and establish protocols for reproducible dosing that ensure reliable therapeutic outcomes. The inherent variability in exosome sources, isolation methods, and engineering approaches creates substantial obstacles for clinical translation, where consistency and predictability are paramount [5] [1]. This comparison guide objectively analyzes the performance of engineered versus natural exosomes in chronic wound models, with a specific focus on strategies to mitigate heterogeneity and standardize dosing protocols.
Exosome heterogeneity stems from multiple factors throughout the production pipeline. The biological source of exosomes significantly influences their characteristics; mesenchymal stem cells (MSCs) from different tissues (adipose, bone marrow, umbilical cord) produce exosomes with distinct cargo profiles and therapeutic potentials [64]. The physiological state of parent cells—affected by age, metabolic conditions, and culture environment—further contributes to heterogeneity, as variations in factors like oxygen tension (hypoxia) or inflammatory priming alter exosome content and yield [5] [64].
Technical aspects of production introduce additional variability. Isolation methods such as ultracentrifugation, tangential flow filtration, size-exclusion chromatography, and polymer-based precipitation yield preparations with different purity profiles, potency, and recovery rates [63]. The culture system employed (2D vs. 3D) also significantly impacts output; research demonstrates that 3D culture systems can increase exosome yield and concentration in conditioned medium by 2.4-fold and 3.2-fold, respectively, compared to conventional 2D protocols [63].
Functional Consequences in Wound Healing Applications
In chronic wound models, this heterogeneity manifests functionally through variable performance across key healing parameters. Inconsistent anti-inflammatory effects may arise from fluctuating levels of immunomodulatory cargos (e.g., miR-146a, IL-10) between batches, potentially disrupting the precise balance required to transition wounds from chronic inflammation to proliferation [5] [65]. Similarly, variable pro-angiogenic capacity due to differential loading of vascular endothelial growth factor (VEGF) or pro-angiogenic miRNAs can lead to unpredictable neovascularization, compromising healing in ischemic wounds [66].
Divergent fibroblast modulation represents another concern, as inconsistent cargo may fail to properly coordinate the shift from fibroblast proliferation to maturation, potentially resulting in either delayed healing or excessive scar formation [5]. These functional implications underscore why addressing heterogeneity is not merely a manufacturing concern but a fundamental prerequisite for therapeutic efficacy.
Engineered vs. Natural Exosomes: A Comparative Analysis of Consistency
Performance Consistency in Chronic Wound Models
Table 1: Comparative Analysis of Engineered vs. Natural Exosomes in Chronic Wound Applications
| Parameter | Natural Exosomes | Engineered Exosomes | Experimental Evidence |
|---|---|---|---|
| Cargo Consistency | High batch-to-batch variability in miRNA, protein, and lipid profiles | Precisely controlled cargo loading via parental cell modification or direct loading | Natural exosome cargo depends on parent cell status; engineered exosomes allow standardized loading of specific miRNAs (e.g., miR-126 for angiogenesis) [67] [64] |
| Dosing Precision | Variable potency requiring frequent re-calibration; ~10-200 µg exosomal protein needed per mouse in preclinical studies | More predictable dose-response relationships due to standardized bioactive cargo | Studies report natural exosome dosing requires 109-1010 particles per treatment in mice with high variability; engineered versions show more consistent efficacy at similar particle counts [63] [68] |
| Targeting Efficiency | Limited inherent targeting; predominantly reliant on passive uptake | Enhanced active targeting via surface modifications (peptides, antibodies) | Engineered exosomes with RGD peptides show ~3.2-fold increased retention in wound beds compared to natural counterparts in rodent diabetic ulcer models [67] [66] |
| Production Yield | Generally low yield; challenging to scale | Improved production possible via engineered parent cells and optimized protocols | 3D culture systems with specialized media (VSCBIC-3) increase yield by 2.4-fold; engineering can further enhance production [63] |
| Functional Outcomes in Wounds | Variable effects on re-epithelialization, angiogenesis, and collagen deposition | More consistent and potent promotion of healing phases in diabetic, venous, and pressure ulcer models | Engineered exosomes with specific "4-pro and 5-anti" effects show more reproducible promotion of skin regeneration across batches [5] |
Experimental Evidence from Preclinical Models
Quantitative assessments in diabetic wound models demonstrate that engineered exosomes exhibit superior consistency in key healing parameters. In studies evaluating angiogenic potency, engineered exosomes loaded with specific pro-angiogenic miRNAs (miR-126, miR-210) consistently induced capillary density increases of 45-60% across multiple batches, whereas natural exosomes showed batch-dependent variation ranging from 15-55% enhancement [67]. Similarly, in re-epithelialization metrics, engineered versions promoted more predictable wound closure rates (85-90% reduction in wound area by day 14) compared to natural exosomes (60-85% reduction) in murine models [1].
The consistency of immunomodulatory effects further distinguishes these platforms. Engineered exosomes surface-modified to target macrophages reliably induced M2 polarization with a consistent 2.8-3.2-fold increase in anti-inflammatory markers (IL-10, Arg-1) across batches. Natural exosomes, in contrast, exhibited variable performance (1.5-3.0-fold increase) dependent on donor cell status and isolation methods [65]. This functional reproducibility positions engineered exosomes as a more reliable platform for clinical translation.
Strategic Approaches for Enhancing Batch Consistency
Production Protocol Standardization
Establishing robust, standardized production protocols is foundational to mitigating exosome heterogeneity. Research indicates that implementing defined culture conditions significantly enhances consistency. The use of serum-free, chemically-defined media such as the VSCBIC-3 solution prevents introduction of foreign vesicles from fetal bovine serum and maintains cell viability and morphology during exosome production, directly improving batch consistency [63].
Advanced 3D culture systems utilizing microcarriers increase production scale while enhancing purity and bioactivity. Compared to conventional 2D platforms, 3D systems demonstrate more uniform nutrient distribution and waste removal, creating a more homogeneous environment that yields exosomes with reduced batch-to-batch variation [63]. Combining 3D culture with tangential flow filtration (TFF) isolation further improves consistency by providing superior separation efficiency and reproducibility compared to traditional ultracentrifugation methods [63].
Table 2: Research Reagent Solutions for Consistent Exosome Production
| Reagent/Category | Specific Examples | Function in Consistency Management |
|---|---|---|
| Defined Culture Media | VSCBIC-3, SF-DMEM, dEx-DMEM | Eliminate serum-derived variability; maintain stable nutrient composition for reproducible exosome production [63] |
| 3D Culture Systems | Microcarriers (Cytodex, Synthemax), Bioreactors | Provide scalable, homogeneous culture environment; enhance yield and reduce heterogeneity through uniform cell-environment interactions [63] [68] |
| Isolation & Purification | Tangential Flow Filtration, Size-Exclusion Chromatography | Enable high-recovery, consistent isolation with minimal damage; improve batch-to-batch comparability [63] |
| Characterization Tools | NTA, Western Blot, miRNA Profiling | Quantify particle size, concentration, and specific markers (CD63, CD81, TSG101); verify batch consistency through multi-parameter assessment [63] [64] |
| Engineering Modification Tools | Lentiviral Vectors, Electroporation, Surface Conjugation | Introduce uniform therapeutic cargo (miRNAs, lncRNAs) or targeting ligands; standardize functional capacity across batches [67] [66] |
Engineering Solutions for Standardization
Exosome engineering provides powerful tools to actively counteract heterogeneity through precise cargo control. Parent cell engineering via genetic modification to overexpress specific therapeutic molecules (e.g., lncRNAs, miRNAs) ensures consistent cargo loading across production batches. For instance, engineering mesenchymal stem cells to overexpress the lncRNA KLF3-AS1 results in exosomes that reliably promote angiogenesis through consistent modulation of the VEGFA pathway [65].
Post-isolation modification approaches offer alternative standardization pathways. Direct loading of predetermined therapeutic cargo quantities into natural exosomes via electroporation or sonication enables precise dosing control, though potential impacts on vesicle integrity must be carefully managed [66]. Similarly, surface engineering with targeting ligands (RGD peptides, antibodies) confers consistent tissue homing properties independent of natural variations in surface protein composition [67] [66].
These engineering strategies transform exosomes from highly variable biological entities into more consistent therapeutic products with controlled characteristics and predictable biological effects—essential qualities for clinical development and regulatory approval.
Methodologies for Establishing Reproducible Dosing
Quantitative Dosing Protocols
Establishing reproducible dosing requires standardized quantification methods that correlate with therapeutic efficacy. Research indicates that a multi-parameter approach provides the most reliable dosing foundation. While particle concentration measured by nanoparticle tracking analysis (NTA) offers fundamental quantification, supplementing with protein content (μg/mL via BCA/ Bradford assay) and specific potency markers (e.g., therapeutic miRNA copies/vesicle) creates a more robust dosing framework [63] [68].
Experimental data from preclinical wound models provides guidance for dosing ranges. In murine diabetic wound studies, effective dosing typically falls between 10⁹-10¹⁰ particles or 10-200 μg exosomal protein per application [63] [68]. For engineered exosomes with enhanced potency, the lower end of this spectrum often suffices, potentially reducing manufacturing requirements. Importantly, dosing frequency requires optimization based on wound environment dynamics; chronic wounds with elevated protease activity may necessitate more frequent application (e.g., every 48-72 hours) to maintain therapeutic levels [1].
Bioactivity Standardization Assays
Beyond physical quantification, functional potency assays are critical for ensuring reproducible biological effects. Standardized in vitro bioassays measuring specific wound-relevant activities provide essential quality control. These include:
- Fibroblast migration assays (scratch/wound closure models) quantifying concentration-dependent enhancement of cell migration [63]
- Angiogenesis protocols (tube formation assays with HUVECs) measuring pro-angiogenic potential [66]
- Macrophage polarization assays flow cytometric analysis of M1/M2 marker expression following exosome treatment [65]
Establishing correlation between in vitro potency metrics and in vivo efficacy enables development of potency units based on biological activity rather than solely physical characteristics. For instance, engineering exosomes to deliver consistent copies of specific lncRNAs (e.g., MEG3 for anti-fibrotic effects or DUXAP10 for rejuvenation) per vesicle provides a quantifiable biological standard for dosing [65].
Experimental Workflows for Consistency Assessment
Comprehensive Characterization Pipeline
The following diagram illustrates the key experimental workflow for assessing exosome batch consistency:
Diagram 1: Experimental workflow for comprehensive exosome batch consistency assessment
Detailed Experimental Protocols
Protocol 1: Standardized Production Using 3D Culture Systems
This protocol, adapted from established upscaling methodologies, enhances batch consistency through controlled culture conditions [63]:
- Cell Culture: Expand adipose-derived mesenchymal stem cells (AD-MSCs) in defined growth media to 80% confluence
- 3D Seeding: Transfer 1×10⁷ cells to 100 mL spinner flask containing 0.5 g/L microcarriers (e.g., Cytodex 3) in VSCBIC-3 serum-free medium
- Conditioning: After 24 hours attachment, replace medium with exosome-collecting solution (VSCBIC-3) and culture for 48 hours
- Harvesting: Collect conditioned medium and separate cells/debris via centrifugation at 2,000 × g for 30 minutes
- Concentration: Concentrate supernatant 20-50× using tangential flow filtration with 100 kDa cutoff membranes
- Isolation: Purify exosomes using size-exclusion chromatography (qEV columns) or ultracentrifugation at 100,000 × g for 70 minutes
- Characterization: Quantify yield via NTA and BCA protein assay; verify markers (CD63, CD81, TSG101) by western blot
Protocol 2: Potency Assessment Through Fibroblast Migration Assay
This functional assay evaluates batch consistency through biological activity measurement [63]:
- Cell Preparation: Culture human dermal fibroblasts in DMEM with 10% FBS to 90% confluence
- Scratch Creation: Create uniform scratch wound using pipette tip on confluent monolayer
- Treatment: Apply exosomes at standardized concentration (1×10⁹ particles/mL) in serum-free medium
- Imaging: Capture images at scratch creation (0h) and 24h post-treatment using phase-contrast microscopy
- Quantification: Calculate migration area using ImageJ software with formula: Migration Area = (A₀ - A₂₄)/A₀ × 100%
- Acceptance Criteria: Consistent batches demonstrate migration enhancement of 40-60% relative to untreated controls
The journey toward clinically viable exosome therapies for chronic wounds necessitates a fundamental shift from variable biological products to standardized pharmaceutical agents. Engineered exosomes demonstrate distinct advantages in achieving the batch-to-batch consistency and reproducible dosing required for reliable therapeutic outcomes. Through integrated strategies combining production protocol standardization, precision engineering, and comprehensive characterization, researchers can systematically address the challenges of heterogeneity. The continued refinement of these approaches will ultimately enable the full therapeutic potential of exosome technology, transforming wound care and regenerative medicine through consistent, predictable, and efficacious treatments.
Head-to-Head: Validating Therapeutic Efficacy in Preclinical and Clinical Chronic Wound Models
This comparison guide provides an objective analysis of experimental data and methodologies for evaluating bioactivity in key wound healing processes. We systematically compare the effects of various therapeutic candidates, including engineered exosomes, natural exosomes, and other bioactive molecules, on fibroblast proliferation, keratinocyte migration, and macrophage polarization in vitro. Designed for researchers, scientists, and drug development professionals, this guide synthesizes quantitative data, detailed protocols, and molecular mechanisms to facilitate informed decision-making in chronic wound research, with particular emphasis on the evolving context of engineered versus natural exosomes.
Successful wound healing relies on the coordinated functions of multiple cell types. Fibroblasts, keratinocytes, and macrophages constitute a fundamental cellular triad that drives the repair process through proliferation, migration, and phenotypic polarization, respectively. In vitro assessment of these cellular activities provides critical predictive data for therapeutic efficacy before advancing to complex in vivo models.
The emergence of exosome-based therapies has introduced promising approaches for modulating wound healing pathways. Exosomes are small, endosome-derived membrane vesicles (30-150 nm in diameter) that play key roles in intercellular communication by transferring bioactive cargo such as proteins, lipids, and nucleic acids [17]. While natural exosomes derived from sources like mesenchymal stem cells (MSCs) exhibit inherent therapeutic potential, engineered exosomes are being developed with enhanced targeting specificity and optimized cargo loading to overcome limitations of their natural counterparts [5].
This guide establishes standardized frameworks for comparing therapeutic interventions across these essential wound healing assays, with particular attention to the differential effects of natural and engineered exosome preparations.
Fibroblast Proliferation Assays
Quantitative Comparison of Pro-Fibrotic Agents
Fibroblast proliferation is a cornerstone of tissue repair, driving granulation tissue formation and extracellular matrix (ECM) deposition. The following table summarizes quantitative data for various compounds that enhance fibroblast proliferation in vitro.
Table 1: Comparison of Pro-Fibrotic Agents in Fibroblast Proliferation Assays
| Therapeutic Agent | Cell Type | Assay Method | Key Proliferation Findings | Signaling Pathways Activated |
|---|---|---|---|---|
| Collectin-11 (CL-11) [69] | Primary renal fibroblasts | EdU assay, PCNA Western blot | Significant proliferation increase | ERK, AKT/mTOR, STAT3, SMAD2 |
| PCL-Zn-ECM (PZE) Scaffold [70] | NIH3T3 fibroblasts | Metabolic activity (Alamar Blue), LDH assay | Enhanced cell viability and proliferation vs. PCL alone | Not specified |
| MSC-derived Exosomes [1] | Human dermal fibroblasts | CCK-8 assay, BrdU incorporation | Dose-dependent proliferation increase | TGF-β/SMAD, ERK |
Experimental Protocol: EdU Proliferation Assay
The EdU (5-ethynyl-2'-deoxyuridine) assay provides a sensitive, non-radioactive method for detecting proliferating cells [69]. This protocol is adapted from CL-11 research:
- Cell Seeding: Plate fibroblasts in complete medium at 5×10³ cells/well in 96-well plates and incubate overnight.
- Treatment Application: Replace medium with serum-free medium containing test compounds (e.g., recombinant CL-11 at 1-5 μg/mL) or vehicle control.
- EdU Labeling: Add EdU solution to culture medium (final concentration 10 μM) and incubate for 6-24 hours.
- Cell Fixation: Remove medium, rinse with PBS, and fix cells with 4% paraformaldehyde for 15 minutes.
- Detection: Permeabilize cells with 0.5% Triton X-100, then apply Alexa Fluor 488-azide using the Click-iT EdU kit per manufacturer instructions.
- Counterstaining and Analysis: Stain nuclei with DAPI, visualize with fluorescence microscopy, and quantify EdU-positive cells.
Alternative Method: PCNA Western blot analysis can provide complementary proliferation data through protein expression levels [69].
Research Reagent Solutions: Fibroblast Proliferation
Table 2: Essential Reagents for Fibroblast Proliferation Assays
| Reagent / Kit | Function / Application | Example Source / Catalog |
|---|---|---|
| Click-iT EdU Kit [69] | Fluorescent detection of DNA synthesis in proliferating cells | Thermo Fisher Scientific (C10337) |
| Anti-PCNA Antibody [69] | Western blot detection of proliferation marker | Santa Cruz Biotechnology (sc-56) |
| Recombinant Collectin-11 [69] | Stimulates fibroblast proliferation and activation | Novoprotein |
| PCL-Zn Composite [70] | Biomaterial scaffold enhancing fibroblast growth | Fabricated via electrospinning |
Keratinocyte Migration Assays
Quantitative Comparison of Pro-Migratory Agents
Keratinocyte migration is essential for re-epithelialization during wound healing. The following table compares interventions that enhance this process.
Table 3: Comparison of Pro-Migratory Agents in Keratinocyte Migration Assays
| Therapeutic Agent | Cell Type | Assay Method | Key Migration Findings | Signaling Pathways |
|---|---|---|---|---|
| FOSL1 [71] | HaCaT keratinocytes | Transwell, Scratch assay | Promoted migration via IL-17 pathway | IL-17 signaling |
| TRPV3 activation [72] | Mouse keratinocytes | Scratch assay | Accelerated migration in vitro (not in vivo) | EGFR-mediated mechanism |
| MSC-derived Exosomes [1] | Human keratinocytes | Scratch assay | Enhanced migration rate | Not specified |
Experimental Protocol: Oris Pro Gap Closure Assay
The Oris Pro cell migration assay provides a standardized, high-throughput compatible method for quantifying cell migration without mechanical wound induction [73]. This protocol is adapted from fibroblast research with applications for keratinocytes:
- Plate Preparation: Use Oris Pro 96-well plates containing dissolvable plugs in the center of each well.
- Cell Seeding: Trypsinize and resuspend keratinocytes (e.g., HaCaT cells) in serum-free medium. Seed at 5×10⁴ cells/well (100% confluent) and incubate for 3 hours for cell attachment.
- Plug Dissolution and Baseline Imaging: Confirm plug dissolution (approximately 30 minutes) creating a uniform cell-free detection zone. Acquire phase-contrast images at 0-hour timepoint using microscope with built-in incubator chamber.
- Treatment Application: Add pre-warmed treatment media (containing test compounds, conditioned media, or controls) to appropriate wells. Ensure consistent serum concentration across all wells.
- Timepoint Imaging: Capture images at designated intervals (e.g., 24h, 48h) for each well, maintaining identical positioning.
- Image Analysis:
- Use Fiji/ImageJ software for processing
- Invert image colors to enhance border clarity
- Manually select cell-free zones using circular selection tool
- Measure area at each timepoint
- Calculate migration area: (Area 0h - Area 24h)
- Normalize data to control conditions
Figure 1: Keratinocyte Migration Experimental Workflow
FOSL1-IL17 Signaling Pathway in Keratinocyte Migration
Research demonstrates that the transcription factor FOSL1 promotes keratinocyte migration through IL-17 signaling pathway modulation [71]. The following diagram illustrates this molecular mechanism:
Figure 2: FOSL1-IL17 Signaling in Keratinocyte Migration
Macrophage Polarization Assays
Standardized Protocol for M2-like Macrophage Induction
Macrophage polarization toward M2-like phenotypes is crucial for inflammation resolution and tissue repair. The following generalized protocol enables efficient, reproducible induction of M2-like macrophages from mouse and rat bone marrow mononuclear cells (BMNCs) [74]:
BMNC Isolation:
- Euthanize mice/rats following institutional guidelines
- Excise femurs and tibiae under sterile conditions
- Sterilize bones in 70% ethanol (5 min) → cold PBS (5 min) → cold DMEM (5 min)
- Flush bone marrow with cold PBS using 22G/24G needle
- Filter cell suspension through 70 μm strainer
- Isolate BMNCs using Histopaque-1083 density gradient centrifugation
M2-like Macrophage Differentiation:
- Culture BMNCs in high-glucose DMEM with 10% FBS
- Add species-specific recombinant Mcsf (e.g., 25 ng/mL for mouse)
- Incubate for 72 hours to generate M0 macrophages
- Stimulate with recombinant IL-4 (20 ng/mL) for 48 hours to induce M2 polarization
Phenotype Validation:
- Flow Cytometry: Analyze M2 markers (CD206, CD163, Arg1)
- Gene Expression: qRT-PCR for M2 markers (Arg1, Retnla, Igf1, Il10)
- Functional Assays: Phagocytosis, cytokine secretion profiling
Research Reagent Solutions: Macrophage Polarization
Table 4: Essential Reagents for Macrophage Polarization Assays
| Reagent | Function / Application | Example Source / Catalog |
|---|---|---|
| Recombinant Mcsf [74] | Differentiation of BMNCs to macrophages | Peprotech (400-28 for rat, 315-02 for mouse) |
| Recombinant IL-4 [74] | Polarization to M2-like phenotype | Peprotech (400-04 for rat, 214-14 for mouse) |
| Anti-CD206 Antibody [74] | Flow cytometry detection of M2 marker | Bio Legend (141710) |
| Histopaque-1083 [74] | BMNC isolation by density gradient | Sigma Aldrich (10831) |
Engineered vs Natural Exosomes in Macrophage Polarization
Engineered exosomes (eExo) represent an advanced therapeutic platform with enhanced capabilities compared to natural exosomes. The following table highlights key differences:
Table 5: Engineered vs Natural Exosomes in Wound Healing Applications
| Characteristic | Natural Exosomes | Engineered Exosomes |
|---|---|---|
| Targeting Specificity | Limited inherent tropism | Enhanced via surface modification [5] |
| Cargo Loading | Natural, unmodified content | Precisely controlled therapeutic cargo [5] [17] |
| Production Consistency | Batch-to-batch variability | Improved standardization potential [5] |
| Therapeutic Effects | "4-pro": pro-angiogenic, pro-proliferation, pro-migration, pro-matrix remodeling [5] | Enhanced "4-pro" plus "5-anti": anti-inflammatory, anti-oxidative, anti-apoptotic, anti-microbial, anti-fibrotic [5] |
| Manufacturing Scalability | Moderate challenges | Advanced but with remaining hurdles [17] |
Figure 3: Engineered Exosome Modification Strategies
Comparative Analysis and Research Applications
Cross-Assay Correlation and Predictive Value
Integrating data from proliferation, migration, and polarization assays provides comprehensive insights into therapeutic potential. The most promising wound healing candidates demonstrate consistent efficacy across all three assay types:
- Engineered exosomes show particular promise by simultaneously promoting fibroblast proliferation and keratinocyte migration while modulating macrophage polarization toward regenerative M2 phenotypes [5].
- Multi-target therapies addressing all three cellular processes typically demonstrate superior in vivo efficacy compared to single-target approaches.
- Biomaterial scaffolds such as PZE composites enhance fibroblast responses while potentially serving as delivery platforms for exosomal therapies [70].
Technical Considerations for Assay Selection
When designing experiments to assess bioactivity in wound healing contexts:
- Proliferation Assays: Combine EdU staining with metabolic activity assays (e.g., Alamar Blue) to distinguish true proliferation from enhanced metabolic activity.
- Migration Assays: Utilize both gap-closure (Oris Pro) and transmembrane (Boyden chamber) approaches to assess different migration modes.
- Polarization Assays: Employ multiple validation methods (surface markers, gene expression, cytokine secretion) to comprehensively characterize macrophage phenotypes.
- Exosome Characterization: Implement nanoparticle tracking analysis (NTA), transmission electron microscopy (TEM), and Western blotting for CD81, CD63, CD9 to verify exosome quality [17].
This comparison guide provides a standardized framework for assessing bioactivity across fundamental wound healing processes. The integrated data demonstrates that while natural exosomes and other bioactive compounds show significant potential, engineered exosomes represent a promising advancement with enhanced targeting specificity and controllable therapeutic cargo. The continued refinement of in vitro assessment protocols, particularly those enabling high-throughput screening of engineered exosome candidates, will accelerate the development of effective therapies for chronic wounds and fibrotic conditions.
Researchers should consider implementing the described methodologies and comparative frameworks to systematically evaluate novel therapeutic candidates, with particular attention to their differential effects across fibroblast proliferation, keratinocyte migration, and macrophage polarization assays.
Chronic wounds, including diabetic, venous, and pressure ulcers, represent a significant clinical challenge with limited effective treatment options. Exosome therapy has emerged as a promising regenerative medicine approach, leveraging these nanoscale extracellular vesicles for their innate role in intercellular communication and wound repair processes [75] [76]. Exosomes are broadly categorized into two groups: natural exosomes, which are isolated directly from cell cultures without further modification, and engineered exosomes (eExo), which are purposefully modified to enhance their therapeutic properties through cargo loading or surface modifications [75] [5]. Within the context of a broader thesis on chronic wound research, this guide provides an objective comparison of the preclinical efficacy of these two exosome types across different chronic wound models, supported by experimental data and detailed methodologies to inform researchers, scientists, and drug development professionals.
Comparative Efficacy Data in Preclinical Models
The therapeutic potential of exosomes has been evaluated across various animal models of chronic wounds. The tables below summarize key quantitative findings from preclinical studies, comparing the performance of natural and engineered exosomes.
Table 1: Efficacy of Natural Exosomes in Preclinical Chronic Wound Models
| Wound Model | Exosome Source | Key Efficacy Metrics | Reported Outcomes | Citation |
|---|---|---|---|---|
| Diabetic Ulcer | Adipose-derived Stem Cells (ASCs) | - Wound closure rate- Granulation tissue formation- Angiogenesis | - Visible granulation within 2 weeks- Complete closure in 3/4 cases (94 days median) | [46] |
| Diabetic Ulcer | Mesenchymal Stem Cells (MSCs) | - Inflammation modulation- Re-epithelialization- Collagen remodeling | - Shortened inflammatory phase- Accelerated angiogenesis and cell migration | [77] [78] |
| Venous Ulcer | MSC-derived | - Arterial Resistive Index (RI)- Venous reflux time | - RI decreased from 0.93 to 0.77- Reflux time fell from 2.8s to 1.4s | [46] |
| Diabetic Peripheral Neuropathy | Multiple Sources (MSC, Schwann cell) | - Nerve Conduction Velocity (NCV)- Intraepidermal Nerve Fiber Density (IENFD) | - Significant improvement in MCV and SCV- Trend toward restored IENFD | [79] |
Table 2: Enhanced Efficacy of Engineered Exosomes in Preclinical Models
| Engineering Strategy | Wound Model | Key Efficacy Metrics | Reported Outcomes vs. Natural Exosomes | Citation |
|---|---|---|---|---|
| Ligand modification for targeted delivery | Diabetic Ulcer | - Target cell specificity- Wound closure rate- Therapeutic cargo delivery | Improved targeting specificity and enhanced cellular uptake | [75] [5] |
| Cargo optimization (e.g., specific miRNAs, growth factors) | Chronic Non-Healing Wound | - Angiogenesis- Re-epithelialization- Collagen deposition | Superior pro-angiogenic and immunomodulatory effects | [5] [7] |
| Biomimetic engineering | Pathological Scarring | - Scar tissue formation- Collagen fiber organization | Enhanced anti-scarring inhibition effects | [5] |
| "Stealth" coatings for improved biocompatibility | Diabetic Ulcer | - Circulation half-life- Immunogenicity | Improved stability and reduced clearance | [75] |
Detailed Experimental Protocols
To ensure reproducibility and provide a clear technical framework, this section outlines standardized methodologies for key experiments cited in the comparative data.
Protocol for Topical Exosome Application in Rodent Diabetic Ulcer Model
This protocol is adapted from a clinical case series [46] and common preclinical practices.
- Animal Model Induction: Utilize streptozotocin (STZ)-induced diabetic rodents (e.g., C57BL/6J mice or Sprague-Dawley rats). Confirm stable hyperglycemia (blood glucose >300 mg/dL) for 2 weeks prior to wounding.
- Wound Creation: Create full-thickness excisional wounds (e.g., 6-8 mm diameter) on the dorsal skin under anesthesia. Apply a silicone splint to prevent natural contraction and model human healing.
- Exosome Preparation and Application:
- Natural Exosomes: Isolate exosomes from human adipose-derived stem cell (ADSC) culture supernatant via sequential ultracentrifugation. Characterize by nanoparticle tracking analysis (size/concentration) and Western blot (CD63, CD81, TSG101).
- Application: Resuspend exosomes in PBS at a concentration of 1 × 10^12 particles/mL. Apply topically at a dose of 0.1 mL/cm² of wound area on day 0 (post-wounding) and then weekly.
- Control Group: Apply an equivalent volume of PBS.
- Wound Assessment and Monitoring:
- Closure Rate: Capture digital images every 2-3 days. Use planimetry software to calculate wound area and determine percentage closure over time.
- Histological Analysis: Harvest wound tissue at endpoints (e.g., days 7, 14). Process for H&E staining to assess re-epithelialization and granulation tissue thickness, and Masson's trichrome staining to evaluate collagen deposition and maturity.
- Immunohistochemistry: Stain for CD31 to quantify capillary density (angiogenesis) and for specific macrophages (e.g., iNOS for M1, CD206 for M2) to assess immune modulation.
Protocol for Engineering Exosomes via Cargo Loading
This protocol describes a common method for loading therapeutic miRNAs into exosomes [5] [7].
- Parent Cell Pre-treatment:
- Culture ADSCs to 70% confluency.
- Transfect cells with miRNA mimics (e.g., miR-126-3p for angiogenesis) or inhibitors using a suitable transfection reagent. Use a non-targeting miRNA sequence as a negative control.
- Incubate for 48 hours to allow for miRNA overexpression and subsequent loading into nascent exosomes.
- Engineered Exosome Isolation and Validation:
- Collect the conditioned media and isolate exosomes via ultracentrifugation as in section 3.1.
- Validate successful miRNA loading using quantitative RT-PCR. Extract total RNA from the purified exosomes and measure the levels of the target miRNA relative to a control (e.g., U6 snRNA).
- Functional Validation in Vitro:
- Perform a tube formation assay using Human Umbilical Vein Endothelial Cells (HUVECs).
- Treat HUVECs with engineered exosomes, natural exosomes (from non-transfected cells), and PBS control.
- Quantify parameters like total tube length, number of branches, and number of meshes to confirm the enhanced pro-angiogenic effect of the engineered exosomes.
Signaling Pathways in Exosome-Mediated Wound Repair
Exosomes derived from MSCs promote healing in chronic wounds by modulating key signaling pathways across different phases of repair. The diagram below illustrates the primary mechanisms of action in target cells like fibroblasts, endothelial cells, and macrophages.
The diagram illustrates how MSC-derived exosomes deliver functional cargo (proteins, miRNAs, lipids) to key skin cells, facilitating diabetic wound repair through multiple coordinated mechanisms [77] [78] [76]. In fibroblasts, exosomal TGF-β/Smad signaling activation promotes collagen synthesis and improves the MMP/TIMP balance for better extracellular matrix (ECM) remodeling [76]. In endothelial cells, exosomes activate PI3K/Akt and other pathways to upregulate VEGF and FGF signaling, thereby stimulating angiogenesis which is crucial for nutrient delivery in ischemic wounds [77] [78]. For macrophages, exosomal cues modulate the NF-κB pathway to shift polarization from pro-inflammatory M1 to pro-healing M2 phenotypes, resolving chronic inflammation characteristic of diabetic wounds [77] [76].
The Scientist's Toolkit: Research Reagent Solutions
The following table details essential materials and reagents commonly used in exosome research for chronic wound healing, along with their primary functions in experimental workflows.
Table 3: Key Research Reagents and Materials for Exosome Studies
| Reagent/Material | Function in Research | Specific Examples & Notes |
|---|---|---|
| Mesenchymal Stem Cells (MSCs) | Source of natural and engineered exosomes. | Adipose tissue (ADSCs), Bone Marrow (BMSCs), and Umbilical Cord (UCMSCs) are common sources. UCMSCs are cost-effective and have high self-renewal capacity [80]. |
| Sequential Ultracentrifugation | Standard method for isolating exosomes from cell culture media. | Involves differential spins to remove cells, debris, and larger vesicles, followed by a high-speed pelletization of exosomes [46]. |
| Nanoparticle Tracking Analysis (NTA) | Characterizes exosome size distribution and concentration. | Instruments like Malvern Nanosight quantify particles per milliliter and confirm vesicle size (typically 30-150 nm) [46] [78]. |
| Tetraspanin Antibodies | Confirms exosome identity via surface markers. | Antibodies against CD63, CD81, and CD9 are used in Western Blot or flow cytometry for characterization [75] [80]. |
| miRNA Mimics/Inhibitors | For engineering exosomes with enhanced or suppressed miRNA cargo. | Used to transfect parent cells to load specific miRNAs (e.g., miR-126 for angiogenesis) into exosomes [5] [7]. |
| Hydrogel-based Dressings | Biomaterial scaffold for sustained exosome delivery. | Chitosan or hyaluronic acid hydrogels can extend exosome residency and provide a moist wound environment [78] [5]. |
| Streptozotocin (STZ) | Chemical for inducing Type 1 Diabetes in rodent models. | Creates a hyperglycemic environment essential for studying diabetic wound healing impairments [79]. |
Preclinical data robustly demonstrates the therapeutic potential of both natural and engineered exosomes in enhancing healing across diabetic, venous, and pressure ulcer models. Natural exosomes function as multifaceted signaling packages, effectively modulating inflammation, promoting angiogenesis, and encouraging tissue regeneration [77] [46] [76]. Engineered exosomes (eExo), however, consistently show a superior efficacy profile by leveraging targeted delivery systems, optimized therapeutic cargo, and enhanced biocompatibility to address the complex pathophysiology of chronic wounds more precisely [75] [5]. The future clinical translation of these therapies hinges on overcoming challenges related to scalable GMP-compliant production, establishing rigorous quality control standards, and validating long-term safety and efficacy through large-scale clinical trials [75]. The ongoing refinement of engineering strategies promises to usher in a new era of precision medicine for chronic wound management.
The therapeutic application of exosomes, particularly in managing chronic wounds, represents a paradigm shift in regenerative medicine. Chronic wounds, characterized by impaired vascularity, persistent inflammation, and extracellular matrix dysfunction, present a significant clinical challenge with limited effective treatments [46]. Exosomes—nanoscale extracellular vesicles secreted by cells—have emerged as promising acellular therapeutic agents. They mediate intercellular communication by transporting bioactive molecules such as proteins, lipids, and nucleic acids, influencing various aspects of wound healing including angiogenesis, inflammation modulation, and cell proliferation [67] [13].
A critical distinction exists between naturally secreted exosomes and engineered exosomes (eExo). Natural exosomes derive their cargo composition passively from their parent cells, whereas eExo are deliberately modified to enhance therapeutic properties. Engineering strategies aim to improve cargo loading, targeting specificity, tissue penetration, and environmental stability, thereby potentially overcoming limitations of natural exosomes for complex wound environments [81] [82]. This review systematically evaluates clinical and preclinical evidence comparing the efficacy of natural and engineered exosomes, focusing on two critical wound healing parameters: wound closure rates and perfusion improvement.
Clinical Case Evidence: Natural Exosomes
A recent clinical case series provides compelling human data on the efficacy of natural, non-engineered exosomes for treating refractory chronic wounds [46]. The study involved four patients with chronic lower-extremity ulcers of varying etiologies (venous, diabetic, arterial) that had persisted for at least 6 months and failed conventional therapies including compression, debridement, and topical care.
Therapeutic Protocol and Outcomes
Patients received monthly topical applications of adipose-derived stem-cell exosomes (Exo-HL) at a concentration of 1×10^12 particles/mL, applied at 0.1 mL/cm² wound area. Outcomes were assessed through serial wound measurements and Doppler ultrasonography at baseline and 3-month intervals.
Table 1: Clinical Outcomes from Natural Exosome Case Series [46]
| Case | Age | Wound Type | Baseline Area (cm²) | Time to Closure (days) | Arterial Resistive Index (Pre/Post) | Venous Reflux Time (Pre/Post seconds) |
|---|---|---|---|---|---|---|
| 1 | 58 | Venous Ulcer | Not specified | Improved (not closed) | 0.89 → 0.72 | 2.8 → 1.2 |
| 2 | 62 | Venous Ulcer | Not specified | 60 | 0.92 → 0.78 | Improved |
| 3 | 42 | Venous Ulcer | Not specified | Nearly closed by 7 months | Improved | Improved |
| 4 | Not specified | Mixed | 12.4 (median) | 94 (median for 3 cases) | 0.93 → 0.77 (mean) | 2.8 → 1.4 (mean) |
Key Clinical Findings
The case series demonstrated consistent wound healing progression across all patients [46]:
- Rapid Granulation: All wounds showed visible granulation tissue formation within 2 weeks of initial application.
- Closure Achievement: Three of four achieved complete wound closure after a median of 94 days.
- Perfusion Enhancement: Doppler studies confirmed significant microcirculatory improvements, with mean arterial resistive index decreasing from 0.93±0.04 to 0.77±0.03, indicating improved distal flow.
- Venous Function Improvement: Venous reflux time fell from 2.8±0.3 to 1.4±0.2 seconds, suggesting improved venous valve function.
This clinical evidence, though limited by sample size and lack of control group, provides direct human data supporting natural exosome therapy for wound healing and perfusion improvement.
Preclinical Evidence: Engineered vs. Natural Exosomes
Preclinical studies offer controlled environments to directly compare engineered and natural exosomes, providing insights into their mechanistic differences and relative efficacies.
Meta-Analysis of Preclinical Studies
A comprehensive meta-analysis of 83 preclinical studies evaluated mesenchymal stem cell-derived extracellular vesicles (MSC-EVs) in wound healing and skin regeneration [16]. The analysis included various EV types, including small EVs (sEVs, often referred to as exosomes) and apoptotic EVs (ApoEVs).
Table 2: Preclinical Meta-Analysis: Efficacy Comparison by Exosome Characteristics [16]
| Factor Analyzed | Subgroup | Relative Efficacy in Wound Healing | Notes |
|---|---|---|---|
| EV Type | Apoptotic sEVs (ApoSEVs) | Best for wound closure, collagen deposition | Compared to sEVs and ApoBDs |
| Small EVs (sEVs)/Exosomes | Best for revascularization | Superior to ApoEVs in angiogenesis | |
| Administration Route | Subcutaneous Injection | Superior for closure, collagen, revascularization | Compared to topical dressing/covering |
| MSC Source | Adipose-derived (ADSCs) | Best for wound closure rate, collagen deposition | |
| Bone Marrow (BMMSCs) | Best for revascularization |
Engineering Strategies and Enhanced Efficacy
Engineering approaches significantly enhance exosome therapeutic potential through multiple strategies [81] [82]:
Parent Cell Modification: Genetically engineering parent cells to produce exosomes enriched with specific therapeutic molecules (e.g., miRNA-31-5p, HOTAIR lncRNA) [82].
Direct Cargo Loading: Using electroporation, ultrasound, or incubation to directly load therapeutic agents (e.g., miRNA-21-5p, curcumin) into isolated exosomes [82].
Surface Modification: Altering exosome surface proteins to enhance tissue-specific targeting.
Biomaterial Integration: Combining exosomes with hydrogels or scaffolds to improve retention and controlled release at wound sites [83].
Diagram: Enhanced Therapeutic Mechanisms of Engineered vs. Natural Exosomes in Wound Healing. Engineered exosomes utilize multiple strategic advantages to improve therapeutic outcomes across various wound healing parameters.
Advanced Engineering: Integrated Delivery Systems
The most sophisticated engineering approaches combine exosomes with advanced biomaterials to create integrated therapeutic systems. A notable example is the development of exosome-coated oxygen nanobubble-laden hydrogel [83].
Technology Design and Workflow
This system addresses a critical limitation in chronic wound environments: hypoxia-induced compromise of exosome uptake [83]. The engineering involves:
- Oxygen Nanobubble (ONB) Core: Glycosylated protein conjugates encapsulating nanoscale oxygen bubbles for sustained oxygen release.
- Exosome Coating: ADSC-derived exosomes coated onto ONB surface through ultrasonication.
- Hydrogel Matrix: Polyvinyl alcohol/gelatin-borax hybrid hydrogel providing self-healing properties, hemostasis, and anti-inflammatory activity.
Diagram: Integrated Exosome Engineering Workflow. This multi-step process creates a sophisticated delivery system combining oxygen nanobubbles, exosomes, and hydrogel matrix to address multiple wound healing barriers simultaneously.
Enhanced Efficacy of Engineered Systems
In vivo studies in a rat full-thickness wound model demonstrated the superior performance of this engineered system compared to natural exosomes [83]:
- Accelerated Wound Closure: Significant reduction in wound size compared to natural exosome treatment.
- Improved Exosome Delivery: Enhanced intracellular exosome delivery in hypoxic wound tissue.
- Hypoxia Amelioration: Sustained oxygen release countered wound hypoxia.
- Anti-inflammatory Effects: Borate bonds in hydrogel decomposed hydrogen peroxide, reducing oxidative stress.
This integrated approach represents a significant advancement over natural exosome applications, simultaneously addressing multiple pathological aspects of chronic wounds.
Experimental Protocols and Methodologies
Standardized Exosome Isolation and Characterization
Robust methodology is essential for valid experimental outcomes in exosome research. Based on the analyzed studies, key protocols emerge:
Isolation Methods [46] [48] [83]:
- Sequential Ultracentrifugation: The most common method, involving centrifugation at increasing speeds (300×g for cell removal, 2000×g for debris, 100,000×g for exosome pelleting).
- Size-Exclusion Chromatography: Alternative method providing high purity exosome preparations.
- Precipitation-Based Kits: Commercial kits offering convenience but potential impurity co-precipitation.
Characterization Requirements [16] [48]:
- Nanoparticle Tracking Analysis (NTA): For determining exosome size distribution (typically 40-160nm) and concentration.
- Transmission Electron Microscopy (TEM): For visualizing cup-shaped spherical morphology.
- Western Blotting: For detecting exosome surface markers (CD9, CD63, CD81).
- Protein Quantification: Bicinchoninic acid (BCA) assay for standardizing dosages.
In Vivo Wound Healing Assessment
Standardized protocols enable comparison across studies [46] [16] [48]:
Animal Models:
- Diabetic models (db/db mice, STZ-induced) for chronic wound simulation.
- Full-thickness excisional wounds (dorsal, diabetic foot, leg).
- Burn, scleroderma, and photoaging models for specific wound types.
Outcome Measures:
- Wound Closure Rate: Serial measurement of wound area using digital photography with scale.
- Histological Analysis: Collagen deposition, granulation tissue formation, re-epithelialization.
- Perfusion Assessment: Doppler ultrasonography for arterial resistive index and venous reflux time.
- Angiogenesis Quantification: Immunohistochemistry for CD31+ blood vessels (microvessel density).
Table 3: Key Research Reagent Solutions for Exosome Wound Healing Studies
| Reagent/Category | Specific Examples | Function/Application | Evidence Source |
|---|---|---|---|
| Exosome Source | Adipose-Derived Stem Cells (ADSCs) | Superior for wound closure, collagen deposition | [16] [48] |
| Umbilical Cord MSCs | Promote angiogenesis, reduce scarring | [82] | |
| Isolation Kits | Exo-HL (Primoris International) | Clinical-grade exosome production | [46] |
| Characterization Tools | Nanoparticle Tracking Analysis (NTA) | Size distribution and concentration | [48] [83] |
| Transmission Electron Microscopy | Morphological verification | [48] [83] | |
| Delivery Systems | PVA/Gelatin-Borax Hydrogel | Self-healing, antioxidative dressing | [83] |
| Oxygen Nanobubbles (ONB) | Hypoxia amelioration in wounds | [83] | |
| Animal Models | db/db Mice | Type 2 diabetes chronic wound model | [16] [48] |
| STZ-Induced Diabetic Mice | Type 1 diabetes wound model | [16] |
The evidence reviewed demonstrates significant potential for both natural and engineered exosomes in enhancing wound closure rates and perfusion improvement. Clinical case studies establish the real-world efficacy of natural exosomes, showing complete wound closure in 3 of 4 refractory cases with simultaneous improvement in arterial and venous perfusion parameters [46].
Preclinical evidence indicates that engineered exosomes hold promise for superior therapeutic outcomes through enhanced cargo delivery, targeted specificity, and improved tissue penetration [81] [82]. Advanced engineering strategies that integrate exosomes with oxygen-delivery systems and bioactive hydrogels demonstrate particularly robust effects, addressing multiple pathological aspects of chronic wounds simultaneously [83].
Future research directions should focus on standardizing isolation and characterization protocols, conducting larger controlled clinical trials, and developing more sophisticated engineering approaches that can dynamically respond to the wound microenvironment. The evolving field of exosome engineering represents a promising frontier for developing effective therapies for complex chronic wounds that remain challenging to manage with current treatment modalities.
Chronic wounds, characterized by a failure to proceed through an orderly and timely healing process within three months, represent a significant global health challenge due to their complex pathophysiology and resistance to conventional treatments [5] [11]. In this landscape, exosome-based therapies have emerged as promising regenerative solutions. Natural exosomes are nanoscale extracellular vesicles (30-150 nm) secreted natively by cells, serving as crucial intercellular communicators by transporting bioactive cargoes like proteins, lipids, and nucleic acids [1] [17]. These vesicles originate from the endosomal system, forming through inward budding of endosomal membranes to create multivesicular bodies that subsequently fuse with the plasma membrane for release [36].
Engineered exosomes represent the next evolutionary step in vesicular therapeutics - natural exosomes purposefully modified through bioengineering techniques to enhance their therapeutic properties [7]. The fundamental distinction lies in intentional modification: while natural exosomes possess inherent biological activity, engineered exosomes are strategically optimized for improved targeting specificity, enhanced cargo loading, increased stability, and superior therapeutic outcomes in the challenging chronic wound microenvironment [5] [7]. This analysis directly compares these two therapeutic approaches across critical performance parameters in chronic wound models, providing researchers with evidence-based insights for therapeutic development.
Comparative Therapeutic Mechanisms and Action
Mechanisms of Action in Wound Healing
Table 1: Comparative Mechanisms of Natural vs. Engineered Exosomes in Wound Healing
| Healing Phase | Natural Exosome Mechanisms | Engineered Exosome Enhancements |
|---|---|---|
| Inflammation | Modulate macrophage polarization from M1 to M2 phenotype via miRNAs (e.g., miR-146a, miR-223); suppress NF-κB signaling [11]. | Surface-modified exosomes with targeting peptides for enhanced recruitment to inflammatory cells; anti-inflammatory cytokines loaded for localized suppression [5] [7]. |
| Angiogenesis | Transfer pro-angiogenic miRNAs (e.g., miR-21, miR-29a) and proteins (VEGF, FGF-2) to endothelial cells [11] [1]. | Engineered to overexpress specific angiogenic factors (VEGF, HIF-1α); modified for targeted delivery to endothelial cells with prolonged retention [5] [24]. |
| Proliferation | Promote fibroblast proliferation, keratinocyte migration, and re-epithelialization through inherent cargo [1] [53]. | Loaded with specific growth factors (EGF, KGF) or miRNAs to directly enhance cellular proliferation mechanisms; surface engineering improves penetration through fibrotic tissue [7] [36]. |
| Remodeling | Regulate collagen synthesis and ECM organization via TGF-β signaling modulation [5] [1]. | Engineered to precisely control collagen I:III ratio through targeted miRNA delivery; modified to suppress TGF-β1 pathway activation to reduce scarring [5] [7]. |
Quantitative Efficacy Comparison in Chronic Wound Models
Table 2: Experimental Efficacy Data from Preclinical Chronic Wound Models
| Parameter | Natural Exosomes | Engineered Exosomes | Model System | Citation |
|---|---|---|---|---|
| Angiogenesis Capacity | ~1.8-fold increase in capillary density | ~3.2-fold increase in capillary density | Diabetic mouse model | [7] [24] |
| Healing Rate | 40-50% wound closure by day 7 | 70-85% wound closure by day 7 | Diabetic rat full-thickness wound | [5] [7] |
| Targeting Efficiency | Limited tissue retention (<24 hours) | Significantly enhanced retention (>72 hours) | Murine burn model | [7] [36] |
| Inflammatory Modulation | Moderate reduction in TNF-α, IL-6 | Dramatic reduction (≥80%) in pro-inflammatory cytokines | Chronic ulcer mouse model | [5] [11] |
| Collagen Deposition | Improved collagen organization | Optimal collagen I:III ratio with superior tensile strength | Porcine wound model | [1] [7] |
Experimental Methodologies and Engineering Strategies
Engineering Approaches for Enhanced Performance
The superior performance of engineered exosomes stems from sophisticated modification strategies that address specific limitations of natural exosomes:
Surface Engineering enhances targeting specificity and tissue retention. Cellular-level modification involves transfecting parent cells with plasmids encoding targeting ligands (e.g., RGD peptides, GE11 peptides) that become incorporated into exosome membranes [23] [7]. Post-isolation modification utilizes click chemistry or hydrophobic insertion to conjugate homing peptides directly onto purified exosome surfaces, enabling precise targeting to specific wound cell types like endothelial cells or fibroblasts [23] [36].
Cargo Loading strategies significantly augment therapeutic potency. Active loading methods include electroporation, sonication, extrusion, and freeze-thaw cycles to incorporate therapeutic molecules (drugs, RNAs, proteins) into pre-isolated exosomes [23] [17]. Passive loading approaches involve incubating donor cells with desired cargo or transfecting them with plasmids encoding therapeutic miRNAs/mRNAs, resulting in naturally loaded exosomes [23]. Genetic modification of parent cells using CRISPR/Cas9 or lentiviral vectors enables stable production of exosomes overexpressing specific therapeutic factors [23] [53].
Experimental Workflow for Comparative Studies
Diagram 1: Comprehensive experimental workflow for comparative exosome studies, spanning from source preparation to functional evaluation in chronic wound models.
Signaling Pathway Engineering for Enhanced Wound Healing
Diagram 2: Engineered exosomes modulate multiple signaling pathways to enhance critical processes in wound healing, demonstrating their multi-target therapeutic advantage.
Research Reagent Solutions for Exosome Studies
Table 3: Essential Research Reagents for Exosome Engineering and Evaluation
| Reagent/Category | Specific Examples | Research Function | Application Context |
|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Kit, ExoQuick-TC | Polymer-based precipitation for exosome isolation from cell media or biological fluids | Initial isolation step; suitable for multiple sample types but may co-precipitate contaminants [23] [36] |
| Characterization Antibodies | Anti-CD63, Anti-CD81, Anti-CD9, Anti-Tsg101, Anti-Alix | Western blot detection of exosome-specific markers for identity verification | Essential for characterization according to MISEV guidelines; confirms vesicle identity and purity [17] [36] |
| Engineering Tools | Lactadherin, Streptavidin, CP05 peptide, pH-sensitive lipids | Surface modification reagents for enhancing targeting and cellular uptake | Enables precise engineering of exosome surfaces for improved targeting to specific wound cell types [23] [7] |
| Cargo Loading Reagents | Electroporation buffers, Sonication equipment, Lipofectamine, Transfection reagents | Facilitate loading of therapeutic miRNAs, drugs, or proteins into exosomes | Critical for creating engineered exosomes with enhanced therapeutic payloads [23] [17] |
| Cell Culture Supplements | MSC-specific media, Exosome-depleted FBS, Hypoxia chambers | Optimize cell culture conditions for enhanced exosome production and specific cargo loading | Preconditioning strategies significantly influence exosome yield and biological activity [23] [53] |
| Animal Model Reagents | Streptozotocin, Imiquimod, Full-thickness wound devices | Induce chronic wound conditions (diabetes, inflammation) in preclinical models | Essential for validating therapeutic efficacy in physiologically relevant chronic wound environments [5] [24] |
Discussion: Clinical Translation and Future Directions
The comparative analysis demonstrates that engineered exosomes hold distinct advantages in targeting precision, therapeutic potency, and functional outcomes for chronic wound treatment. The engineered approach enables precise control over therapeutic cargo and targeting specificity, addressing fundamental limitations of natural exosomes in the complex chronic wound microenvironment [5] [7].
However, significant translation challenges remain for both platforms. Natural exosomes face issues of heterogeneity, limited targeting capability, and rapid clearance, while engineered exosomes present manufacturing complexities, scalability challenges, and regulatory considerations for modified biological products [23] [36]. Current research is addressing these limitations through advanced biomaterial-assisted delivery systems, such as exosome-laden hydrogels and 3D-bioprinted scaffolds that enhance retention and provide controlled release at the wound site [1] [24].
Future development will likely focus on precision engineering strategies that incorporate multiple modification approaches - surface targeting, optimized cargo loading, and enhanced stability - within a single vesicle platform. As manufacturing technologies advance and standardized protocols emerge, engineered exosomes are poised to transition from research tools to clinical therapeutics, potentially revolutionizing the management of chronic wounds that currently defy conventional treatments [7] [36]. The continued integration of biomaterial science with vesicle engineering will further enhance the therapeutic potential of these nanoscale systems, moving the field closer to clinically viable regenerative solutions for chronic wound patients.
Conclusion
The transition from natural to engineered exosomes represents a paradigm shift in the therapeutic approach to chronic wounds. While natural exosomes provide a powerful foundational platform with inherent biocompatibility and multi-faceted healing properties, engineered exosomes offer a path to precision medicine by overcoming limitations in targeting, retention, and potency. The future of this field hinges on resolving critical challenges in manufacturing standardization, rigorous safety profiling, and the execution of large-scale controlled clinical trials. Success will depend on interdisciplinary collaboration to translate these sophisticated nanotherapies from a promising bench-side discovery into a reliable, effective, and accessible bedside treatment, ultimately transforming the standard of care for patients with debilitating chronic wounds.
References
- 1. Exosome as emerging therapeutic platform for wound ... [https://www.sciencedirect.com/science/article/abs/pii/S0040816625004604]
- 2. Extracellular vesicles: biogenesis mechanism and impacts ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12412255/]
- 3. Extracellular vesicle-based drug overview [https://www.nature.com/articles/s41392-025-02312-w]
- 4. The role of exosomes in immunopathology and potential ... [https://www.nature.com/articles/s41423-025-01323-5]
- 5. Precision exosome engineering for enhanced wound ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12103044/]
- 6. Exosome biogenesis: machinery, regulation, and therapeutic ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-022-01671-0]
- 7. Advancements in engineered exosomes for wound repair [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2023.1301362/full]
- 8. Latest developments in advanced exosome engineering ... [https://link.springer.com/article/10.1007/s44395-025-00023-3]
- 9. The Application of Exosomes From Different Sources Loaded ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12531312/]
- 10. Exosomes As A Revolutionary Tool In Wound Healing And ... [https://www.jneonatalsurg.com/index.php/jns/article/view/8374]
- 11. Therapeutic Potential of Stem Cell-Derived Exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12383377/]
- 12. Insights into optimizing exosome therapies for acute skin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11283324/]
- 13. Clinical applications of stem cell-derived exosomes [https://www.nature.com/articles/s41392-023-01704-0]
- 14. Promotion of angiogenesis and suppression ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11634875/]
- 15. Microenvironmental cue-regulated exosomes as ... - Nature [https://www.nature.com/articles/s41427-022-00419-y]
- 16. Mesenchymal stem cells-derived small extracellular vesicles ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-024-05744-0]
- 17. Engineered exosomes: a promising drug delivery platform ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2025.1583992/full]
- 18. MSC-derived exosomes injectable hyaluronic acid ... [https://www.sciencedirect.com/science/article/pii/S0168365925006066]
- 19. Major Common Hallmarks and Potential Epigenetic Drivers ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12428840/]
- 20. Emerging Therapies in Chronic Wound Healing [https://pmc.ncbi.nlm.nih.gov/articles/PMC12619873/]
- 21. Cellular and Molecular Mechanisms of Wound Repair [https://www.mdpi.com/2073-4409/14/23/1850]
- 22. Microengineered diabetic wound-on-a-chip model for ... [https://pubs.rsc.org/en/content/articlehtml/2025/lc/d5lc00500k]
- 23. Exosome Source Matters: A Comprehensive Review from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11858990/]
- 24. Biological Nanoparticles for Enhancing Chronic Wound ... [https://www.mdpi.com/2073-4409/14/20/1637]
- 25. Extracellular vesicles as tools and targets in therapy for ... [https://www.nature.com/articles/s41392-024-01735-1]
- 26. Extracellular vesicle microRNA cargoes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12551403/]
- 27. Scalable continuous-flow electroporation platform enabling ... [https://www.nature.com/articles/s41598-023-33941-2]
- 28. Electroporation- and Liposome-Mediated Co-Transfection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12297979/]
- 29. Engineered exosomes: a promising drug delivery platform ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12098103/]
- 30. Comparing chemical transfection, electroporation, and ... [https://bmcmolcellbiol.biomedcentral.com/articles/10.1186/s12860-024-00511-x]
- 31. Design of Functional RGD Peptide-Based Biomaterials for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9967156/]
- 32. RGD-Binding Integrins Revisited - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC8038522/]
- 33. Design and applications of self-assembled polypeptide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12375629/]
- 34. RGDSP-functionalized peptide hydrogel stimulates growth ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11513332/]
- 35. Elucidating the Surface Functionality of Biomimetic RGD ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2020.567693/full]
- 36. From bench to bedside: the research status and application ... [https://jnanobiotechnology.biomedcentral.com/articles/10.1186/s12951-025-03461-4]
- 37. Optimizing mesenchymal stem cell extracellular vesicles ... [https://www.sciencedirect.com/science/article/pii/S2352320424001081]
- 38. Enhancing wound healing with nanoparticle-infused ... [https://link.springer.com/article/10.1007/s44340-025-00030-1]
- 39. The utilization of exosomes in hydrogels - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC12486605/]
- 40. Different exosomes are loaded in hydrogels for the application ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11911322/]
- 41. Engineered exosomes and composite biomaterials for tissue ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10945329/]
- 42. 3D-printed advanced scaffold armed with exosomes ... [https://www.sciencedirect.com/science/article/pii/S2452199X25001768]
- 43. Harnessing the potential of hydrogels for advanced ... [https://www.nature.com/articles/s41392-024-01852-x]
- 44. preconditioning strategies for MSC-derived extracellular ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1509418/full]
- 45. Exosomes derived from hypoxia-preconditioned M2 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12247280/]
- 46. Exosome Therapy for Chronic Wound Healing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12494051/]
- 47. Hypoxia preconditioned MSC exosomes attenuate high- ... [https://www.sciencedirect.com/science/article/pii/S2452199X25002464]
- 48. Wound healing effects of exosomes from hypoxia [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04636-4]
- 49. From bench to bedside: the research status and application ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12103788/]
- 50. Recent advances in scalable exosome production [https://www.sciencedirect.com/science/article/pii/S2096691125000196]
- 51. A comprehensive review on recent advances in exosome ... [https://www.sciencedirect.com/science/article/pii/S1936523324002481]
- 52. Five Exosome Biotechs to Watch: July 2025 [https://www.pharmasalmanac.com/articles/five-exosome-biotechs-to-watch-july-2025]
- 53. Cell-engineered technologies for wound healing and ... [https://www.nature.com/articles/s44385-025-00042-w]
- 54. Recent advances and future prospects of engineered ... [https://www.sciencedirect.com/science/article/abs/pii/S1773224725000991]
- 55. Challenges and opportunities in exosome research ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6481742/]
- 56. Exosomes as natural vectors for therapeutic delivery of ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1485769/full]
- 57. Exosome-Based Drug Delivery: A Next-Generation Platform ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12567338/]
- 58. Biodistribution of Exosomes and Engineering Strategies for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8325750/]
- 59. A comprehensive review of challenges and advances in ... [https://pubs.rsc.org/en/content/articlehtml/2024/na/d4na00501e]
- 60. Recent advances in the application of engineered ... [https://www.sciencedirect.com/science/article/abs/pii/S0014299924009269]
- 61. A strategy for risk mitigation of antibodies with fast clearance [https://pmc.ncbi.nlm.nih.gov/articles/PMC3502242/]
- 62. Role of Exosomes in Dermal Wound Healing: A Systematic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9400548/]
- 63. Potential upscaling protocol establishment and wound ... [https://www.nature.com/articles/s41598-025-93219-7]
- 64. Adipose-derived stem cell exosomes - Biomarker Research [https://biomarkerres.biomedcentral.com/articles/10.1186/s40364-025-00801-2]
- 65. Therapeutic potential of exosomal lncRNAs derived from stem ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04200-0]
- 66. Native and engineered extracellular vesicles for wound ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2022.1053217/full]
- 67. Advancements in engineered exosomes for wound repair [https://pmc.ncbi.nlm.nih.gov/articles/PMC10682480/]
- 68. Extracellular vesicles: From large-scale production and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12048759/]
- 69. Collectin-11 promotes fibroblast proliferation and ... [https://www.frontiersin.org/articles/10.3389/fimmu.2025.1592921/full]
- 70. Enhanced Cell Proliferation, Migration, and Fibroblast ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11822518/]
- 71. FOSL1 promotes keratinocyte migration and wound repair ... [https://www.nature.com/articles/s41598-025-99128-z]
- 72. TRPV3 activation accelerates keratinocyte migration in ... [https://www.sciencedirect.com/science/article/pii/S0026895X25157441]
- 73. Fibroblast Gap-closure Assay-Microscopy-based in vitro ... [https://en.bio-protocol.org/en/bpdetail?id=3333&type=0]
- 74. A generalized protocol for the induction of M2-like ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11964487/]
- 75. The emerging role of extracellular vesicles in diabetes and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12537412/]
- 76. Mesenchymal stem cell-derived exosomes: The dawn of diabetic wound healing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9791572/]
- 77. Exosomes in Diabetic Wound Healing: Mechanisms ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12377389/]
- 78. Advances of exosomes in diabetic wound healing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11836438/]
- 79. Cell-derived exosome therapy for diabetic peripheral ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04432-0]
- 80. Exosomes derived from mesenchymal stem cells in ... [https://www.nature.com/articles/s41419-024-06659-w]
- 81. Precision exosome engineering for enhanced wound healing and scar revision | Journal of Translational Medicine | Full Text [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06578-0]
- 82. Engineered stem cell exosomes for oral and maxillofacial ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2022.1038261/full]
- 83. Exosome-coated oxygen nanobubble-laden hydrogel ... [https://www.nature.com/articles/s41467-024-47696-5]