In Vivo Yamanaka Factor Delivery: Methods, Applications, and Safety for Therapeutic Reprogramming
This article provides a comprehensive analysis of current methods for delivering Yamanaka factors (OCT4, SOX2, KLF4, c-MYC) for in vivo reprogramming, a transformative approach in regenerative medicine.
In Vivo Yamanaka Factor Delivery: Methods, Applications, and Safety for Therapeutic Reprogramming
Abstract
This article provides a comprehensive analysis of current methods for delivering Yamanaka factors (OCT4, SOX2, KLF4, c-MYC) for in vivo reprogramming, a transformative approach in regenerative medicine. Tailored for researchers and drug development professionals, it explores the foundational principles of cellular reprogramming and its potential to reverse age-related phenotypes and enhance tissue regeneration. The scope covers the full spectrum of viral and non-viral delivery systems, their mechanisms, and application-specific selection. It further details strategies for optimizing efficiency and safety, including cyclic induction and AI-engineered factors, and concludes with rigorous validation frameworks and comparative analyses of different methodologies to guide preclinical and clinical translation.
The Science of Cellular Reprogramming: From Yamanaka Factors to In Vivo Rejuvenation
The discovery that somatic cells can be reprogrammed to a pluripotent state represents a paradigm shift in developmental biology and regenerative medicine. Cellular reprogramming is the process that allows the conversion of differentiated cells back into a pluripotent state, fundamentally challenging the long-held belief that cell differentiation is an irreversible process [1] [2]. This groundbreaking achievement, pioneered by Shinya Yamanaka in 2006, demonstrated that the forced expression of four specific transcription factors—OCT4, SOX2, KLF4, and c-MYC (collectively known as the OSKM or Yamanaka factors)—could revert specialized adult cells to induced pluripotent stem cells (iPSCs) [2] [3]. The implications of this discovery extend far beyond basic science, opening new avenues for regenerative medicine, disease modeling, and the emerging field of epigenetic rejuvenation [1] [4].
Over the past decade, research has revealed that transient expression of these nuclear reprogramming factors can combat age-related deterioration at cellular, tissue, and organismal levels without completing the full reprogramming cycle to pluripotency [1]. This partial reprogramming approach has demonstrated remarkable potential for rejuvenation—restoring cellular or organismal functions to a more youthful state while retaining differentiated cell identity [1] [3]. The progressive nature of aging and its influence by external factors suggests the process likely stems from failures in maintenance mechanisms that ultimately impact epigenetic regulation and gene expression [1]. Among the hallmarks of aging, the loss of epigenetic information has been proposed as a critical cause that precedes many other aspects of age-related deterioration [1].
The Molecular Basis of Epigenetic Reset
Epigenetic Landscape of Aging and Reprogramming
Aging is characterized by progressive epigenetic alterations, including changes in DNA methylation patterns, histone modifications, and chromatin organization [1] [3]. These changes disrupt normal gene expression patterns and cellular function, contributing to the phenotypic manifestations of aging. The epigenetic clock, a biomarker of aging based on DNA methylation patterns, accurately predicts biological age across tissues and cell types [3]. During natural embryonic development and in cellular reprogramming, this epigenetic information is reset to a more youthful state [4] [3].
The fundamental principle underlying epigenetic reset is that somatic cell identity is dictated primarily by epigenetic changes rather than alterations in genomic DNA sequence [3]. When cells are reprogrammed to pluripotency, either via somatic cell nuclear transfer (SCNT) or induced pluripotency, aged epigenetic signatures are erased and reset to a ground state [3]. iPSCs generated through OSGM-mediated reprogramming show complete epigenetic rejuvenation, with epigenetic clocks reset to zero and telomeres elongated to lengths comparable to embryonic stem cells [3]. This resetting occurs even when reprogramming cells from patients with accelerated aging conditions like Hutchinson-Gilford progeria syndrome (HGPS) and from supercentenarians [3].
Key Epigenetic Modifications in Reprogramming
DNA demethylation is a crucial step in epigenetic reset during reprogramming. Genome-wide DNA demethylation erases somatic cell memory and allows for the establishment of a new pluripotent or rejuvenated epigenetic state [5]. This process involves both passive replication-dependent demethylation and active enzymatic demethylation mechanisms. The ten-eleven translocation (TET) family of proteins, particularly TET1, plays a vital role in active DNA demethylation during germline epigenetic reprogramming and somatic cell reprogramming [5].
Histone modifications also undergo comprehensive remodeling during reprogramming. Changes in histone methylation (e.g., H3K9me2/3, H3K27me3), acetylation, and other post-translational modifications contribute to the chromatin state transition from a somatic to a more plastic or pluripotent configuration [1] [4]. OCT4 directly recruits chromatin remodeling complexes like the BAF complex to promote a euchromatic state and binds enhancers of Polycomb-repressed genes to induce conversion of their associated promoters from monovalent to bivalent domains [1]. These coordinated epigenetic changes enable the dramatic transcriptional reorganization required for identity reset.
The OSKM Factors: Molecular Mechanisms and Functions
OCT4: The Master Regulator of Pluripotency
OCT4 (encoded by the POU5F1 gene) is a POU-family transcription factor widely regarded as the master regulator of epigenetic reprogramming [1]. It is expressed in every cell during early stages of murine and human development, where it upregulates genes related to pluripotency, self-renewal, and stem cell maintenance [6]. During reprogramming, OCT4 operates through multiple distinct mechanisms. First, it directly recruits the BAF chromatin remodeling complex to promote a euchromatic state that enhances binding access for other reprogramming factors [1]. Second, OCT4 binds enhancers of Polycomb group-repressed genes to induce conversion of their associated promoters [1]. Third, it binds regulatory regions of pluripotency network genes to establish an autoregulatory pluripotency network [1]. Finally, OCT4 directly upregulates histone demethylases KDM3A and KDM4C, which remove repressive H3K9 methylation marks at pluripotency gene promoters [1]. Optimal reprogramming requires a threefold excess of OCT4 relative to other factors, underscoring its pivotal role [1]. Remarkably, OCT4 overexpression alone can induce pluripotency when other canonical reprogramming factors are endogenously expressed or in the presence of chromatin-remodeling chemical factors [1] [7].
SOX2: The Chromatin Priming Partner
SOX2 is a high-mobility group (HMG) box transcription factor essential for early development and the maintenance of pluripotency [1] [6]. During reprogramming, most studies focus on its heterodimerization and cooperative function with OCT4 [1]. Single-molecule imaging reveals that SOX2 engages chromatin first and primes target sites for subsequent OCT4 binding [1]. This pioneering function is supported by in vivo studies showing SOX2 alone can open chromatin and bind target DNA sites before OCT4 arrival [1]. OCT4/SOX2 shared binding sites show the most profound increase in accessibility during early reprogramming, and this partnership is critical for inducing pluripotency [1]. SOX2 promotes ectodermal gene expression while reducing mesodermal gene expression during the reprogramming process [2].
KLF4: The Dual-Function Transcriptional Regulator
KLF4 is a zinc-finger transcription factor containing both activator and repressor domains, conferring dual functionality during cell differentiation and reprogramming [1]. While OCT4 and SOX2 primarily increase chromatin accessibility during reprogramming, KLF4 drives the first wave of transcriptional activation [1]. Co-immunoprecipitation and ChIP-seq studies reveal that OCT4-SOX2 binding increases KLF4 binding by several folds, predominantly in chromatin regions that are closed in human fibroblasts [1]. KLF4 upregulates expression of NANOG, a critical pluripotency factor [6]. The stoichiometry of KLF4 expression significantly impacts reprogramming efficiency, with precise levels required for optimal results [2].
c-MYC: The Reprogramming Amplifier
In contrast to the other three factors, c-MYC does not function as a pioneer factor during reprogramming but serves as a potent amplifier of the process [1]. Although not strictly required for reprogramming initiation, c-MYC substantially increases efficiency and kinetics [1] [7]. The presence of c-MYC increases OSK binding by two-fold, and this modulatory relationship is bidirectional—MYC binding increases by 40-fold in the presence of OSK [1]. c-MYC can upregulate up to 15% of genes in the human genome through chromatin structure modification and is involved in numerous molecular pathways [6]. Its strong pro-proliferative effects contribute to its oncogenic potential, necessitating caution for in vivo applications [1] [7]. Alternative factors like L-MYC or N-MYC can substitute for c-MYC with reduced tumorigenic risk [7].
Table 1: Core OSKM Reprogramming Factors and Their Molecular Functions
| Factor | Gene Family | Key Functions in Reprogramming | Molecular Mechanisms | Essentiality |
|---|---|---|---|---|
| OCT4 | POU transcription factor | Master regulator of pluripotency | Recruits BAF chromatin remodeler; establishes autoregulatory pluripotency network; induces permissive chromatin state | Essential |
| SOX2 | HMG-box transcription factor | Chromatin priming partner for OCT4 | Opens chromatin for OCT4 binding; forms heterodimers with OCT4; regulates ectodermal genes | Essential |
| KLF4 | Zinc-finger transcription factor | Dual activator/repressor; early transcriptional wave | Activated by OCT4-SOX2; upregulates NANOG; binds closed chromatin regions | Non-essential but enhances efficiency |
| c-MYC | Basic helix-loop-helix transcription factor | Reprogramming amplifier; proliferation driver | Modifies chromatin structure; enhances OSK binding; promotes metabolic switching | Non-essential |
Partial Reprogramming for Epigenetic Rejuvenation
Conceptual Framework and Definition
Partial reprogramming represents a modified approach to cellular reprogramming where cells are exposed to Yamanaka factors for sufficient duration to produce epigenetic rejuvenation but not long enough to complete dedifferentiation to pluripotency [8] [3]. This strategy aims to separate the rejuvenative properties of reprogramming from the loss of cellular identity, creating a potential therapeutic window for anti-aging interventions [3]. During partial reprogramming, cells undergo epigenetic remodeling that restores youthful patterns of gene expression while maintaining their differentiated state and function [1] [8].
The theoretical basis for partial reprogramming stems from observations that aging and reprogramming represent opposing epigenetic trajectories [4] [3]. If aging involves progressive accumulation of epigenetic errors and loss of epigenetic information, then reprogramming factors might reverse this process by restoring more youthful epigenetic patterns [1]. Partial reprogramming approaches typically involve transient expression of OSKM factors using doxycycline-inducible systems or non-integrating delivery methods in short cycles that prevent full dedifferentiation [4].
Evidence from Experimental Models
Multiple studies have demonstrated the feasibility of partial reprogramming for epigenetic rejuvenation. In progeric mouse models, cyclic induction of OSKM factors extended median lifespan by 33% and ameliorated age-related physiological changes without causing teratomas [4]. Similarly, in wild-type aged mice, long-term cyclic OSKM induction restored youthful transcriptomic, lipidomic, and metabolomic profiles across multiple tissues including spleen, liver, skin, kidney, lung, and skeletal muscle [4]. This regimen also promoted functional regeneration, improving wound healing and reducing fibrosis [4].
At the cellular level, partial reprogramming has been shown to reverse epigenetic age as measured by DNA methylation clocks in human cells [3]. Transcriptomic analyses reveal that partially reprogrammed cells adopt gene expression profiles characteristic of younger cells while retaining lineage-specific markers [1] [4]. These changes are associated with functional improvements including enhanced mitochondrial function, reduced oxidative stress, and restoration of nuclear architecture [4] [3].
Chemical Reprogramming Alternatives
Recent advances in chemical reprogramming offer a non-genetic alternative for epigenetic rejuvenation [4]. Small molecule cocktails can induce partial reprogramming without exogenous transcription factor expression, potentially enhancing safety profiles for therapeutic applications [4] [7]. For example, a two-chemical reprogramming procedure extended C. elegans lifespan by 42.1% while reducing DNA damage and ameliorating epigenetic age-related marks [4]. Another study demonstrated that a 7c chemical cocktail could rejuvenate mouse fibroblasts at a multi-omics scale, reversing both transcriptomic and epigenomic aging clocks [4].
Notably, chemical reprogramming appears to operate through distinct mechanisms from OSKM-mediated approaches. While OSKM reprogramming typically downregulates the p53 pathway, chemical reprogramming with the 7c cocktail upregulates p53 activity [4]. Furthermore, chemical reprogramming achieves epigenetic rejuvenation without increasing cell proliferation rates, suggesting that active epigenetic remodeling rather than passive dilution through cell division underlies the rejuvenation effects [4].
Experimental Protocols for In Vivo Reprogramming
Transgenic Mouse Models for Inducible Reprogramming
The most established method for studying in vivo reprogramming utilizes transgenic mice with doxycycline-inducible OSKM expression cassettes [4]. These models enable precise temporal control over reprogramming factor expression through administration of doxycycline in drinking water or food. The following protocol represents a standardized approach for in vivo partial reprogramming studies:
Materials:
- TRE-OSKM transgenic mice (often with reverse tetracycline-controlled transactivator, rtTA)
- Doxycycline hyclate
- Standard mouse chow or drinking water delivery system
Cyclic Induction Protocol:
- Administration Phase: Administer doxycycline (typically 0.1-2 mg/mL in drinking water with 1-5% sucrose) for 1-3 days to induce OSKM expression.
- Withdrawal Phase: Remove doxycycline for 4-7 days to allow transgene silencing and cellular recovery.
- Cycle Repetition: Repeat this cycle for predetermined durations (e.g., 10-35 cycles) depending on experimental goals.
- Monitoring: Regularly assess health parameters, including weight, activity, and blood markers, to detect potential adverse effects.
This cyclic approach has been successfully employed in multiple studies demonstrating partial reprogramming benefits while minimizing the risk of teratoma formation [4]. The specific cycle duration and total treatment period must be optimized for each experimental context and desired outcome.
Viral Vector-Mediated Delivery Systems
For translation to potential therapeutic applications, viral vector systems offer an alternative to transgenic models. Adeno-associated viruses (AAVs), particularly AAV9 with its broad tissue tropism, have been successfully used for in vivo delivery of reprogramming factors [4]. The following protocol outlines this approach:
Materials:
- AAV vectors encoding OSKM factors (often with TRE promoter)
- AAV encoding rtTA (if using inducible system)
- Appropriate viral vector controls
- Injection equipment (syringes, needles, etc.)
Delivery Protocol:
- Vector Preparation: Package OSKM factors into AAV9 capsids for optimal tissue distribution. Exclusion of c-Myc reduces tumorigenic risk [4].
- Systemic Administration: Administer AAV vectors via intravenous injection (typical dose: 10^11-10^12 vector genomes per mouse).
- Induction Cycles: If using inducible system, administer doxycycline cycles as described above.
- Efficiency Assessment: Monitor transduction efficiency and transgene expression using appropriate reporters or molecular assays.
This approach has demonstrated efficacy in aged wild-type mice, with one study reporting 109% extension of remaining lifespan in 124-week-old mice treated with OSK (excluding c-Myc) delivered via AAV9 [4].
Table 2: In Vivo Reprogramming Delivery Systems and Parameters
| Delivery Method | Key Features | Advantages | Limitations | Typical Induction Protocol |
|---|---|---|---|---|
| Transgenic (dox-inducible) | Genetically integrated OSKM with rtTA | Precise temporal control; reproducible expression | Not translatable to humans; potential background effects | 2-day dox ON, 5-day dox OFF (cyclic) |
| AAV Vectors | Viral delivery of OSKM genes | Applicable to any genetic background; potential clinical translation | Immune response concerns; variable transduction efficiency | Single injection with or without cyclic dox induction |
| Sendai Virus | RNA-based viral vector | Non-integrating; high efficiency | Transient expression; potential immunogenicity | Single injection (transient expression) |
| Chemical Reprogramming | Small molecule cocktails | Non-genetic; tunable; potentially safer | Lower efficiency; complex optimization | Continuous or cyclic treatment |
Research Reagent Solutions
Table 3: Essential Research Reagents for OSKM Reprogramming Studies
| Reagent Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Reprogramming Factors | OCT4, SOX2, KLF4, c-MYC (OSKM) | Induction of pluripotency and epigenetic reset | Can be delivered as genes, mRNAs, or proteins; optimal stoichiometry critical |
| Delivery Vectors | Retrovirus, Lentivirus, Sendai virus, AAV, Episomal plasmids | Vehicle for factor delivery | Non-integrating methods (Sendai, episomal) preferred for clinical translation |
| Induction Systems | Doxycycline, rtTA, TRE promoter | Temporal control of expression | Allows cyclic induction for partial reprogramming |
| Small Molecule Enhancers | Valproic acid, 5'-azacytidine, RepSox, CHIR99021 | Epigenetic modulators and signaling pathway inhibitors | Improve efficiency; some can replace transcription factors |
| Cell Culture Media | Advanced RPMI, DMEM/F12, StemFlex | Support cell growth during reprogramming | Often supplemented with specific growth factors |
| Assessment Tools | DNA methylation clocks, RNA sequencing, Immunofluorescence markers | Evaluation of reprogramming efficiency and rejuvenation | Multi-omics approaches provide comprehensive assessment |
Signaling Pathways in Epigenetic Reset
The following diagram illustrates the key signaling pathways and molecular interactions involved in OSKM-mediated epigenetic reset:
OSKM-Mediated Epigenetic Reset Signaling Pathway
This diagram illustrates the core signaling cascade initiated by OSKM expression, leading to coordinated epigenetic remodeling, transcriptional reprogramming, and ultimately cellular rejuvenation. The process begins with OSKM factors binding to target sequences and initiating widespread epigenetic changes through chromatin remodeling, DNA demethylation, and histone modifications [1]. These epigenetic changes enable transcriptional reprogramming characterized by silencing of somatic genes, activation of pluripotency networks, and metabolic shifting toward glycolysis [2]. The cumulative effect of these molecular changes is the improvement of key cellular phenotypes associated with aging, including mitochondrial function, telomere maintenance, and reduced senescence [3].
The OSKM factors represent powerful tools for epigenetic reset with significant potential for both basic research and therapeutic applications. Their ability to remodel the epigenetic landscape and reverse age-associated changes positions them uniquely in the arsenal of regenerative medicine strategies. However, significant challenges remain in translating these findings into safe and effective therapies.
Future research directions should focus on several key areas. First, achieving precise spatiotemporal control over reprogramming factor expression will be essential to minimize risks such as teratoma formation and loss of cellular identity [4]. Second, development of non-integrating delivery methods and chemical alternatives to genetic reprogramming will enhance safety profiles for potential clinical translation [4] [7]. Third, a deeper understanding of the molecular mechanisms underlying partial reprogramming and epigenetic rejuvenation will enable more targeted and efficient approaches [1] [3].
As the field advances, the core principles of OSKM-mediated epigenetic reset continue to provide a foundation for innovative strategies to combat age-related decline and disease. The careful balance between harnessing the rejuvenative potential of these factors while maintaining cellular identity represents the next frontier in reprogramming research, with profound implications for the future of regenerative medicine and healthy aging.
The discovery that somatic cells could be reprogrammed to pluripotency using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) represented a fundamental breakthrough in regenerative medicine [7]. This process, which produces induced pluripotent stem cells (iPSCs), initially offered unprecedented potential for disease modeling and cell therapy. However, the clinical application of full reprogramming faced significant barriers, particularly the risk of teratoma formation and the complete loss of cellular identity [6] [8].
The paradigm has since evolved from achieving pluripotency to utilizing partial reprogramming—a controlled, transient exposure to reprogramming factors that rejuvenates cells without altering their identity [4]. This approach leverages the remarkable discovery that OSKM expression can reset epigenetic aging signatures while maintaining cellular differentiation, offering a potential strategy to combat age-related degeneration and enhance tissue repair [9] [4]. This Application Note frames these advances within the context of Yamanaka factor delivery methods for in vivo reprogramming research, providing researchers with quantitative comparisons, detailed protocols, and essential resource guidance.
Quantitative Analysis of In Vivo Reprogramming Approaches
The efficacy and safety of in vivo reprogramming are highly dependent on the methodology, including the specific factors used, delivery system, and induction regimen. The tables below summarize key quantitative data from foundational studies.
Table 1: Comparison of In Vivo Reprogramming Factor Configurations
| Factor Combination | Key Features & Rationale | Efficiency & Safety Notes | Reported Outcomes |
|---|---|---|---|
| OSKM (Oct4, Sox2, Klf4, c-Myc) | Original Yamanaka factors; potent reprogramming [7] | High efficiency but significant tumorigenic risk due to c-Myc [7] [4] | Extends lifespan in progeria mice by ~33%; teratoma formation with continuous induction [9] |
| OSK (Oct4, Sox2, Klf4) | Exclusion of oncogenic c-Myc for improved safety [4] | Reduced tumorigenic risk; lower efficiency than OSKM [7] [4] | Extends remaining lifespan in aged wild-type mice by 109%; improves frailty index [4] |
| OSKMLN (OSKM, LIN28, NANOG) | Expanded factor set to enhance reprogramming [9] | Used in selective regeneration studies; safety profile under investigation [9] | Promotes rejuvenation capacity in mouse models [9] |
| Chemical Cocktails | Non-genetic integration; small molecule delivery [4] | Favorable safety profile; multi-step process with lower potency [4] | Rejuvenates fibroblasts on multi-omics scale; increases C. elegans lifespan by 42.1% [4] |
Table 2: Analysis of Delivery Systems and Induction Regimens for In Vivo Reprogramming
| Delivery Method | Induction Regimen | Animal Model | Key Results & Safety Findings |
|---|---|---|---|
| Dox-Inducible Transgene (Tet-O system) | Cyclic (2 days ON, 5 days OFF) [9] | Progeria mice (HGPS model) [9] | Lifespan Extension: 33%Safety: No teratomas with cyclic induction [9] |
| Dox-Inducible Transgene (Tet-O system) | Long-term (7-10 months) & Short-term (1 month) cyclic [4] | Wild-type mice [4] | Rejuvenation: Younger transcriptome, lipidome, metabolome in multiple tissues; improved skin regeneration; no teratomas [4] |
| AAV9 Gene Therapy | Cyclic (1 day ON, 6 days OFF) [4] | Aged wild-type mice (124 weeks old) [4] | Lifespan Extension: 109% increase in remaining lifespanHealthspan: Improved frailty index score (6.0 vs. 7.5 in controls) [4] |
| Continuous Induction | Constitutive OSKM expression for several weeks [9] | 4Fj, 4Fk, 4F-A, 4F-B mouse models [9] | Safety Risk: Teratoma formation in multiple organs; dysplasia in pancreas, liver, kidney [9] |
Experimental Protocols for Key In Vivo Applications
Protocol: Cyclic Partial Reprogramming in a Progeria Mouse Model
This protocol is adapted from the landmark study that demonstrated partial reprogramming can ameliorate age-related phenotypes in vivo [9].
Objective: To extend healthspan and lifespan in a HGPS mouse model via cyclic, transient expression of OSKM factors without inducing teratomas.
Materials:
- Animal Model: Progeroid LAKI mice harboring a Dox-inducible polycistronic OSKM cassette (e.g., 4Fj or 4Fk models) [9].
- Inducing Agent: Doxycycline hyclate (Dox) administered in drinking water (e.g., 2 mg/mL with 1% sucrose) or via diet.
- Control Groups: Progeria mice without the transgene, and progeria mice with the transgene but without Dox administration.
Methodology:
- Induction Schedule: Begin cyclic induction at weaning or upon onset of phenotypic signs. A representative effective schedule is a 2-day ON / 5-day OFF cycle, repeated weekly for the duration of the study [9].
- Dox Administration: Provide Dox-medicated water ad libitum during the "ON" phases. Replace with normal water during "OFF" phases. Shield water and cages from light to maintain Dox stability.
- Health Monitoring: Weigh animals weekly and monitor for established progeria phenotypes (e.g., spine curvature, skin integrity, overall activity).
- Endpoint Analysis:
- Lifespan: Record survival to determine median lifespan extension.
- Tissue Collection: Harvest tissues (e.g., skin, cardiovascular, liver) for histopathological analysis. Key assessments include:
Protocol: AAV9-Mediated OSK Delivery for Rejuvenation in Aged Wild-Type Mice
This protocol details a gene therapy approach for organism-wide rejuvenation, excluding the oncogene c-Myc [4].
Objective: To deliver OSK factors via AAV9 to aged wild-type mice and evaluate the impact on healthspan and lifespan using a cyclic induction system.
Materials:
- Animal Model: Aged wild-type mice (e.g., 124 weeks old) [4].
- Viral Vectors:
- AAV9-TRE-OSK: AAV9 vector containing a Tet-On responsive element (TRE) driving expression of OSK.
- AAV9-rtTA: AAV9 vector expressing the reverse tetracycline-controlled transactivator (rtTA).
- Inducing Agent: Doxycycline.
Methodology:
- Vector Delivery: Co-administer AAV9-TRE-OSK and AAV9-rtTA via systemic injection (e.g., intravenous or intraperitoneal) to the aged mice. Allow 2-4 weeks for widespread vector distribution and transgene expression stabilization.
- Cyclic Induction: Initiate a chronic cyclic induction regimen, such as 1 day ON / 6 days OFF, via oral Dox administration (e.g., in food or water) [4].
- Phenotypic Assessment:
- Frailty Index: Calculate a non-invasive frailty index based on standard criteria (e.g., coat condition, gait, hearing loss) at regular intervals.
- Functional Tests: Conduct behavioral assays relevant to aging (e.g., grip strength, rotarod, cognitive tests).
- Molecular Analysis: Upon study completion, perform multi-omics analysis (e.g., transcriptomics, epigenomics) on tissues like liver, spleen, and skin to confirm a shift toward younger molecular signatures [4].
Visualization of Key Concepts and Workflows
Diagram 1: Full vs. Partial Reprogramming Cell Fate. Partial reprogramming avoids pluripotency to rejuvenate somatic cells.
Diagram 2: In Vivo Delivery Method Trade-offs. Each delivery platform offers distinct advantages and limitations for research and translation.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for In Vivo Reprogramming Research
| Reagent / Tool | Function & Application | Key Considerations |
|---|---|---|
| Dox-Inducible Mouse Models (4Fj, 4Fk, 4F-A, 4F-B) [9] | Provides a genetically defined system for spatiotemporal control of OSKM expression via Dox administration. | Ideal for proof-of-concept and mechanistic studies. Choice of model (integration locus) can affect expression levels and patterns [9]. |
| AAV Vectors (e.g., AAV9, AAV-DJ) | Enables gene delivery in wild-type animals; AAV9 offers broad tissue tropism [4]. | Crucial for translational research. Requires careful titer optimization. Immune response and potential integration are key safety considerations. |
| Tet-On System Components (rtTA, TRE Promoter) | Allows controlled, dose-dependent transgene expression in combination with Dox [9] [4]. | The core of controllable systems. Leaky expression without Dox can be an issue; newer generations (rtTA2, rtTA3) offer improved specificity. |
| Small Molecule Cocktails (e.g., 7c) [4] | Provides a non-genetic method for inducing partial reprogramming, enhancing clinical safety. | Efficiency can be lower than OSKM. Mechanisms may differ (e.g., p53 pathway activation vs. OSKM-mediated downregulation) [4]. |
| Alternative Reprogramming Factors (L-Myc, N-Myc, LIN28, NANOG) [7] | Used to replace oncogenic c-Myc or enhance reprogramming efficiency and safety. | L-Myc is a common, less tumorigenic substitute for c-Myc [7]. OSKMLN combination is used for enhanced rejuvenation effects [9]. |
| Epigenetic Age Clocks (e.g., DNA methylation clocks) [4] | Quantitative biomarkers to assess the degree of cellular rejuvenation following partial reprogramming. | Critical for validating efficacy. Multi-omic clocks (transcriptomic, epigenomic, metabolomic) provide a more comprehensive picture [4]. |
Partial reprogramming has firmly established itself as a potent strategy for reversing age-related cellular decline and enhancing regenerative capacity in vivo. The successful application of cyclic OSKM or OSK induction, via either transgenic models or AAV delivery, to extend lifespan in progeroid and aged wild-type mice provides compelling evidence for its therapeutic potential [9] [4].
Future research must prioritize the refinement of delivery systems to achieve precise spatiotemporal control, minimizing the risks of tumorigenesis and loss of cellular identity [10] [9]. The development of efficient, non-integrating delivery methods and the exploration of chemical reprogramming cocktails represent the most promising paths toward clinical translation [7] [4]. As these technologies mature, partial reprogramming is poised to revolutionize the treatment of age-related diseases and regenerative medicine.
Aging is characterized by a progressive decline in physiological function, driven by a set of interconnected cellular and molecular processes known as the hallmarks of aging [11] [12]. These include genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, mitochondrial dysfunction, and cellular senescence, among others. In recent years, cellular reprogramming has emerged as a transformative strategy to combat these hallmarks. This approach, particularly using the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC, collectively OSKM), can reverse age-related deterioration at the molecular and cellular level [4] [13].
This Application Note delineates the molecular mechanisms by which reprogramming, especially in vivo partial reprogramming, counteracts key hallmarks of aging. We provide a detailed framework for researchers and drug development professionals, focusing on practical protocols, key signaling pathways, and essential reagents for investigating reprogramming-based rejuvenation in the context of in vivo delivery systems.
Molecular Interplay: Reprogramming and the Hallmarks of Aging
Partial reprogramming does not erase cellular identity but applies a transient, controlled pressure that resets aged cells to a more youthful state, effectively targeting multiple hallmarks of aging simultaneously [8] [13]. The table below summarizes the mechanistic counteractions.
Table 1: How Partial Reprogramming Counters Hallmarks of Aging
| Hallmark of Aging | Effect of Partial Reprogramming | Key Molecular Changes & Evidence |
|---|---|---|
| Epigenetic Alterations | Reversal of age-related epigenetic changes and restoration of youthful gene expression patterns. | Restoration of youthful DNA methylation patterns and histone marks (e.g., H3K9me3); resetting of epigenetic clocks [4] [9] [13]. |
| Cellular Senescence | Reduction in senescent cell burden and suppression of the Senescence-Associated Secretory Phenotype (SASP). | Downregulation of p53/p21 pathways; reduction in SASP factor release; reversal of nucleocytoplasmic compartmentalization (NCC) defects [13]. |
| Mitochondrial Dysfunction | Improvement in mitochondrial function and restoration of energy metabolism. | Enhancement of oxidative phosphorylation; reduction in mitochondrial ROS [4] [13]. |
| Stem Cell Exhaustion | Rejuvenation of tissue-specific stem and progenitor cells, enhancing regenerative capacity. | Improved function of muscle stem cells; enhanced tissue regeneration in skin, pancreas, and muscle after injury [4] [9]. |
| Altered Intercellular Communication | Amelioration of chronic, age-related inflammation. | Reduction in pro-inflammatory signaling; reshaping of the extracellular matrix (ECM) and secretome [4] [14]. |
| Genomic Instability | Potential indirect effects via improved DNA repair and redox homeostasis. | Reduction of DNA damage markers; though noted, this is not a primary mechanism and reprogramming cannot repair DNA damage [8]. |
Figure 1: Logical flow depicting how partial reprogramming interventions target and counteract specific hallmarks of aging through distinct molecular mechanisms, leading to a rejuvenated state.
Quantitative Evidence from Key In Vivo Studies
In vivo studies have demonstrated the efficacy of partial reprogramming in extending healthspan and lifespan, with outcomes highly dependent on the delivery and expression protocol.
Table 2: Summary of Key In Vivo Reprogramming Studies in Aging Models
| Animal Model / Delivery Method | Reprogramming Factors & Regimen | Key Quantitative Outcomes | Reference |
|---|---|---|---|
| Progeric LAKI Mice(Dox-inducible transgenic) | OSKMCyclic: 2-day ON, 5-day OFF | 33% increase in median lifespanReduction of mitochondrial ROSRestoration of H3K9me levelsNo teratomas after 35 cycles | [4] |
| Wild-Type Mice(AAV9 gene therapy delivery) | OSKCyclic: 1-day ON, 6-day OFF | 109% extension of remaining lifespan (from 124 weeks)Frailty index score: 6.0 (Treated) vs. 7.5 (Untreated) | [4] |
| Wild-Type Mice(Dox-inducible transgenic) | OSKMLong-term (7-10 months) & short-term (1 month) induction | Rejuvenation of transcriptome, lipidome, metabolome in multiple tissuesIncreased skin regeneration capacityNo teratoma formation | [4] |
| C. elegans(Chemical reprogramming) | Two-chemical cocktailNot specified | 42.1% lifespan extensionReduced DNA damage, oxidative stressAmelioration of H3K9me3/H3K27me3 | [4] |
Core Signaling Pathways in Reprogramming-Induced Rejuvenation
The rejuvenating effects of partial reprogramming are mediated through specific molecular pathways. Understanding these is critical for optimizing protocols and developing targeted therapies.
The HGF/MET/STAT3 Signaling Axis
Single-cell transcriptomics and secretome analysis of high-efficiency reprogramming in microfluidic devices have identified the HGF/MET/STAT3 axis as a critical pathway. This pathway functions as a cell-non-autonomous mechanism where specific sub-populations of reprogramming cells secrete HGF (Hepatocyte Growth Factor), which accumulates in the confined extracellular environment and signals through the MET receptor tyrosine kinase on other cells, activating STAT3. This extrinsic signaling enhances the overall efficiency of the reprogramming process and contributes to the rejuvenation of the cellular population [14].
TET-DNA Demethylase Pathway
The restoration of youthful DNA methylation patterns is a hallmark of epigenetic rejuvenation. Research has shown that the vision-improving effects of OSK expression in aged and glaucomatous mice require the activity of TET DNA demethylases [13]. This indicates that the active, enzymatic process of DNA demethylation is a crucial mechanism downstream of Yamanaka factor expression for resetting the epigenetic clock and restoring tissue function.
Figure 2: Key signaling pathways in reprogramming, including the extrinsic HGF/MET/STAT3 axis and the intrinsic TET-mediated DNA demethylation pathway.
Detailed Experimental Protocols
This section provides a foundational methodology for conducting in vivo reprogramming studies, adaptable for various research objectives.
Protocol: Cyclic Induction of Yamanaka Factors in Dox-Inducible Mouse Models
Application: For long-term rejuvenation studies in progeria and wild-type aged mice. Background: This protocol is based on the seminal study by Ocampo et al. (2016) and follow-up work, which demonstrated lifespan extension and multi-tissue rejuvenation without teratoma formation [4] [9].
Materials:
- Animal Model: Reprogrammable mouse model with Dox-inducible OSKM or OSK cassette (e.g., 4F-B, i4F-B, or similar [9]).
- Inducing Agent: Doxycycline hyclate (Dox) dissolved in drinking water (e.g., 2 mg/mL with 1-5% sucrose) or administered via diet.
- Control: Age-matched littermates receiving normal water/diet.
Procedure:
- Animal Weaning & Genotyping: Wean mice at 3-4 weeks and genotype to confirm the presence of the transgene.
- Baseline Characterization: Prior to induction, collect baseline data (e.g., body weight, frailty index, blood samples for omics analysis).
- Cyclic Dox Administration:
- Induction Cycle: Administer Dox-water/diet for a defined "ON" phase (e.g., 2 days).
- Washout Cycle: Replace with regular water/diet for a defined "OFF" phase (e.g., 5 days).
- Repetition: Repeat this cycle for the study duration (e.g., weekly cycles for up to 10 months in wild-type mice [4]).
- In-Life Monitoring: Monitor mice weekly for weight, signs of distress, and tumor formation. Conduct functional tests (e.g., grip strength, rotarod) periodically.
- Tissue Collection & Analysis: At endpoint, collect tissues for:
- Histology: Assess tissue architecture, fibrosis (e.g., Masson's Trichrome), and senescence (e.g., SA-β-Gal staining).
- Molecular Analysis: Perform RNA-seq, whole-genome bisulfite sequencing (WGBS), and metabolomics/lipidomics to evaluate transcriptomic and epigenetic age reversal [4].
Safety Note: Continuous induction of OSKM for more than a week can lead to teratoma formation and organ failure [9]. Cyclic induction with adequate OFF periods is critical for safety.
Protocol: Assessing Rejuvenation Markers in Human Cells Using a Nucleocytoplasmic Compartmentalization (NCC) Assay
Application: High-throughput screening for chemical rejuvenation cocktails or genetic interventions. Background: The breakdown of nucleocytoplasmic compartmentalization (NCC) is a conserved hallmark of aging and senescence. This assay quantifies NCC integrity as a proxy for cellular youth [13].
Materials:
- Cell Lines: Human fibroblasts from young and old donors, or replicatively senescent fibroblasts.
- Reporter Construct: Lentivirus encoding an NCC reporter (e.g., mCherry-NLS and eGFP-NES).
- Induction System: Lentivirus with Tet-On inducible OSK/OSKM polycistronic construct or chemical cocktails.
- Imaging: Automated fluorescence microscope and image analysis software (e.g., ImageJ).
Procedure:
- Cell Culture & Senescence Induction: Culture young and old fibroblasts in low-serum conditions to suppress cell division. Generate replicatively senescent cells by serial passaging until growth arrest.
- Stable Cell Line Generation: Transduce fibroblasts with the NCC reporter and the inducible reprogramming construct. Select with appropriate antibiotics.
- Intervention:
- Genetic: Add Dox to the medium to induce OSK expression for a partial reprogramming duration (e.g., 3-7 days).
- Chemical: Treat cells with candidate small molecule cocktails.
- Image Acquisition & Quantification:
- Image live cells using an automated microscope.
- Quantify the Pearson correlation coefficient between mCherry (nuclear) and eGFP (cytoplasmic) signals. A lower correlation indicates better NCC integrity and a more youthful state [13].
- Validation: Correlate NCC findings with other aging biomarkers, such as transcriptomic aging clocks and SASP factor secretion (e.g., IL-6, IL-8).
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Research Reagents for In Vivo Reprogramming Studies
| Reagent / Tool | Function & Utility in Reprogramming Research | Example Application |
|---|---|---|
| Dox-Inducible Mouse Models(e.g., 4F-B, i4F-B) | Enables precise temporal control of OSKM expression in vivo via oral Dox administration. | Long-term cyclic rejuvenation studies; tissue-specific regeneration assays [4] [9]. |
| AAV9 Vectors | Efficient in vivo gene delivery vehicle for widespread transduction across tissues, including the CNS. | Delivery of OSK factors in a gene therapy approach for lifespan studies [4] [15]. |
| Sendai Virus (SeV) Vectors | Non-integrating, cytoplasmic RNA virus for high-efficiency reprogramming of human cells in vitro. | Generating high-quality, patient-specific iPSCs with low risk of genomic integration [16]. |
| Chemical Cocktails (e.g., 7c) | Non-genetic method to induce partial reprogramming and rejuvenation. | Reversing transcriptomic age in human fibroblasts; exploring alternative rejuvenation pathways [4] [13]. |
| Tet-On System | Inducible gene expression system allowing controlled timing and dosage of transgene expression. | Controlled OSK expression in stable cell lines for in vitro rejuvenation assays [13]. |
| scRNA-seq & Secretome Analysis | Tools for resolving cellular heterogeneity and characterizing the extracellular protein environment. | Identifying key signaling pathways (e.g., HGF/MET) and intermediate cell states during reprogramming [14]. |
The discovery that somatic cells can be reprogrammed using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) has opened transformative avenues for regenerative medicine. While initially developed for in vitro applications, this technology has evolved to include in vivo reprogramming, a strategy with profound implications for treating age-related diseases, including progeroid syndromes. This application note synthesizes key proof-of-concept studies conducted in both progeria and wild-type mouse models, providing a detailed examination of the protocols, quantitative outcomes, and essential reagents that underpin this promising field. The content is framed within a broader thesis investigating Yamanaka factor delivery methods, highlighting the critical transition from in vitro validation to in vivo therapeutic application.
Core Concepts and Signaling Pathways
Figure 1 illustrates the primary molecular mechanism of Hutchinson-Gilford Progeria Syndrome (HGPS) and the conceptual framework of the in vivo reprogramming strategy discussed in this note.
Figure 1: Molecular Pathogenesis of HGPS and In Vivo Reprogramming Strategy. The diagram illustrates the dominant-negative effect of the LMNA G608G mutation, which leads to progerin production and nuclear dysfunction [6]. Conversely, transient induction of Yamanaka factors (OSKM) promotes partial reprogramming, enabling epigenetic rejuvenation without complete dedifferentiation [9].
Hutchinson-Gilford Progeria Syndrome (HGPS) is a rare genetic disorder characterized by accelerated aging. It is predominantly caused by a de novo point mutation (c.1824 C>T; p.G608G) in the LMNA gene, which encodes nuclear lamins A and C [6]. This mutation activates a cryptic splice site, leading to the production of a truncated, permanently farnesylated protein called progerin. Progerin accumulation causes nuclear envelope abnormalities, genomic instability, and premature cellular senescence [6] [17]. Cardiovascular complications stemming from these defects are the primary cause of mortality.
In vivo reprogramming using Yamanaka factors offers a compelling strategy to counteract these effects. Unlike full reprogramming, which aims to create induced pluripotent stem cells (iPSCs), partial reprogramming involves the transient expression of OSKM. This brief exposure is sufficient to reverse age-associated epigenetic marks, such as DNA methylation patterns and heterochromatin loss, without erasing cellular identity or forming teratomas [8] [9]. The conceptual pathway from the HGPS mutation to its potential rescue via OSKM is outlined in Figure 1.
Key In Vivo Studies and Quantitative Outcomes
Research in animal models has provided crucial proof-of-concept evidence for the efficacy and therapeutic potential of in vivo reprogramming. The following table summarizes the designs and key quantitative findings from landmark studies in both progeria and wild-type mouse models.
Table 1: Summary of Key In Vivo Reprogramming Studies in Mouse Models
| Animal Model | Intervention | Key Quantitative Outcomes | Reference |
|---|---|---|---|
| HGPS Mouse Model(LmnaG609G/G609G) | Cyclic OSKM expression(2 days ON / 5 days OFF) | • Lifespan extension: Significant• Epigenetic reset: Restoration of H3K9me3 markers• Functional improvement: Better skin integrity, reduced spine curvature, enhanced cardiovascular function [9]. | |
| HGPS Mouse Model(Homozygous for human LMNA c.1824 C>T) | AAV9-delivered ABE(Single injection at P14) | • Mutation correction: 20-60% across organs at 6 months• Lifespan extension: Median lifespan increased from 215 to 510 days (2.4-fold)• Vascular rescue: Normalized VSMC counts, prevented fibrosis [17]. | |
| Wild-Type C57BL/6 Mice(Aged) | Cyclic OSKM expression(Long-term cycling) | • Molecular rejuvenation: Restored youthful DNA methylation, transcriptomic, and lipidomic profiles in spleen, liver, skin, kidney, lung, muscle.• Tissue regeneration: Enhanced muscle repair and wound healing, reduced fibrosis [8] [9]. | |
| Wild-Type C57BL/6 Mice(Aged, 55 weeks old) | Single-cycle OSKM expression(1 week duration) | • Systemic epigenetic impact: DNA methylation reprogramming in pancreas, liver, spleen, and blood [8]. | |
| Reprogrammable i4F-B Mice(Cerebral Ischemia) | Doxycycline-inducible OSKM(7-day infusion) | • Neural regeneration: Generation of new neurons (NeuN+)• Behavioral recovery: Improved performance in rotarod and ladder walking tests [15]. |
The data from these studies robustly demonstrate that in vivo reprogramming can mitigate complex aging phenotypes. In progeria models, the strategy not only extends lifespan but also rescues specific pathological hallmarks, particularly at the vascular level [17]. In wild-type aged mice, the approach reverses molecular signatures of aging and enhances innate regenerative capacity across multiple tissues [8] [9].
Detailed Experimental Protocols
This section provides detailed methodologies for two pivotal types of experiments referenced in Table 1: the induction of partial reprogramming in progeria mice and the assessment of its effects in wild-type aged mice.
Protocol 1: Cyclic In Vivo Reprogramming in a Progeria Mouse Model
This protocol is adapted from the landmark study by Ocampo et al. (2016) that demonstrated lifespan extension in a progeroid model [9].
A. Animal Model and Genetic Tools
- Animals: Use progeroid LmnaG609G/G609G mice or transgenic mice harboring a doxycycline-inducible OKSM cassette (e.g., 4F-A, 4F-B models with OSKM integrated at the Col1a1, Neto2, or Pparg loci) [9].
- Genotyping: Confirm genotype via PCR from tail-tip DNA using primers specific for the LmnaG609G allele or the transgenic OSKM cassette [18].
B. Induction of Partial Reprogramming
- Compound: Administer doxycycline (Dox) in the drinking water (e.g., 2 mg/mL with 1-5% sucrose) or via diet.
- Cyclic Regimen: Implement a cyclic schedule of 2 consecutive days of Dox administration, followed by 5 days without Dox. Repeat this cycle weekly, starting from weaning until endpoint [9].
- Control Groups: Include both progeroid and wild-type littermates receiving standard water/diet without Dox.
C. Tissue Harvesting and Analysis (Endpoints)
- Lifespan Monitoring: Monitor survival daily. Record age at death or humane endpoint.
- Tissue Collection: At specified timepoints or endpoint, harvest tissues (e.g., skin, aorta, liver, spleen) for analysis.
- For histology: Immersion-fix in 4% PFA or 3% NBF for 2-24 hours depending on tissue, then cryoprotect in 30% sucrose before embedding and cryo-sectioning [18].
- For molecular analysis: Snap-freeze tissue in liquid nitrogen and store at -80°C.
- Phenotypic Scoring: Quantify age-related phenotypes such as spine curvature (via X-ray), body weight, and dorsal skin integrity.
- Molecular Analysis:
- Immunofluorescence: Stain tissue sections for H3K9me3, progerin, and lamin A/C to assess nuclear lamina integrity and epigenetic marker restoration [9].
- DNA Methylation Analysis: Isolate genomic DNA and perform whole-genome bisulfite sequencing (WGBS) or targeted analysis to assess epigenetic age reversal [8].
Protocol 2: Assessing Rejuvenation in Wild-Type Aged Mice
This protocol outlines the procedure for inducing and analyzing the effects of partial reprogramming in physiologically aged, wild-type mice [8] [9].
A. Animal Model and Induction
- Animals: Use aged (e.g., 18-24 month old) wild-type C57BL/6 mice. For targeted reprogramming, use transgenic models (e.g., i4F-B) with inducible OSKM.
- Reprogramming Induction:
B. Functional Regeneration Assays
- Muscle Injury and Repair:
- One week after a Dox cycle, induce muscle injury in the tibialis anterior muscle via intramuscular injection of cardiotoxin (e.g., 50 µL of 10 µM solution).
- Harvest muscles at 5-, 10-, and 15-days post-injury.
- Analyze regeneration by quantifying centrally nucleated myofibers on H&E-stained sections and performing IF for embryonic myosin heavy chain (eMyHC) [9].
- Skin Wound Healing:
- Create full-thickness skin wounds (e.g., 4 mm biopsy punch) on the dorsum.
- Monitor wound closure daily via digital photography and planimetric analysis.
- Harvest wound tissue at various stages for analysis of re-epithelialization, collagen deposition (Masson's Trichrome stain), and reduction in fibrosis [9].
C. Molecular Phenotyping
- Multi-Omics Analysis:
- DNA Methylation: Use liver/spleen DNA to profile methylation clocks via WGBS or array-based platforms (e.g., Illumina MethylationEPIC) [8].
- Transcriptomics: Perform RNA-seq on tissues like liver, skin, and skeletal muscle to assess reversion to youthful gene expression profiles [8] [9].
- Lipidomics: Conduct mass spectrometry-based lipidomic profiling on plasma and liver tissue to evaluate restoration of youthful lipid composition [8].
Figure 2 visualizes the core workflow of these protocols, from animal preparation to data analysis.
Figure 2: General Workflow for In Vivo Reprogramming Studies. The diagram outlines the key stages of a typical in vivo reprogramming experiment, from the initial animal model preparation to the final data analysis, as described in the protocols above.
The Scientist's Toolkit: Research Reagent Solutions
Successful execution of in vivo reprogramming studies relies on a specific set of research reagents and tools. The following table catalogs essential items for building this experimental pipeline.
Table 2: Essential Research Reagents for In Vivo Reprogramming Studies
| Reagent / Tool | Function / Application | Examples & Notes |
|---|---|---|
| Inducible OSKM Mouse Models | Enables spatiotemporally controlled expression of Yamanaka factors in vivo. | • 4Fj, 4Fk (Col1a1 locus)• 4F-A/4FsA (Neto2 locus)• 4F-B/4FsB (Pparg locus) [9]. |
| Adeno-Associated Virus (AAV) | Efficient in vivo delivery vector for reprogramming factors or editors. | • AAV9: Broad tropism, crosses blood-brain barrier. Used for ABE delivery [17]. |
| Doxycycline (Dox) | Inducer for Tet-On systems; activates OSKM transgene expression. | Administered in drinking water (2 mg/mL with sucrose) or diet [9]. |
| Adenine Base Editor (ABE) | Corrects the point mutation causing HGPS (A•T to G•C) without double-strand breaks. | ABE7.10max-VRQR: Used to correct LMNA c.1824 C>T in HGPS mice [17]. |
| Histology & Staining Reagents | For tissue fixation, processing, and phenotypic analysis. | • Fixatives: 4% PFA, 3% NBF, Mirsky's fixative [18].• Cryoprotectant: 30% sucrose [18].• Staining: X-Gal solution for reporter mice (e.g., Hmox1, p21) [18]. |
| Antibodies for Phenotyping | Detection of key proteins to assess reprogramming efficacy and safety. | • Progerin/Lamin A/C: Nuclear integrity.• H3K9me3: Heterochromatin marker for epigenetic age.• NeuN: Neuronal differentiation [15] [9] [17]. |
| Reporter Mouse Models | Early detection of cellular stress and DNA damage in vivo. | • Hmox1 reporter: Oxidative stress/inflammation.• p21 reporter: DNA damage/senescence [18]. |
Critical Safety Considerations and Risk Mitigation
The translational potential of in vivo reprogramming is currently balanced against significant safety considerations. The powerful nature of the Yamanaka factors necessitates rigorous control to avoid adverse outcomes. Key risks identified in the literature include:
- Teratoma Formation: Continuous, unregulated expression of OSKM can lead to the formation of teratomas in multiple organs [8] [9]. This is the most significant risk associated with full reprogramming.
- Tissue Dysfunction and Cancer: In some studies, OSKM induction has been linked to dysplastic changes and cancer in organs like the pancreas, liver, and kidney. For instance, in the context of existing mutations (e.g., Kras), OSKM can drive cancer development through dedifferentiation-associated epigenetic changes [9].
- Loss of Cellular Identity: Prolonged exposure to reprogramming factors can cause cells to lose their specialized identity, leading to tissue dysfunction [8].
Risk Mitigation Strategies:
- Cyclic Induction: The use of intermittent, short-cycle induction (e.g., 2 days ON/5 days OFF) has proven effective in achieving rejuvenation benefits, such as lifespan extension in progeria mice, without triggering teratoma formation [9].
- Alternative Factors: Exploring the use of fewer factors (e.g., OSK without c-MYC) or non-integrating delivery systems (e.g., AAV, mRNA) can reduce oncogenic potential [6].
- Tissue-Specific Targeting: Utilizing tissue-specific promoters to restrict reprogramming activity to targeted cell types can minimize off-target effects in sensitive organs [19] [17].
- Novel Editing Approaches: The use of base editing (ABE) to correct the root cause of HGPS, as demonstrated by Koblan et al., represents a highly specific alternative that avoids the pleiotropic effects of OSKM overexpression [17].
The proof-of-concept studies detailed in this application note firmly establish in vivo reprogramming as a potent strategy with demonstrable efficacy in mitigating complex aging phenotypes. Key experiments in progeria models show that both cyclic OSKM expression and targeted genetic correction can significantly extend lifespan and rescue tissue pathology. Parallel studies in wild-type aged mice confirm the ability of partial reprogramming to restore youthful epigenetic and transcriptomic profiles and enhance tissue regeneration. The provided protocols and reagent toolkit offer a foundational roadmap for researchers aiming to replicate and build upon these findings. As the field progresses, the central challenge remains the refinement of delivery and control systems to maximize the therapeutic benefits of reprogramming while unequivocally minimizing its risks, thereby paving the way for clinical translation.
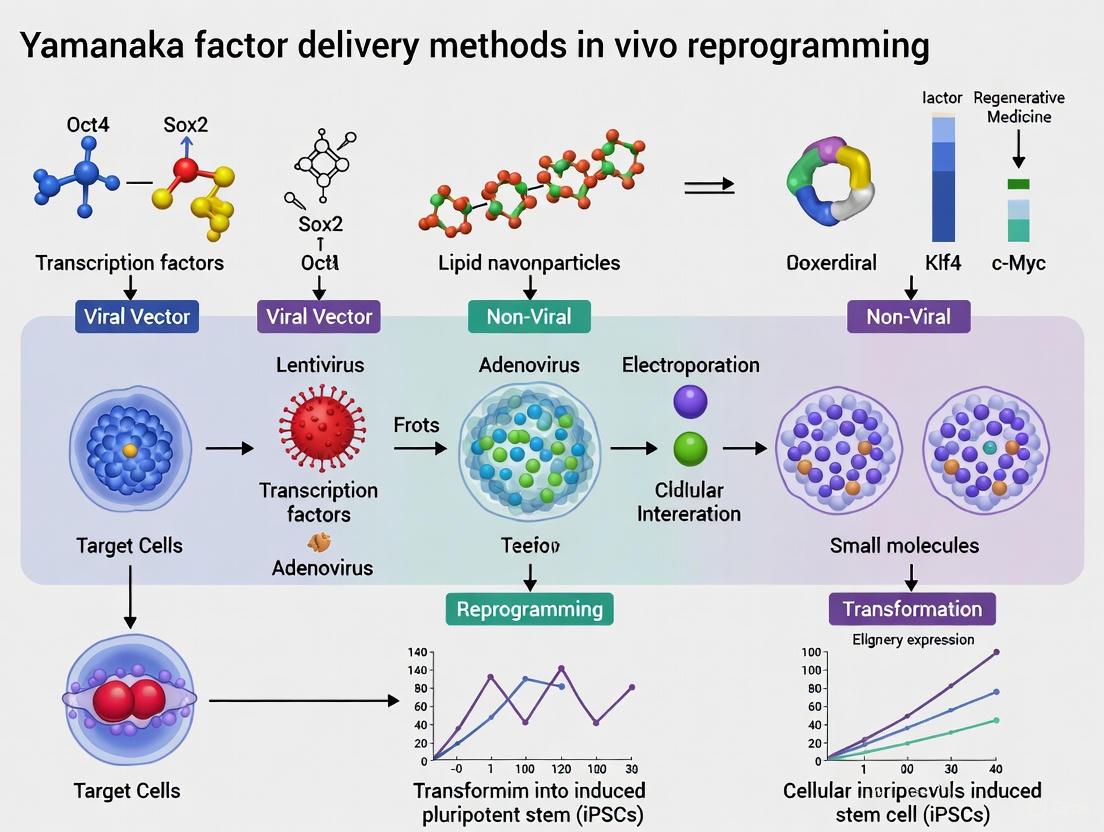
Delivery Systems for In Vivo Reprogramming: Viral Vectors, mRNA, and Chemical Approaches
The delivery of reprogramming factors, such as the Yamanaka cocktail (Oct4, Sox2, Klf4, c-Myc, or OSKM), is a foundational step in in vivo reprogramming and rejuvenation research [4] [20]. The choice of viral vector is critical, as it determines the efficiency, durability, and safety of cellular reprogramming. Adeno-associated viruses (AAVs), lentiviruses (LVs), and adenoviruses (Ad) are the most prominently used vectors, each with distinct advantages and limitations concerning packaging capacity, tropism, duration of expression, and immunogenicity [21] [22] [23]. This application note provides a structured comparison of these viral vectors and detailed protocols for their use in delivering reprogramming factors for in vivo applications.
Vector Comparison and Selection Criteria
Selecting the appropriate viral vector requires a careful assessment of the experimental goals, including the desired expression kinetics, size of the genetic payload, and target tissue. The following tables summarize the key characteristics and selection criteria for the three major viral vector systems.
Table 1: Fundamental Characteristics of Viral Vectors for Factor Expression
| Feature | Adeno-Associated Virus (AAV) | Lentivirus (LV) | Adenovirus (Ad) |
|---|---|---|---|
| Genome Material | Single-stranded DNA (ssDNA) | Single-stranded RNA (ssRNA) | Double-stranded DNA (dsDNA) |
| Packaging Capacity | ~4.5 - 5.0 kb [24] [23] | ~8 kb [21] [23] | Up to ~36 kb [23] |
| Integration Profile | Predominantly non-integrating (episomal) [24] [22] | Integrates into host genome [21] [22] | Non-integrating [21] |
| Expression Duration | Long-term (months to years) [24] [23] | Long-term (stable integration) [21] [23] | Short-term/Transient (days to weeks) [21] [23] |
| Typical In Vivo Use | In vivo gene delivery [22] | Ex vivo cell modification [22] | In vivo vaccination, transient expression [21] [23] |
| Primary Safety Concerns | Pre-existing immunity, dose-dependent liver toxicity [24] | Risk of insertional mutagenesis [21] [22] | Strong innate immune response [21] [23] |
Table 2: Application-Based Selection Guide for Reprogramming Research
| Criterion | Adeno-Associated Virus (AAV) | Lentivirus (LV) | Adenovirus (Ad) |
|---|---|---|---|
| Ideal for Long-Term Expression | Excellent - Sustained episomal expression in non-dividing cells [24] [21] | Excellent - Stable genomic integration [21] [23] | Poor - Transient expression due to immune clearance [21] |
| Ideal for Large Genetic Payloads | Poor - Limited to ~5 kb [24] [23] | Good - Capacity up to ~8 kb [21] [23] | Best - Can accommodate very large inserts [23] |
| Immunogenicity | Low immunogenicity [21] [22] | Low immunogenicity [25] | High immunogenicity [21] [23] |
| Key Advantage for Reprogramming | High safety profile and specificity via serotype tropism (e.g., AAV9 for systemic CNS delivery) [24] [4] | Ability to deliver large or multiple factor cassettes and create stable engineered cell lines ex vivo [21] [22] | High transduction efficiency and rapid onset of expression for proof-of-concept studies [21] [23] |
Experimental Protocols for In Vivo Reprogramming
AAV-Mediated In Vivo Delivery of Yamanaka Factors
This protocol details the methodology for systemic in vivo reprogramming using AAV vectors, based on studies that have demonstrated efficacy in murine models [26] [4].
Key Reagents:
- Plasmids: AAV transfer plasmid(s) containing the gene(s) of interest (e.g., OSK or OSKM) under a constitutive or inducible promoter (e.g., CMV, CAG), flanked by AAV Inverted Terminal Repeats (ITRs) [24] [26].
- Packaging System: AAV rep/cap plasmid (serotype-specific, e.g., AAV8 or AAV9 for broad tropism) and adenoviral helper plasmid [24] [26].
- Cells: HEK293T cells for vector production.
- Transfection Reagent: Polyethylenimine (PEI) or commercial equivalent.
- Purification Materials: Iodixanol gradient solutions, Amicon ultra-concentrator columns.
- Animals: Adult wild-type or disease model mice.
- Optional: Doxycycline (dox) for inducible systems (e.g., Tet-On) [4].
Procedure:
- Vector Production: Co-transfect HEK293T cells with the AAV transfer plasmid, rep/cap plasmid, and helper plasmid using PEI transfection [26].
- Harvest and Lysis: Collect cells 48-72 hours post-transfection. Lyse cells via freeze-thaw cycles and release viral particles using benzonase nuclease to degrade unpackaged nucleic acids.
- Purification: Purify AAV vectors using iodixanol density gradient ultracentrifugation. Recover the virus-containing fraction from the 40% iodixanol layer.
- Concentration and Buffer Exchange: Concentrate and desalt the viral preparation using Amicon ultra-concentrator columns with PBS-MK buffer (PBS with 1 mM MgCl₂ and 2.5 mM KCl).
- Titration: Determine the genomic titer (vector genomes/mL, vg/mL) of the purified AAV via quantitative PCR (qPCR) against a standard curve.
- In Vivo Administration: Administer AAV vectors systemically via intravenous (IV) injection (e.g., tail vein) into adult mice. A common dose for systemic expression ranges from (1 \times 10^{11}) to (1 \times 10^{12}) vg per mouse [26]. For an inducible system, also deliver an AAV vector expressing rtTA.
- Induction of Expression (for Inducible Systems): Initiate transgene expression by administering doxycycline (dox) in the drinking water (e.g., 2 mg/mL with sucrose) or via chow. Cyclic induction protocols (e.g., 2-days on/5-days off) have been used to promote rejuvenation while minimizing teratoma risk [4].
- Validation: Analyze reprogramming efficiency and functional outcomes through immunohistochemistry, RNA sequencing, and behavioral assays several weeks post-induction.
Lentiviral Vector Production for Ex Vivo Cell Engineering
While less common for direct in vivo reprogramming, lentiviral vectors are powerful tools for ex vivo engineering of cells, such as fibroblasts or stem cells, which can subsequently be used in vivo.
Key Reagents:
- Plasmids: Second or third-generation lentiviral packaging system (e.g., psPAX2, pMD2.G for VSV-G pseudotyping), and transfer plasmid containing the gene of interest [22] [23].
- Cells: HEK293T cells for production; target cells (e.g., fibroblasts, stem cells) for transduction.
- Transfection Reagent: PEI or commercial equivalent.
- Concentration Reagents: Lenti-X concentrator or ultracentrifugation equipment.
- Safety: All work must be conducted in appropriate biosafety level (BSL-2) containment.
Procedure:
- Vector Production: Co-transfect HEK293T cells with the lentiviral transfer plasmid and packaging plasmids (psPAX2, pMD2.G) [23].
- Harvest Supernatant: Collect the virus-containing supernatant at 48 and 72 hours post-transfection. Pool and filter the supernatant through a 0.45 µm filter to remove cellular debris.
- Concentration: Concentrate the lentiviral particles using Lenti-X Concentrator solution according to the manufacturer's instructions, or via ultracentrifugation.
- Titration: Determine the functional titer (Transducing Units/mL, TU/mL) on target cells (e.g., HEK293T) by serial dilution and measurement of reporter expression (e.g., fluorescence) after 48-72 hours.
- Ex Vivo Transduction: Isolate and culture primary target cells. Transduce cells with the lentiviral vector in the presence of a transduction enhancer like Polybrene (e.g., 8 µg/mL).
- Selection and Expansion: If the vector contains a selectable marker (e.g., puromycin resistance), apply selection pressure 48 hours post-transduction to eliminate untransduced cells and expand the polyclonal population.
- Validation and Implantation: Validate reprogramming factor expression in vitro and then transplant the engineered cells back into the animal model for further study.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Viral Reprogramming Experiments
| Reagent / Material | Function / Application | Examples / Notes |
|---|---|---|
| HEK293T Cells | Production workhorse for AAV, LV, and Ad vectors due to high transfection efficiency and provision of adenoviral E1 genes. | Critical for high-titer virus production; requires rigorous quality control to prevent contamination [26] [23]. |
| Polyethylenimine (PEI) | Cationic polymer used for transient transfection of packaging cells with viral plasmids. | Cost-effective and scalable alternative to commercial transfection reagents [23]. |
| Iodixanol Gradient | Medium for density-based purification of AAV vectors via ultracentrifugation. | Effective method for separating full AAV capsids from empty capsids and cellular impurities [26]. |
| Lenti-X Concentrator | A commercial solution that precipitates lentiviral particles from large volumes of supernatant. | Simplifies LV concentration without the need for ultracentrifugation [23]. |
| Doxycycline (Dox) | Small molecule inducer for Tet-On/Tet-Off inducible gene expression systems. | Enables temporal control over Yamanaka factor expression, crucial for partial reprogramming and safety [4] [20]. |
| qPCR Reagents | For absolute quantification of viral vector genomic titer (vg/mL). | Essential for standardizing the dose administered in experiments [26]. |
Pathway and Workflow Visualization
The following diagram illustrates the critical steps in the AAV vector transduction pathway, from cellular binding to transgene expression, which underlies its application in reprogramming.
The delivery of reprogramming factors, such as the Yamanaka factors (Oct4, Sox2, Klf4, c-Myc, collectively OSKM), represents a cornerstone of modern in vivo reprogramming research for regenerative medicine. While viral vectors have been widely used, their clinical application faces significant challenges including robust immunogenicity, insertional mutagenesis risks, and complex manufacturing processes [27]. Non-viral delivery systems have emerged as promising alternatives with superior safety profiles and scalable manufacturing characteristics [27] [28]. This application note provides detailed protocols and frameworks for utilizing mRNA and episomal plasmid-based delivery systems specifically for Yamanaka factor delivery in in vivo reprogramming research, enabling safer therapeutic development for neurodegenerative disorders, age-related conditions, and tissue regeneration.
Technology Platforms: Comparative Analysis
The selection of an appropriate delivery vehicle depends on research goals, target cell type, and required expression duration. The table below summarizes the key characteristics of mRNA and episomal plasmid systems for Yamanaka factor delivery.
Table 1: Platform Comparison for Yamanaka Factor Delivery
| Characteristic | mRNA-LNP Platform | Episomal Plasmid Platform |
|---|---|---|
| Mechanism of Action | Cytoplasmic translation; no nuclear entry required | Nuclear entry required; episomal replication |
| Onset of Expression | Hours | Days |
| Expression Duration | Transient (days to weeks) | Prolonged (weeks to months) |
| Risk of Genomic Integration | Nonexistent | Very low (non-integrating by design) |
| Immunogenicity | Moderate to high (can be modulated via nucleoside modifications) | Low to moderate (depends on CpG content) |
| Cargo Capacity | Limited by LNP size constraints | High (can accommodate multiple gene cassettes) |
| Key Safety Advantages | No risk of insertional mutagenesis; precise temporal control | No viral elements; reduced genotoxicity risk compared to integrating vectors |
| Primary Challenges | Controlled LNP biodistribution; managing innate immune response | Low delivery efficiency in vivo; ensuring mitotic stability in dividing cells |
| Ideal Application in Reprogramming | Partial reprogramming; cyclic induction for rejuvenation | Stable reprogramming; long-term factor expression requirements |
mRNA-LNP Platform: Application Notes and Protocols
The mRNA-LNP platform encapsulates in vitro transcribed (IVT) mRNA encoding reprogramming factors within lipid nanoparticles that protect the nucleic acid and facilitate cellular delivery [29] [30]. After cellular uptake via endocytosis, the ionizable lipids within LNPs become protonated in the acidic endosomal environment, promoting endosomal membrane disruption and release of mRNA into the cytoplasm for translation [29]. This system is ideal for transient expression of Yamanaka factors, enabling controlled partial reprogramming strategies that have shown promise in reversing age-related cellular changes without complete dedifferentiation [4].
Critical Reagent Specifications
- Ionizable Lipids: Critical for encapsulation and endosomal escape. Modern designs (e.g., DLin-MC3-DMA, SM-102, ALC-0315) are biodegradable with improved safety profiles [29] [30].
- mRNA Construct: Must include 5' cap structure (ARCA or trimeric cap), 5' and 3' untranslated regions (UTRs), coding sequence for a single Yamanaka factor, and poly(A) tail [30]. Nucleoside modifications (e.g., N1-methylpseudouridine) significantly enhance translation and reduce immunogenicity [30].
- Helper Lipids: Typically include phospholipids (e.g., DSPC), cholesterol, and PEGylated lipids to form stable, serum-stable nanoparticles [29].
Detailed Protocol: mRNA-LNP Formulation and In Vivo Delivery
Protocol 1: LNP Formulation via Microfluidic Mixing
This protocol describes the preparation of LNPs encapsulating mRNA encoding a single Yamanaka factor (e.g., Oct4). For complete reprogramming, formulate separate LNPs for each factor and mix prior to administration or administer sequentially.
Materials:
- mRNA Solution: 0.1 mg/mL mRNA in 10 mM citrate buffer (pH 3.0)
- Lipid Mixture: Ionizable lipid, DSPC, cholesterol, DMG-PEG 2000 (50:10:38.5:1.5 molar ratio) dissolved in ethanol
- Microfluidic Device (e.g., NanoAssemblr, Precision NanoSystems)
- Dialysis Membrane (MWCO 100 kDa)
- PBS (pH 7.4)
Procedure:
- Prepare Solutions: Filter both mRNA solution and lipid mixture through 0.22 µm filters.
- Microfluidic Mixing: Load the aqueous (mRNA) and organic (lipid) phases into separate syringes. Pump through a microfluidic device at a flow rate ratio of 3:1 (aqueous:organic) with a total flow rate of 12 mL/min.
- Dialyze: Immediately collect the LNP formulation and dialyze against a 100-fold volume of PBS (pH 7.4) for 18 hours at 4°C to remove ethanol.
- Concentrate and Characterize: Concentrate using centrifugal filters if necessary. Characterize particles for size (70-100 nm expected), polydispersity index (<0.2 expected), encapsulation efficiency (>90% expected), and endotoxin levels.
Protocol 2: In Vivo Administration for Partial Reprogramming
This protocol utilizes a cyclic induction strategy to achieve transient Yamanaka factor expression for cellular rejuvenation without teratoma formation, as demonstrated in progeria and wild-type mouse models [4].
Materials:
- Formulated LNPs: A mixture of Oct4-, Sox2-, Klf4-, and c-Myc mRNA-LNPs in PBS. (Note: c-Myc may be excluded to reduce oncogenic risk [4]).
- Animal Model: Reprogrammable inducible mice (e.g., i4F-B) or wild-type mice receiving AAV9 delivery vectors for rtTA expression [4].
- Doxycycline: 2 mg/mL in drinking water with 2% sucrose.
Procedure:
- Prime the System: For transgenic models, administer doxycycline water to activate the inducible system.
- Systemic LNP Administration: Inject mRNA-LNP mixture intravenously via tail vein at a dose of 0.5 mg mRNA per kg body weight.
- Cyclic Induction Regimen: Follow a precise cycle of induction. A typical protocol for rejuvenation is a 2-day pulse of LNP administration with doxycycline, followed by a 5-day chase period without treatment [4]. Repeat this cycle for the desired duration (e.g., 35 cycles over 10 months in wild-type mice [4]).
- Monitor Outcomes: Assess reprogramming efficacy via epigenetic clock analysis, transcriptomic profiling, and functional tissue improvement. Monitor closely for teratoma formation.
The diagram below illustrates the key molecular pathway and workflow for mRNA-mediated in vivo reprogramming.
Episomal Plasmid Platform: Application Notes and Protocols
Episomal plasmid vectors are non-integrating DNA molecules that persist in the nucleus through replication mechanisms independent of the host chromosome [31]. These systems typically employ elements from viruses (e.g., Epstein-Barr Virus' oriP/EBNA1) or cellular components (e.g., Scaffold/Matrix Attachment Regions - S/MAR) to enable long-term persistence in dividing cells without integration [31] [32]. This makes them particularly valuable for reprogramming applications requiring sustained expression of Yamanaka factors over multiple cell divisions while minimizing genotoxic risks associated with integrating vectors.
Critical Reagent Specifications
- Replication Origin: oriP from EBV for latent replication or S/MAR sequences for chromatin attachment and replication initiation [31] [32].
- Nuclear Retention Elements: Family of Repeats (FR) from EBV or S/MAR sequences for mitotic stability [31].
- Transgene Cassette: Multiple Yamanaka factors can be delivered on a single plasmid, separated by self-cleaving 2A peptides or internal ribosome entry sites (IRES) [31].
- Selection System: Optional antibiotic resistance genes for in vitro work, though these should be excluded from clinical preparations.
Detailed Protocol: Episomal Plasmid Delivery for Stable Reprogramming
Protocol 1: Plasmid Design and Preparation for Yamanaka Factor Expression
This protocol covers the design and preparation of S/MAR-based episomal plasmids, which are preferred over viral-element plasmids due to their completely non-viral nature and reduced safety concerns [31] [32].
Materials:
- Backbone Vector: S/MAR-containing plasmid backbone (e.g., pEPI)
- Yamanaka Factor Genes: cDNA sequences for Oct4, Sox2, Klf4, c-Myc
- Promoter: Ubiquitous promoter (e.g., CAGGS) or tissue-specific promoter
- Restriction Enzymes and T4 DNA Ligase
- Competent E. coli (e.g., Stbl3 for unstable inserts)
Procedure:
- Vector Design: Clone Yamanaka factor genes into the episomal backbone. For single-plasmid delivery, connect factors using P2A self-cleaving peptide sequences.
- Bacterial Transformation: Transform competent E. coli with the constructed plasmid using heat shock or electroporation.
- Plasmid Amplification: Culture transformed bacteria in selective medium (e.g., containing appropriate antibiotic) for 16-18 hours at 32°C for S/MAR vectors to minimize recombination.
- Plasmid Purification: Use an endotoxin-free maxiprep kit to purify plasmid DNA. Determine concentration and purity (A260/A280 ratio >1.8).
Protocol 2: In Vivo Delivery and Analysis of Reprogramming
This protocol describes the delivery of episomal plasmids to the central nervous system for in vivo reprogramming of glial cells to neurons, a promising approach for neurodegenerative diseases [15].
Materials:
- Purified Episomal Plasmid: S/MAR-based plasmid encoding all four Yamanaka factors
- Complexation Agent: Linear polyethylenimine (PEI, 25 kDa) or commercial transfection reagent suitable for in vivo use
- Animal Model: Mouse model of neurodegenerative disease (e.g., Parkinson's disease, brain injury model)
- Stereotactic Injection Apparatus
Procedure:
- Plasmid Complex Formation: Complex 5 µg of episomal plasmid with PEI at an N/P ratio of 8 in 5% glucose solution. Vortex and incubate for 15 minutes at room temperature.
- Stereotactic Intracranial Injection: Anesthetize and secure animal in stereotactic frame. Inject plasmid complexes into target brain regions (e.g., cerebral cortex, lateral ventricle) at a rate of 0.2 µL/min. Multiple injection sites may be used for larger areas.
- Post-Procedure Monitoring: Monitor animals for behavior changes and sacrifice at predetermined time points (e.g., 2, 4, 8 weeks post-injection).
- Tissue Analysis: Analyze reprogramming efficiency via immunohistochemistry for neuronal markers (NeuN, Map2), synaptic markers, and absence of glial markers (GFAP) [15]. Assess functional integration via electrophysiology.
The diagram below illustrates the key mechanism and workflow for episomal plasmid-based reprogramming.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagents for Non-Viral Yamanaka Factor Delivery
| Reagent Category | Specific Examples | Function in Reprogramming |
|---|---|---|
| Ionizable Lipids | DLin-MC3-DMA, SM-102, ALC-0315 | LNP self-assembly and endosomal escape for mRNA delivery [29] |
| mRNA Modifications | N1-methylpseudouridine (m1Ψ), 5-methylcytidine | Reduce immunogenicity and enhance translational efficiency [30] |
| Episomal Elements | S/MAR sequences, EBV oriP/EBNA1 | Nuclear retention and replication without integration [31] [32] |
| Promoter Systems | CAGGS, Dox-inducible (TRE), tissue-specific promoters | Control timing and specificity of Yamanaka factor expression |
| Polymer Transfection | Linear PEI (25 kDa), Polyamidoamine (PAMAM) dendrimers | DNA complexation and endosomal buffering for plasmid delivery [28] |
| Reprogramming Reporters | Nanog-GFP, Oct4-GFP, SSEA-1 antibody | Track reprogramming progression and pluripotency acquisition |
| Plasmid Selection | Antibiotic resistance (in vitro), PiggyBac transposase (integrating control) | Select for transfected cells and compare to integrating methods |
Concluding Remarks
The non-viral delivery platforms detailed in these application notes provide researchers with powerful tools for safe and controlled delivery of Yamanaka factors in in vivo reprogramming studies. The mRNA-LNP platform offers precise temporal control ideal for partial reprogramming and rejuvenation approaches, while episomal plasmid systems enable sustained expression necessary for complete cellular reprogramming without genotoxic risks associated with viral vectors. As these technologies continue to evolve, they hold tremendous promise for developing transformative therapies for neurodegenerative diseases, age-related conditions, and tissue regeneration. Researchers are encouraged to carefully select the platform that best aligns with their specific reprogramming goals, duration requirements, and safety considerations.
The discovery of induced pluripotent stem cells (iPSCs) via the introduction of the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC) represented a paradigm shift in regenerative medicine [7] [33]. However, the clinical translation of this technology, particularly for in vivo reprogramming, faces significant hurdles related to the delivery and safety of genetic vectors. These challenges include the risk of insertional mutagenesis, potential immunogenicity, and the oncogenic risk associated with factors like c-MYC [33] [4]. Chemical reprogramming, which utilizes defined combinations of small molecules to alter cell fate, has emerged as a promising non-genetic alternative. These small molecules offer advantages such as easy delivery, precise dose and timing control, non-immunogenicity, and reversibility, thereby presenting a potentially safer profile for therapeutic applications [34] [33] [35]. This document outlines the core principles, key cocktails, and detailed protocols for employing small molecule cocktails in reprogramming research, providing a framework for their use as an alternative to genetic Yamanaka factor delivery in in vivo studies.
Key Small Molecule Cocktails and Their Applications
Research has identified several effective small molecule combinations, ranging from complex cocktails to refined, minimal formulations. These cocktails target specific signaling pathways and epigenetic mechanisms to orchestrate cellular reprogramming, rejuvenation, and transdifferentiation.
Table 1: Key Small Molecule Cocktails for Reprogramming and Rejuvenation
| Cocktail Name | Core Components | Primary Application | Reported Outcomes | Key Mechanisms of Action |
|---|---|---|---|---|
| 7c Cocktail [36] [4] | CHIR99021, DZNep, Forskolin, TTNPB, Valproic Acid, RepSox, Tranylcypromine | In vitro partial reprogramming and rejuvenation | Improved molecular hallmarks of aging in human cells; restored mitochondrial function; reversed transcriptomic and epigenomic aging clocks [4]. | GSK-3 inhibition (CHIR99021), histone methylation inhibition (DZNep), adenylate cyclase activation (Forskolin), epigenetic modulation (VPA, TCP), TGF-β inhibition (RepSox) [36] [4]. |
| 2c Cocktail [34] [36] | RepSox, Tranylcypromine (TCP) | In vivo healthspan extension; neurological and skeletal health improvement | Increased maximum lifespan in senior mice; improved neurological status, fur, and skeletal health in old mice; extended lifespan in C. elegans [34] [36]. | TGF-β pathway inhibition (RepSox); LSD1/KDM1A inhibition, leading to epigenetic changes (Tranylcypromine) [34]. |
| VCR / VCRF Cocktail [33] | Valproic Acid, CHIR99021, RepSox (VCR); with Forskolin (VCRF) | Fibroblast to neuron reprogramming | Generation of neuron-like cells expressing TUJ1 and other neuronal markers from human fibroblasts [33]. | Histone deacetylase inhibition (VPA), GSK-3 inhibition (CHIR99021), TGF-β pathway inhibition (RepSox), cAMP pathway activation (Forskolin) [33]. |
| LC Cocktail [35] | LDNN193189, CHIR99021 | Astrocyte-to-neuron conversion in vivo | Reprogramming of reactive astrocytes into mature, long-surviving neurons in the injured adult and aged mouse spinal cord [35]. | BMP signaling inhibition (LDN193189); GSK-3 inhibition, Wnt activation (CHIR99021) [35]. |
The molecular mechanisms of these cocktails can be visualized as a network of targeted pathways that work in concert to induce cell fate changes.
Detailed Experimental Protocols
This section provides a step-by-step guide for key in vivo and in vitro experiments demonstrating the efficacy of small molecule reprogramming.
Protocol: In Vivo Reprogramming and Rejuvenation in Mice using the 2c Cocktail
This protocol is adapted from a 2025 study investigating the effects of RepSox and Tranylcypromine (TCP) on aging phenotypes in C3H female mice [34].
Objective: To assess the effects of the 2c cocktail on lifespan, healthspan, and age-related physiological markers in aging mice.
Materials and Reagents:
- Animals: Female C3H mice, divided into "senior" (10-13 months) and "old" (16-20 months) groups.
- Small Molecules: RepSox (MedChemExpress, HY-13912), Tranylcypromine (TCP, MedChemExpress, HY-16599).
- Vehicle: Dimethyl sulfoxide (DMSO), sterile.
- Preparation of Stock Solution:
- Dissolve RepSox in DMSO to a concentration of 5 mg/kg.
- Dissolve TCP in DMSO to a concentration of 3 mg/kg.
- Combine the solutions and mix thoroughly to ensure homogeneity.
- Sterilize the final solution by filtration through a 0.22 µm syringe filter [34].
Procedure:
- Animal Grouping: Randomly divide mice into treatment and control groups (e.g., n=10-11 per group). Control groups receive vehicle (DMSO) injections only.
- Dosing Regimen:
- Administer the 2c cocktail or vehicle via intraperitoneal (IP) injection.
- The injection volume is calculated based on the animal's body weight, which should be measured prior to each injection.
- Maintain a dosing interval of every 72 hours for a total treatment duration of 30 days [34].
- Long-term Monitoring (for lifespan studies): After the 30-day treatment period, continue to monitor the animals for survival without further intervention.
- Physiological and Behavioral Assessments (to be performed during and after treatment):
- Neurological Score: Assess before each injection and then weekly using a standardized scale evaluating motor activity, coordination, reflexes, and muscle tone. A score of 1-5 indicates mild CNS damage [34].
- Open Field Test: Conduct every 2 weeks to evaluate locomotor and exploratory activity. Record the number of entries into central and peripheral squares of the arena over a 5-minute period [34].
- Skeletal Deformation Examination: Visually assess and score spinal curvature (lordosis/kyphosis) and tail position on a scale from 0 (no deformation) to 2 (pronounced deformation) [34].
- Endpoint Analysis:
- Histology: At the experimental endpoint, euthanize animals and perfuse with 4% PFA. Collect organs (brain, liver, heart, kidneys, lungs, muscles) for histological analysis. Tissues can be embedded in paraffin or cryogel for sectioning and staining (e.g., H&E) [34].
- Survival Analysis: Use Kaplan-Meier survival curves and statistical tests like the Gao-Allison test to analyze differences in lifespan and maximum lifespan [34].
Protocol: In Vivo Astrocyte-to-Neuron Conversion using a Two-Molecule Cocktail
This protocol is based on a 2023 study demonstrating neuronal reprogramming in the injured adult mouse spinal cord using small molecules alone [35].
Objective: To chemically reprogram endogenous reactive astrocytes into functional neurons in the context of spinal cord injury (SCI).
Materials and Reagents:
- Animals: Adult (2-3 months) or aged (13-16 months) C57BL/6J mice.
- Small Molecules: LDN193189 (BMP inhibitor), CHIR99021 (GSK-3 inhibitor). Both from MedChemExpress.
- Vehicle: DMSO.
- AAV Vectors: (Optional, for lineage tracing) AAV2/9-GFAP-mCherry to label astrocytes.
- BrdU: For labeling proliferating cells.
Procedure:
- Spinal Cord Injury Model:
- Anesthetize the mouse with 2% pentobarbital (30 mg/kg).
- Perform a laminectomy at the T8 vertebra.
- Induce a crush injury by laterally compressing the T8 spinal cord with forceps for 15 seconds [35].
- Chemical Administration:
- Prepare the chemical cocktail (LC: LDN193189 and CHIR99021) in DMSO and further dilute in sterile saline to the working concentration.
- Administer the cocktail via intraperitoneal injection.
- The treatment should commence shortly after injury (e.g., within 24 hours) and continue based on the established regimen in the study [35].
- Lineage Tracing (Optional):
- To confirm the astrocytic origin of newly formed neurons, inject AAV2/9-GFAP-mCherry via tail vein injection prior to or concurrently with the chemical treatment [35].
- Tissue Collection and Analysis:
- At desired time points (e.g., 1-12 months post-injury), transcardially perfuse mice with ice-cold PBS followed by 4% PFA.
- Dissect and post-fix spinal cords, then cryoprotect in sucrose.
- Section the spinal cord coronally on a cryostat (10 µm thickness).
- Perform immunohistochemistry for neuronal markers (e.g., NeuN, Tuj1, MAP2) and an astrocytic marker (GFAP) to identify converted cells. Co-staining with mCherry (if AAV was used) or other lineage tracing methods confirms the origin from astrocytes [35].
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of chemical reprogramming protocols relies on high-quality, specific reagents. The following table details critical components and their functions.
Table 2: Essential Research Reagents for Small Molecule Reprogramming
| Reagent / Tool | Function / Role in Reprogramming | Example Use Cases |
|---|---|---|
| RepSox (E-616452) | Inhibitor of the TGF-β receptor I (ALK5). Reduces mesenchymal signatures and promotes a more plastic state conducive to reprogramming [34] [33]. | Component of 2c and 7c cocktails for rejuvenation; part of VCR cocktail for neuronal reprogramming. |
| Tranylcypromine (TCP) | Lysine-specific histone demethylase 1 (LSD1/KDM1A) inhibitor. Alters the epigenetic landscape by increasing H3K4 methylation, facilitating gene expression changes [34] [33]. | Component of 2c and 7c cocktails for lifespan extension and epigenetic rejuvenation. |
| CHIR99021 | Highly selective glycogen synthase kinase-3 (GSK-3) inhibitor. Activates the Wnt/β-catenin signaling pathway, which supports self-renewal and reprogramming [33] [35]. | Component of 7c, VCR, and LC cocktails; key for neuronal and astrocytic reprogramming. |
| Valproic Acid (VPA) | Broad-spectrum histone deacetylase (HDAC) inhibitor. Promotes an open chromatin state, allowing access to transcriptional machinery for reprogramming [33]. | Used in the initial 7c cocktail and VCR neuronal reprogramming cocktail. |
| LDN193189 | Selective inhibitor of BMP type I receptors (ALK2/ALK3). Inhibition of BMP signaling promotes a neural fate and is crucial for glia-to-neuron conversion [35]. | Key component of the LC cocktail for in vivo astrocyte-to-neuron conversion. |
| DMSO (Cell Culture Grade) | Universal solvent for dissolving hydrophobic small molecules. Ensures compound bioavailability and is used for preparing vehicle control solutions [34] [35]. | Vehicle for all in vivo and in vitro small molecule delivery in the cited protocols. |
| AAV Vectors (e.g., AAV2/9) | In vivo gene delivery tools for lineage tracing. Allows specific labeling of cell populations (e.g., GFAP-positive astrocytes) to track the origin of reprogrammed cells [35]. | Critical for validating that converted neurons originate from the intended target population (e.g., astrocytes). |
Chemical reprogramming with small molecule cocktails represents a frontier in regenerative medicine, offering a viable and potentially safer alternative to genetic Yamanaka factor delivery for in vivo applications. The progression from complex cocktails like 7c to more refined combinations like 2c highlights a focused effort to improve specificity and reduce toxicity [36]. Proof-of-concept studies have successfully demonstrated in vivo rejuvenation of age-related phenotypes and direct conversion of glial cells to neurons in injury models, showcasing the therapeutic potential of this approach [34] [35].
Future work will need to focus on optimizing delivery strategies to enhance tissue-specific targeting and bioavailability, further refining cocktail compositions to maximize efficacy and safety, and rigorously assessing long-term consequences in higher-order organisms. As the field advances, small molecule reprogramming is poised to become an indispensable tool for probing the mechanisms of cellular plasticity and aging, ultimately accelerating the development of novel regenerative therapies.
The Tetracycline-On (Tet-On) inducible expression system is a powerful genetic tool that enables researchers to control the timing of gene expression with precision using a small molecule. In the context of in vivo reprogramming research, this technology is indispensable for the controlled expression of Yamanaka factors (OCT4, SOX2, KLF4, c-MYC, or OSKM), which are used to reprogram somatic cells into induced pluripotent stem cells (iPSCs) [6] [7]. The core principle of the Tet-On system involves a reverse tetracycline-controlled transactivator (rtTA) that activates transcription from a Tetracycline Response Element (TRE) promoter only in the presence of an inducer molecule, typically doxycycline (Dox) [37] [38]. This allows for precise temporal control over the expression of the reprogramming factors, a critical requirement for avoiding uncontrolled dedifferentiation, teratoma formation, and other safety concerns associated with in vivo reprogramming [6] [8].
The development of advanced generations of the Tet-On system, particularly the Tet-On 3G, has provided significant improvements for reprogramming applications, offering reduced background expression ("leakiness") and increased sensitivity to doxycycline, thereby enabling tighter control and higher induction folds of over 10,000-fold [37] [39]. For research aimed at partial reprogramming—a transient reprogramming state that rejuvenates cellular age without completing the transition to pluripotency—this precise temporal control is not just beneficial but essential [8]. The ability to finely tune the duration and level of Yamanaka factor expression allows researchers to navigate the delicate balance between achieving epigenetic rejuvenation and maintaining cellular identity, making the Tet-On platform a cornerstone of modern in vivo reprogramming methodologies.
System Components and Workflow
The Tet-On system functions through a coordinated interaction between genetically encoded components and a small molecule inducer. The following diagram illustrates the core logical relationship and workflow of this system.
Core Components
- Reverse Tetracycline-Controlled Transactivator (rtTA): A fusion protein that combines a mutated tetracycline repressor (rTetR) with a strong viral transcriptional activation domain (VP16) [37]. In the Tet-On system, the rtTA is typically expressed under a constitutive promoter (e.g., CMV, EF1α, or PGK) to ensure constant presence in the cell.
- Tetracycline Response Element (TRE) Promoter: A synthetic inducible promoter that controls the expression of the gene of interest (GOI), such as the OSKM factors. The classic TRE consists of seven tandem repeats of the tetracycline operator (tetO) sequence placed upstream of a minimal promoter, such as a minimal CMV promoter [37]. Newer versions like pTRE3GS and pTight are engineered for significantly reduced basal leakage and higher induced expression levels [37] [40].
- Inducer Molecule: Doxycycline (Dox), a tetracycline analogue, is the preferred inducer due to its high affinity for rtTA, excellent tissue penetration, and favorable pharmacokinetics [37] [38]. Upon binding Dox, the rtTA undergoes a conformational change that enables it to bind specifically to the TRE, initiating transcription of the downstream GOI.
System Generations and Performance
The Tet-On system has evolved through several generations, each improving upon key performance metrics critical for sensitive applications like reprogramming. The table below summarizes the quantitative performance of different Tet-On system components.
Table 1: Key Metrics of Tet-On System Components for Reprogramming Research
| Component | Key Features | Performance Metrics | Relevance to Reprogramming |
|---|---|---|---|
| rtTA (1st Gen) | Original reverse transactivator | Moderate induction fold | Baseline system |
| rtTA-Advanced (2nd Gen) | Improved sensitivity, reduced background | Higher induction vs. 1st Gen | Improved safety profile |
| Tet-On 3G / rtTA3 | Highest sensitivity, lowest background | >10,000-fold induction [37] [39] | Gold standard for precise temporal control |
| TRE3G / pTRE3GS | Promoter optimized for Tet-On 3G | Very low leakiness, high inducibility | Ideal for viral vector construction [40] |
| All-in-One Vectors | Single vector for rtTA & GOI | Up to 25,000-fold induction reported [40] | Simplifies model generation |
Experimental Protocol for Implementing a Tet-On System
This protocol details the establishment of a Dox-inducible system for controlling Yamanaka factor expression in mammalian cells, a critical step for in vitro reprogramming studies or for pre-testing systems intended for in vivo use.
Material and Reagent Setup
- Tet-On Plasmids: Select a system appropriate for your experimental goal. For maximal induction and tight control, the Tet-On 3G system is recommended [39].
- Option A (Two-Vector System): pCMV-Tet3G (or a variant with a constitutive promoter like EF1α for stem cells) and a separate pTRE3G-[Your Gene of Interest] vector.
- Option B (All-in-One System): A single vector such as pLVX-TetOne-Puro, which expresses both the Tet-On 3G transactivator and the inducible GOI, simplifying delivery [40].
- Cell Line: The protocol is applicable to a wide range of mammalian cell lines, including human fibroblasts—a common starting material for reprogramming.
- Culture Reagents:
- Base Medium: Standard for your cell line (e.g., DMEM for fibroblasts).
- Fetal Bovine Serum (FBS): It is critical to use Tet System Approved FBS [40] [39]. Standard FBS may contain trace tetracyclines that cause unintended gene expression and high background, compromising the system's tight control.
- Doxycycline (Dox) Stock Solution: Prepare a 1-10 mg/mL stock solution in sterile water or DMSO. Filter sterilize (0.22 µm), aliquot, and store at -20°C protected from light. Working concentrations typically range from 100 ng/mL to 1 µg/mL.
- Antibiotics: For selection (e.g., Puromycin) if your vector contains a resistance marker.
Step-by-Step Procedure
Cell Seeding and Transfection:
- Seed the target cells (e.g., human fibroblasts) at an appropriate density (e.g., 50-70% confluency) in a culture dish using Tet-approved medium.
- The following day, transfert the cells with the pCMV-Tet3G plasmid (if using a two-vector system) using your preferred transfection method (e.g., lipofection, electroporation). For an all-in-one system, transfect with the single pLVX-TetOne-Puro-GOI plasmid.
- Incubate for 24-48 hours.
Generation of Stable Cell Lines:
- After 48 hours, begin selection by adding the appropriate antibiotic (e.g., Puromycin at a pre-determined lethal concentration) to the culture medium. For the example plasmids above, this selects for cells that have stably integrated the Tet-On 3G transactivator.
- Maintain the cells under selection for at least 5-7 days, changing the medium with antibiotic every 2-3 days until distinct, resistant colonies appear.
- (Optional) Pick individual colonies and expand them to screen for lines with low basal activity and high inducibility.
Induction of Gene Expression and Validation:
- For the two-vector system, now introduce the pTRE3G-GOI plasmid (e.g., pTRE3G-OSKM) into your stable Tet-On 3G cell line via a second transfection. For the all-in-one system, this step is skipped.
- Split the cells and prepare two main experimental groups: Uninduced (No Dox) and Induced (+Dox, e.g., 1 µg/mL).
- Harvest cells at various time points post-induction (e.g., 24h, 48h, 72h).
- Validate the induction:
- Molecular Analysis: Use qRT-PCR to quantify mRNA levels of your GOI (e.g., OCT4) in induced vs. uninduced cells. A successful setup should show minimal expression in the -Dox group and a strong, time-dependent increase in the +Dox group.
- Protein Analysis: Use western blotting or immunofluorescence to detect the translated proteins of the Yamanaka factors.
Advanced Applications and Protocol for Enhanced Control
While the standard Tet-On 3G system is powerful, some applications demand even stricter control. The CASwitch is a synthetic gene circuit that combines the Tet-On 3G system with the CRISPR-Cas13d endoribonuclease CasRx to achieve near-zero leakiness, which is vital for controlling highly toxic genes or for fine-tuning partial reprogramming protocols [41].
CASwitch System Workflow
The following diagram outlines the mechanism of the CASwitch circuit, which uses mutual inhibition to suppress background expression.
Protocol for Implementing the CASwitch
This protocol adapts the system for inducible Yamanaka factor expression, based on the methodology from [41].
Vector Construction:
- Clone your gene of interest (e.g., the OSKM polycistron) into a pTRE3G vector downstream of the promoter.
- Engineer the 3' untranslated region (3'UTR) of the GOI mRNA to contain one or more direct repeat (DR) sequences recognized by the CasRx endoribonuclease. This creates the target for CasRx-mediated degradation.
Cell Transfection and Induction:
- Co-transfect HEK293T (or your target cell line) with three plasmids:
- pCMV-rtTA3G: For constitutive expression of the transactivator.
- pTRE3G-GOI-DR: For inducible expression of the Yamanaka factors.
- pCMV-CasRx: For constitutive expression of the CasRx endoribonuclease.
- Transfer cells to fresh medium with or without Dox (e.g., 1 µg/mL) 24 hours post-transfection.
- Co-transfect HEK293T (or your target cell line) with three plasmids:
Performance Validation:
- Measure leakiness and induced expression 48-72 hours post-induction.
- Compare the luminescence (if using a reporter) or mRNA levels of the CASwitch system against the standard Tet-On 3G system.
- Expected Outcome: The CASwitch should show a >1-log reduction in basal expression (leakiness) compared to Tet-On 3G, while maintaining strong induced expression upon Dox addition [41].
The Scientist's Toolkit: Essential Reagents for Tet-On Reprogramming
Table 2: Key Research Reagent Solutions for Tet-On Based Reprogramming
| Item | Function/Description | Example Product/Catalog # |
|---|---|---|
| Tet-On 3G Transactivator | Core protein that activates TRE promoter upon Dox binding | pCMV-Tet3G Vector [39] |
| TRE3G Inducible Promoter | Optimized promoter for Tet-On 3G; drives GOI expression | pTRE3G Vector [39] |
| All-in-One Inducible Vector | Single-vector system for simplified delivery | pLVX-TetOne-Puro [40] |
| Tet System Approved FBS | Critical reagent; ensures no background induction from media | Takara Bio Tet System Approved FBS [40] [39] |
| Doxycycline Hyclate | Small-molecule inducer; high-purity grade for consistent results | Sigma D9891 (Example) |
| Lentiviral Preps (Tet-On) | For efficient, stable gene delivery in hard-to-transfect cells | Lenti-X Tet-One System [40] |
The Tet-On platform, especially in its latest Tet-On 3G iteration and when enhanced by circuits like the CASwitch, provides an unparalleled level of precise temporal control for in vivo reprogramming research. By enabling researchers to dictate the exact timing and duration of Yamanaka factor expression, these systems are fundamental to advancing our understanding of partial reprogramming, cellular rejuvenation, and the development of safe therapeutic strategies for age-related diseases [42] [8]. The structured protocols and quantitative data provided here serve as a foundational guide for implementing this critical technology.
Tissue-Specific Considerations and Targeting Strategies
The therapeutic application of Yamanaka factors (OSKM: Oct4, Sox2, Klf4, c-Myc) for in vivo reprogramming represents a frontier in regenerative medicine. Moving beyond proof-of-concept studies requires sophisticated strategies that account for the profound biological differences between tissues and the development of delivery systems capable of targeting specific cell types. Achieving therapeutic efficacy while minimizing off-target effects and tumorigenic risks necessitates a deliberate, tissue-informed approach. This document outlines key tissue-specific considerations and provides detailed protocols for researchers developing targeted in vivo reprogramming methodologies, framed within a broader thesis on advancing delivery methods for clinical translation.
Tissue-Specific Proliferation and Reprogramming Responses
The efficacy and safety of in vivo reprogramming are highly dependent on the target tissue's inherent biological properties. Different tissues exhibit varying susceptibilities to reprogramming factors, influenced by baseline proliferation rates, epigenetic landscapes, and the local microenvironment.
Table 1: Tissue-Specific Responses to OSKM Induction
| Tissue/Cell Type | Key Findings | Experimental Model | Citation |
|---|---|---|---|
| Hepatocytes | - Rapid proliferation (Ki67+ cells increased after 2 days)- Dedifferentiation (loss of Alb, Cyp3a11; gain of Afp, Sox9)- Transient liver dysfunction- Enhanced regenerative capacity in injury models | Hep-4F mouse (Alb-Cre; LSL-rtTA; tetO-OSKM) | [43] |
| Cardiac Fibroblasts | - Conversion to induced cardiomyocyte-like cells (iCMs)- Improved cardiac function and reduced fibrosis post-MI- In vivo microenvironment enhances reprogramming maturity | Retroviral/SeV delivery of GMT (Gata4, Mef2c, Tbx5) in mouse MI models | [44] |
| Dermal Fibroblasts | Rejuvenation on a multi-omics level, including reversal of epigenetic clocks | In vitro human fibroblast cultures with OSKM | [4] |
| Chondrocytes | Requirement for silencing of the master gene SOX9 for successful reprogramming to iPSCs | Time-coursed single-cell RNA-seq of human articular chondrocytes | [45] |
| Aged/Inflamed Cells | Inflammation (e.g., COX-2/PGE2 signaling) acts as a barrier to reprogramming; anti-inflammatory compounds (e.g., Diclofenac) enhance efficiency | Reprogramming of aged mouse tail-tip fibroblasts (TTFs) | [44] |
Targeting and Delivery Strategies
Precise targeting is paramount for safe in vivo reprogramming. Strategies can be categorized into promoter-based, viral serotype-based, and non-viral delivery methods.
Promoter-Driven Targeting
Using tissue-specific promoters to drive the expression of reprogramming factors ensures activity only in the desired cell type.
- Liver: The Albumin (Alb) promoter provides high specificity for hepatocytes [43].
- Heart: The α-myosin heavy chain (αMHC) promoter can target cardiomyocytes, though its utility in converting fibroblasts is limited as the goal is to activate it in the converted cells.
- General Approach: In a common transgenic model, the Cre-loxP system is combined with a tissue-specific Cre driver (e.g., Alb-Cre) to activate rtTA expression. Subsequent administration of doxycycline (Dox) induces OSKM expression via the tetO promoter [43].
Viral Serotype-Dependent Tropism
The choice of viral capsid determines which tissues are infected.
- Adeno-Associated Viruses (AAVs): AAV9 exhibits broad tropism, enabling distribution to many tissues, including heart, liver, and skeletal muscle, after systemic administration [4] [44].
- Sendai Virus (SeV): An RNA virus that does not integrate into the host genome, offering a safer profile. SeV vectors have been successfully used for in vivo delivery of cardiac reprogramming factors [44].
Non-Viral and Chemical Reprogramming
To circumvent the risks associated with viral vector integration, non-genetic methods are under active development.
- Chemical Reprogramming: Cocktails of small molecules (e.g., the "7c" cocktail) can induce partial reprogramming and rejuvenation in mouse fibroblasts, acting through pathways that may be distinct from OSKM [4].
- Polymer-Based Delivery: Synthetic polymers like poly(ethylene glycol) (PEG) can be engineered into hydrogels that present specific ligands (e.g., Laminin, RGD peptides) to enhance the efficiency of direct reprogramming in a controlled microenvironment [46].
Detailed Experimental Protocols
Protocol: Liver-Specific Partial Reprogramming for Enhanced Regeneration
Objective: To transiently induce partial reprogramming in hepatocytes to enhance liver regeneration without tumor formation.
Materials:
- Animal Model: Hep-4F mice (genotype: Alb-Cre; LSL-rtTA; tetO-OSKM).
- Inducing Agent: Doxycycline (Dox) in drinking water (0.1 mg/mL with 1% sucrose) or via chow.
- Control: Littermates lacking the tetO-OSKM transgene treated with the same Dox regimen.
Workflow Diagram: In Vivo Liver Reprogramming
Procedure:
- Induction: Administer Dox-containing water (0.1 mg/mL) to adult Hep-4F mice for a period of 1 day. This short pulse is critical to avoid lethal liver dysfunction [43].
- Chase: Remove Dox and monitor mice for the desired period (e.g., 1-14 days for marker analysis, or longer for regeneration studies).
- Functional Regeneration Assay: To assess enhanced regeneration, subject mice to a standard liver injury model, such as intraperitoneal injection of carbon tetrachloride (CCl4, e.g., 0.5 mL/kg of a 1:10 mixture in olive oil), after the Dox withdrawal period.
- Analysis:
- Lineage Tracing: GFP expression (from the IRES-GFP cassette) marks hepatocytes that have activated the reprogramming circuit.
- Proliferation: Immunostaining for Ki67 on liver sections.
- Dedifferentiation Markers: qRT-PCR for mature hepatocyte markers (Alb, Cyp3a11) and progenitor markers (Afp, Sox9, Gata4, Gata6).
- Functional Recovery: Measure the rate of liver mass restoration and serum albumin levels post-injury compared to controls.
- Tumor Surveillance: Monitor mice for up to 9 months for any signs of tumor formation.
Protocol: In Vivo Direct Cardiac Reprogramming
Objective: To convert cardiac fibroblasts into induced cardiomyocyte-like cells (iCMs) in situ following myocardial infarction (MI).
Materials:
- Animal Model: Adult wild-type mice (e.g., C57BL/6) for MI surgery.
- Reprogramming Factors: GMT (Gata4, Mef2c, Tbx5) or GHMT (plus Hand2).
- Delivery Vector: Sendai virus (SeV) expressing GMT (SeV-GMT). SeV is non-integrating and provides high transduction efficiency [44].
- Controls: MI mice treated with a control vector (e.g., expressing GFP).
Workflow Diagram: Direct Cardiac Reprogramming
Procedure:
- Myocardial Infarction: Induce MI in anesthetized mice via permanent ligation of the left anterior descending (LAD) coronary artery.
- Viral Delivery: Immediately following MI, perform intramyocardial injections of SeV-GMT (e.g., ~1 x 10^8 IFU in 20 µL total volume, distributed across multiple sites in the border zone).
- Analysis:
- Lineage Tracing: Use fibroblast-specific lineage tracers (e.g., Tcf21-MerCreMer-labeled cells) to confirm the fibroblast origin of newly formed iCMs.
- Histology and Immunostaining: Analyze heart sections for expression of cardiac proteins (cTnT, α-actinin) and gap junctions (Connexin 43). Co-staining with a fibroblast lineage marker confirms conversion.
- Functional Assessment: Perform echocardiography at 2, 4, and 8 weeks post-MI to measure ejection fraction and fractional shortening.
- Fibrosis Quantification: Use Masson's Trichrome or Picrosirius Red staining on heart sections to quantify scar size.
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for In Vivo Reprogramming Research
| Reagent / Tool | Function / Purpose | Example & Notes |
|---|---|---|
| Inducible Transgenic Mice | Allows spatial and temporal control of OSKM expression. | Hep-4F mice [43]; "i4F-B" mouse [4]. |
| Tissue-Specific AAVs | For targeted gene delivery in wild-type animals. | AAV9 for broad tropism; other serotypes (AAV8 for liver) for more specific targeting [4] [44]. |
| Non-Integrating Viral Vectors | Safer delivery method, reduces risk of insertional mutagenesis. | Sendai Virus (SeV) [44], Adenovirus. |
| Chemical Reprogramming Cocktails | Non-genetic method to induce rejuvenation/repair. | "7c" cocktail for partial reprogramming of fibroblasts [4]. |
| Small Molecule Enhancers | Improve reprogramming efficiency and maturation. | Diclofenac (inhibits inflammation) [44]; RepSox (replaces Sox2) [7]. |
| Engineered Biomaterials | Provides a controlled microenvironment to direct cell fate. | PEG hydrogels functionalized with Laminin or RGD peptide [46]. |
| Lineage Tracing Systems | Crucial for confirming the origin of reprogrammed cells in vivo. | Cre-loxP systems with cell-type-specific Cre drivers (e.g., Postn-Cre for fibroblasts) [44] [43]. |
Concluding Remarks
The path to clinical translation of in vivo reprogramming hinges on mastering tissue-specificity. The protocols and considerations outlined herein demonstrate that success is not a function of factor delivery alone, but of a holistic strategy that integrates precise genetic control, optimized delivery vehicles, and a deep understanding of the target tissue's biology and pathophysiology. Future work must continue to refine safety, particularly concerning the risks of tumorigenicity from residual pluripotent cells, and develop more sophisticated, clinically viable delivery systems that can be targeted and regulated with high precision in human patients.
Overcoming Hurdles: Safety, Efficiency, and Spatiotemporal Control
The clinical application of in vivo reprogramming, a revolutionary approach in regenerative medicine, is significantly hampered by the oncogenic risk of teratoma formation. This risk is intrinsically linked to the use of the Yamanaka factors, particularly the c-MYC oncogene. Teratomas, which are benign tumors containing tissues from all three germ layers, can form when undifferentiated or partially reprogrammed cells persist after transplantation [47] [48]. The ability to form teratomas is a common property of both embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) [47]. While often classified as benign, these tumors have the potential to metastasize upon interaction with specific microenvironments and niches [47]. This Application Note details the mechanisms by which c-MYC contributes to this risk and provides validated experimental protocols for its mitigation in the context of in vivo reprogramming research, providing a critical resource for scientists and drug development professionals.
The Dual Role of c-MYC in Reprogramming and Tumorigenesis
Biological Functions of c-MYC
c-MYC is a master transcription factor that regulates numerous cellular processes essential for both reprogramming and cancer. Its key functions include:
- Promoting Cell Cycle Progression: c-MYC upregulates cyclins and cyclin-dependent kinases while suppressing CDK inhibitors, thereby driving cells into the S phase [49].
- Regulating Metabolism: It alters cellular metabolism by enhancing lipogenesis, glycolysis, and glutaminolysis to support rapid cell growth [49].
- Enhancing Angiogenesis: c-MYC is essential for vasculogenesis and angiogenesis during development and in tumors. It suppresses the anti-angiogenic factor thrombospondin-1 (TSP-1) and upregulates pro-angiogenic factors like VEGF [50] [49]. c-MYC deficiency in embryos leads to profound defects in vasculature and primitive erythropoiesis [50].
- Inducing a Stem Cell-like State: It promotes epigenetic reprogramming and suppresses lineage-specifying transcription factors, thereby enhancing cellular pluripotency [49].
Mechanisms of c-MYC-Induced Teratoma Risk
The incorporation of c-MYC into reprogramming protocols significantly elevates tumorigenic potential through several interconnected mechanisms:
- Inhibition of Apoptosis: The activation of c-MYC normally triggers apoptosis as a safeguard. In reprogramming, this protective mechanism can be bypassed, allowing the survival of damaged or partially reprogrammed cells with high proliferative potential [50].
- Epigenetic Deregulation: Teratoma formation from human iPSCs is associated with the deregulation of epigenetic modulators like lysine-specific demethylase 1 (LSD1). LSD1 expression is minimal in hiPSCs but strongly upregulated in teratomas, and its overexpression is sufficient to enhance teratoma growth [51].
- Cooperation with Other Oncogenic Lesions: Overexpression of c-MYC alone is rarely sufficient for tumorigenesis; it typically collaborates with other genetic alterations, such as mutations in KRAS or TP53 (p53), which are common in cellular environments [49].
Table 1: Key Oncogenic Pathways Activated by c-MYC in Teratoma Formation
| Pathway | c-MYC-Mediated Regulation | Oncogenic Outcome |
|---|---|---|
| Cell Cycle Control | ↑ Cyclins (A, D1, E); ↑ E2F; ↓ p16, p21, p27 | Uncontrolled cellular proliferation [49] |
| Angiogenic Switch | ↑ VEGF; ↓ Thrombospondin-1 (TSP-1) | Tumor vascularization and growth [50] [49] |
| Apoptosis Evasion | Activates ARF–Mdm2–p53 pathway; requires co-existing anti-apoptotic signals | Survival of genomically unstable cells [50] |
| Stemness Maintenance | ↑ Epigenetic reprogramming; ↓ Lineage specification | Persistence of undifferentiated, teratoma-initiating cells [49] |
Application Notes & Experimental Protocols
Protocol 1: In Vivo Reprogramming with Doxycycline-Inducible Yamanaka Factors
This protocol describes the induction of reprogramming in a controlled, transient manner to minimize the risk of teratoma formation, suitable for use in rodent models [15].
3.1.1 Research Reagent Solutions
Table 2: Essential Reagents for In Vivo Reprogramming
| Reagent | Function/Description | Example |
|---|---|---|
| Doxycycline-Inducible Viral Vector | Allows controlled, transient expression of OKSM factors. | Lentivirus or Retrovirus with TRE3G promoter [15] |
| Stereotactic Injection System | Precise delivery of viral vectors to the target brain region. | Digital Stereotaxic Instrument with 10 µL Hamilton Syringe |
| Osmotic Mini-Pump | For sustained, chronic delivery of doxycycline. | Alzet Pump (Model 1004) [15] |
| c-MYC Inhibitor | Small molecule to suppress c-MYC activity post-reprogramming. | MYCi975 or 10058-F4 [49] |
| LSD1 Inhibitor | Small molecule to prevent teratoma formation via epigenetic modulation. | S2157 or Tranylcypromine (TCP) [51] |
3.1.2 Step-by-Step Workflow
- Vector Preparation: Generate high-titer, replication-incompetent lentiviral particles encoding the doxycycline-inducible OKSM (Oct4, Klf4, Sox2, c-Myc) cassette. Purify and concentrate the virus to a titer of >1 x 10^8 IU/mL.
- Stereotactic Surgery: Anesthetize the animal (e.g., C57BL/6 mouse) and secure it in a stereotactic frame. Using aseptic technique, perform a craniotomy and inject 1-2 µL of the viral preparation into the target region (e.g., dentate gyrus or cerebral cortex) at a rate of 0.2 µL/min. Allow the needle to remain in place for 5 minutes post-injection before slow withdrawal [15].
- Factor Induction: Immediately following surgery, implant a subcutaneous osmotic mini-pump pre-filled with a doxycycline solution (2 mg/mL in saline) to infuse for 7 days. Alternatively, administer doxycycline via drinking water (1-2 mg/mL with 1% sucrose) for a defined period, typically 1-3 weeks [15].
- Withdrawal and Monitoring: Terminate doxycycline administration to halt transgene expression. Monitor animals daily for signs of distress and weekly for tumor formation via in vivo imaging (if reporters are used) or behavioral assessment.
Protocol 2: Assessing and Quantifying Teratoma Formation
This protocol provides a standardized method for evaluating the tumorigenic potential of reprogrammed cells in vivo.
3.2.1 Materials
- Immunodeficient mice (e.g., NOD/SCID)
- 4% Paraformaldehyde (PFA)
- Paraffin embedding station and microtome
- Hematoxylin and Eosin (H&E) stain
- Antibodies for immunohistochemistry: Anti-LSD1, Anti-c-MYC, Anti-PECAM-1 (CD31) for vasculature, and lineage-specific markers (βIII-tubulin for ectoderm, SOX17 for endoderm, α-smooth muscle actin for mesoderm) [51].
3.2.2 Procedure
- Cell Transplantation: Harvest reprogrammed cells or their derivatives. Inject a defined number of cells (as few as 100 can be sufficient [51]) subcutaneously or into an immunologically permissive organ (e.g., kidney capsule) of immunodeficient mice.
- Tumor Monitoring: Palpate weekly for tumor formation over a period of 12-24 weeks. Measure tumor dimensions with calipers.
- Histopathological Analysis: At endpoint, resect tumors and fix in 4% PFA. Process, embed in paraffin, and section at 5 µm thickness. Perform H&E staining to identify the heterogeneous tissues representative of all three germ layers—a hallmark of teratomas [51].
- Immunohistochemical Validation: Stain consecutive sections with antibodies against LSD1 and c-MYC to confirm their upregulation in teratoma tissues. Co-staining with germ layer-specific markers confirms the teratoma composition and demonstrates LSD1 expression across all lineages [51].
Protocol 3: Pharmacological Inhibition of c-MYC and Epigenetic Modulators
This protocol outlines the use of small molecules to mitigate teratoma risk by targeting c-MYC and its associated epigenetic pathways.
3.3.1 Reagent Preparation
- Prepare a 10 mM stock solution of the c-MYC inhibitor MYCi975 in DMSO. Store at -20°C.
- Prepare a 10 mM stock solution of the LSD1 inhibitor S2157 in sterile water. Store at -20°C.
- Immediately before use, dilute stocks in saline or PBS for in vivo administration. The final DMSO concentration should not exceed 5%.
3.3.2 Dosing Regimen Initiate treatment after the reprogramming factors have been withdrawn.
- MYCi975: Administer via intraperitoneal (i.p.) injection at 25 mg/kg, daily for 4 weeks [49].
- LSD1 Inhibitor (S2157): Administer via i.p. injection at 10 mg/kg, three times per week for 4 weeks [51].
- Control groups should receive vehicle-only injections.
3.3.3 Efficacy Assessment
- Monitor and compare tumor incidence and size between treated and control groups.
- At endpoint, analyze explanted tissues via immunoblotting to confirm downregulation of c-MYC, LSD1, and their downstream targets.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Mitigating Teratoma Risk
| Category | Reagent | Specific Function | Experimental Context |
|---|---|---|---|
| c-MYC Targeting | MYCi975 | Disrupts MYC-MAX interaction and downregulates MYC target genes [49]. | In vivo administration post-reprogramming to reduce tumorigenicity. |
| Omomyc / OMO-103 | A dominant-negative MYC peptide inhibitor; prevents MYC transactivation [49]. | Co-expression with reprogramming factors or in vivo delivery. | |
| Epigenetic Targeting | LSD1 Inhibitor (S2157) | Inhibits lysine-specific demethylase 1, preventing teratoma formation from hiPSCs [51]. | In vivo administration to prevent teratoma growth. |
| Prospective Cell Purging | Anti-SSEA-5 Antibody | Cell surface antigen marker for pluripotent stem cells; used for FACS/MACS to remove undifferentiated cells [52]. | Purging of cell populations prior to transplantation. |
| Inducible Systems | Doxycycline-Inducible OKSM | Allows for transient, controlled expression of reprogramming factors to limit oncogenic exposure [15]. | Foundation of in vivo reprogramming models in rodents. |
The integration of transient reprogramming protocols, pharmacological inhibition of c-MYC, and epigenetic modulation represents a powerful, multi-pronged strategy to mitigate the oncogenic risk of teratoma formation in in vivo reprogramming research. The experimental protocols and reagent toolkit detailed herein provide a concrete framework for advancing the safety profile of this promising therapeutic modality. As the field progresses, the development of more specific c-MYC inhibitors and refined delivery systems for the Yamanaka factors will be paramount to realizing the full clinical potential of in vivo reprogramming.
The controlled in vivo expression of Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) represents a transformative approach for combating age-related decline and promoting tissue regeneration [4] [9]. This technique holds the potential to reverse epigenetic aging markers and restore youthful cellular function. However, a significant challenge exists: the narrow window between achieving sufficient rejuvenation and triggering complete dedifferentiation, which can lead to loss of cellular identity and tumorigenesis [6] [9]. Continuous induction of OSKM over weeks has been demonstrated to produce teratomas in multiple organs, underscoring the critical nature of this balance [9].
Cyclic induction has emerged as a pivotal strategy to navigate this therapeutic window. By applying short, pulsed expressions of reprogramming factors followed by extended recovery periods, researchers can promote a partial reprogramming state that rejuvenates cells without erasing their identity [4] [9]. This Application Note provides a detailed framework of optimized cyclic induction protocols, supported by quantitative data and practical methodologies, for researchers developing in vivo reprogramming therapies.
Quantitative Analysis of Cyclic Induction Regimens
The efficacy and safety of partial reprogramming are highly dependent on the specific cyclical regimen employed. The table below summarizes key in vivo protocols and their outcomes from recent studies.
Table 1: Comparative Analysis of In Vivo Cyclic Induction Protocols
| Induction System | Factor Combination | Cycle Structure | Experimental Model | Key Efficacy Outcomes | Tumorigenicity |
|---|---|---|---|---|---|
| Dox-inducible Transgene [4] | OSKM | 2 days ON / 5 days OFF | Progeric LAKI mice | 33% median lifespan increase; Reduced mitochondrial ROS; Restored H3K9me levels [4] | No teratomas reported after 35 cycles [4] |
| AAV9 Gene Therapy [4] | OSK (excludes c-MYC) | 1 day ON / 6 days OFF | 124-week-old wild-type mice | 109% remaining lifespan extension; Improved frailty index score [4] | Reduced risk profile by excluding c-Myc [4] |
| Dox-inducible Transgene [4] | OSKM | Long-term (7-10 month) & short-term (1 month) induction | Wild-type mice | Transcriptome, lipidome, metabolome reversion to younger state; Increased skin regeneration [4] | No teratoma formation reported [4] |
The data highlights that shorter, more frequent pulses (e.g., 1-2 days ON) with longer recovery periods (5-6 days OFF) effectively separate rejuvenation from dedifferentiation. The exclusion of the potent oncogene c-Myc (using OSK instead of OSKM) in one protocol further enhances the safety profile for potential clinical translation [4].
Tissue-Specific Considerations for Protocol Optimization
The response to OSKM induction is not uniform across the body. Different tissues exhibit varying levels of reprogramming factor activation, which must be accounted for in protocol design [9]. The following diagram illustrates the general workflow for establishing a cyclic induction protocol, incorporating tissue-specific considerations.
Diagram 1: A generalized workflow for developing a cyclic induction protocol, highlighting the need for tissue-specific assessment and iterative parameter adjustment.
The chromatin landscape and promoter accessibility vary significantly across organs. Robust OSKM induction is typically observed in the intestine, liver, and skin, while the brain, heart, and skeletal muscle show comparatively lower activation [9]. This intrinsic variability necessitates that systemically delivered protocols may have differing efficacies in different organs, and targeted delivery approaches could be required for tissues with low natural uptake.
Detailed Experimental Protocol: Cyclic Induction for In Vivo Rejuvenation
This section provides a step-by-step methodology for implementing a cyclic OSKM induction protocol in a progeria mouse model, based on the landmark study that extended median lifespan by 33% [4].
Materials and Reagent Solutions
Table 2: Essential Research Reagents for In Vivo Reprogramming
| Reagent / Tool | Function / Description | Example & Notes |
|---|---|---|
| Inducible OSKM System | Genetically engineered system for controlled factor expression. | Col1a1 locus-integrated polycistronic OSKM cassette in 4Fj or 4Fk mouse models [9]. |
| Doxycycline (Dox) | Inducer agent for Tet-On system; triggers OSKM expression. | Administered in drinking water or chow (e.g., 2 mg/mL with 1% sucrose). Prepare fresh weekly, protect from light [4] [9]. |
| AAV9 Delivery Vector | Non-integrating viral vector for in vivo gene delivery. | AAV9-TRE-OSK and AAV9-rtTA for delivery to wild-type mice; broad tissue tropism [4]. |
| Chemical Cocktails | Non-genetic alternative for reprogramming. | 7c cocktail can rejuvenate fibroblasts; different pathway from OSKM (upregulates p53) [4]. |
| Epigenetic Age Clocks | Multi-omics biomarker to quantify biological age reversal. | Measures DNA methylation patterns, transcriptomic, and metabolomic age [4]. |
| Teratoma Assay | Critical safety check for pluripotent cell formation. | Histological analysis of major organs (e.g., liver, pancreas, kidney) for dysplastic growth [9]. |
Step-by-Step Procedure
Animal Model Preparation:
- Utilize a progeric mouse model (e.g., LAKI mice with mutant lamin A) harboring a doxycycline-inducible OSKM transgene (e.g., 4Fj or 4Fk models) [4] [9].
- House mice under standard conditions with ad libitum access to food and water. For the control group, use littermates that lack the inducible transgene or do not receive Dox.
Cyclic Induction Regimen:
- Dox Administration: Add doxycycline hydate to the drinking water at a concentration of 2 mg/mL, supplemented with 1% sucrose to mask the bitter taste. Protect water bottles from light to prevent Dox degradation.
- Cycle Initiation: Begin the cyclic regimen when mice show clear signs of aging or as predetermined by the experimental design.
- Pulse Phase (2 days ON): Provide Dox-containing water to the experimental group for a continuous 48-hour period.
- Chase/Recovery Phase (5 days OFF): Replace the Dox-water with standard drinking water for the next 5 days.
- Repetition: Repeat this 7-day cycle (2 days ON, 5 days OFF) for the desired duration, such as 35 cycles as performed in the referenced study [4]. Monitor animal health and weight weekly.
Sample Collection and Analysis:
- Tissue Harvesting: At experimental endpoints, collect target tissues (e.g., skin, liver, kidney, pancreas) and divide them for various analyses: flash-freezing for omics studies, and fixation in formalin for histology.
- Rejuvenation Assessment:
- Epigenetic Clock Analysis: Isolate genomic DNA from tissues and perform whole-genome bisulfite sequencing. Analyze data using established epigenetic clocks to demonstrate reversal of DNA methylation age [4].
- Functional Tests: Conduct tests relevant to the tissue and model. For progeria models, this includes measuring spine curvature, assessing skin integrity, and performing cardiovascular function tests [9].
- Identity and Safety Verification:
- Histopathology: Process fixed tissues, embed in paraffin, section, and stain with Hematoxylin and Eosin (H&E). A trained pathologist should examine slides for any evidence of dysplasia or teratoma formation in all major organs [9].
- Lineage Marker Analysis: Perform immunofluorescence or RNA in-situ hybridization on tissue sections using antibodies or probes against key differentiation markers for the tissue of interest (e.g., Albumin for hepatocytes, Keratin-14 for skin basal cells) to confirm maintenance of cellular identity.
The strategic application of cyclic induction protocols is paramount to unlocking the therapeutic potential of in vivo reprogramming. The summarized data and detailed methodology provide a robust foundation for researchers to develop rejuvenation interventions that effectively balance the profound benefits of age reversal with the imperative of maintaining cellular identity and organismal safety. Future efforts will focus on refining tissue-specific delivery systems and further shortening induction pulses to enhance the precision and translational viability of this groundbreaking technology.
The advent of cellular reprogramming using Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) represents a groundbreaking approach for regenerative medicine and aging intervention [9]. In vivo reprogramming holds promise for reversing age-related cellular changes and restoring tissue function, with studies demonstrating that transient OSKM expression can ameliorate aging phenotypes and extend lifespan in progeria mouse models [8] [9]. However, a significant barrier to clinical translation remains the substantial safety risks associated with uncontrolled reprogramming, including teratoma formation, loss of cellular identity, and organ dysfunction [8] [9]. These limitations necessitate the development of more precise and safer reprogramming factors. This Application Note details how AI-driven protein engineering is being utilized to design novel, high-efficiency Yamanaka factor variants with enhanced functionality and safety profiles for in vivo reprogramming research.
AI-Driven Protein Design: A Paradigm Shift
Traditional protein engineering methods, such as directed evolution, are constrained by their reliance on existing biological templates and extensive experimental screening [53]. AI-driven de novo protein design transcends these limits by leveraging computational frameworks to create proteins with customized folds and functions from first principles [53] [54]. This approach uses generative models trained on vast biological datasets to establish high-dimensional mappings between protein sequence, structure, and function (SSF), enabling the rapid in silico generation and screening of novel protein sequences [53] [54].
Key AI Models and Their Functions in Protein Design
Table 1: Foundational AI Models for De Novo Protein Design
| Model Name | Core Task | Application in Factor Engineering | Key Feature |
|---|---|---|---|
| RFdiffusion [54] | Generates protein backbones conditioned on specific constraints. | Designing novel TF scaffolds and DNA-binding domains. | Diffusion-based generative model for de novo backbone design. |
| RFdiffusion2 [54] | Atom-level protein backbone generation. | Precise engineering of DNA-binding domains and cofactor placement. | Enhanced, atom-aware control for active-site scaffolding. |
| ProteinMPNN [54] | Designs amino acid sequences for a given protein backbone. | Optimizing sequences for stability and solubility of designed factors. | Graph neural network for rapid and robust sequence design. |
| AlphaFold2/3 [55] [54] | Predicts 3D protein structure from amino acid sequence. | Validating the predicted structure of designed factor variants. | High-accuracy single-chain (AF2) and complex (AF3) structure prediction. |
| ESM3 [54] | Jointly generates sequence, structure, and function. | Exploring the sequence-structure-function landscape of TFs. | Generative language model for multi-scale protein design. |
| Boltz-2 [55] | Predicts protein-ligand structure and binding affinity. | Assessing designed factors' interactions with DNA or co-factors. | Unified structure and affinity prediction in seconds. |
Application Note: Designing Safer Yamanaka Factor Variants
Rationale and Strategic Workflow
The core objective is to engineer OSKM variants that minimize off-target effects and the risk of tumorigenesis while maintaining or enhancing reprogramming efficiency. Key strategies include:
- Reducing Oncogenic Potential: Engineering c-MYC variants with attenuated oncogenic activity or designing KLF4 variants that minimize proliferation induction.
- Enhancing Specificity: Modifying DNA-binding domains (DBDs) to increase binding specificity toward rejuvenation-associated gene promoters.
- Improving Safety Profile: Introducing destabilizing domains or degradation tags for tighter temporal control over factor activity [56] [9].
The following workflow diagram outlines the integrated computational and experimental pipeline for achieving this.
Quantitative Performance Metrics for Engineered Factors
The evaluation of novel factor variants requires a multi-parametric approach. The table below summarizes key performance indicators (KPIs) and target values for assessing engineered factors compared to wild-type OSKM.
Table 2: Key Performance Indicators for Engineered Yamanaka Factor Variants
| Performance Category | Specific Metric | Wild-Type (WT) OSKM Baseline | Engineered Variant Target |
|---|---|---|---|
| Reprogramming Efficiency | iPSC Colony Formation Rate | Set to 1.0 (Reference) | >1.5x improvement |
| Epigenetic Age Reversal (DNAmAge) | Partial reduction | Enhanced & more sustained reduction | |
| Process Speed | Time to Pluripotency Marker Expression | ~2-3 weeks | Reduction by 5-7 days |
| Safety Profile | Teratoma Incidence In Vivo | High with continuous expression [9] | Near-zero incidence |
| Off-Target Gene Activation | Significant | Minimum 50% reduction | |
| Molecular Properties | Protein Stability (Half-life) | WT-specific | Tunable (e.g., reduced for c-MYC) |
| DNA Binding Specificity | WT-specific | Enhanced (lower Kd, higher specificity) |
Experimental Protocols
Protocol 1:In SilicoDesign and Validation of TF Variants
Objective: To computationally design and validate novel Yamanaka factor variants with improved DNA-binding specificity and reduced interaction with oncogenic pathways.
Materials:
- Software: RFdiffusion [54], ProteinMPNN [54], AlphaFold2/3 [55] [54], Boltz-2 [55], molecular dynamics (MD) simulation software.
- Hardware: High-performance computing (HPC) cluster with GPU acceleration.
Methodology:
- Input Parameter Definition:
- Specify the desired structural constraint, such as a DNA motif from a rejuvenation-associated gene promoter (e.g., OCT4 promoter itself).
- Define positional constraints within the RFdiffusion model to scaffold a novel transcription factor (TF) DNA-binding domain (DBD) tailored to this motif [54].
- Backbone Generation:
- Execute RFdiffusion to generate 1,000-10,000 candidate protein backbones conditioned on the input DNA motif and any symmetry constraints.
- Sequence Design:
- For each generated backbone, use ProteinMPNN to design multiple optimal amino acid sequences that stabilize the fold [54].
- Structure Validation:
- Process all designed sequences through AlphaFold2 to predict their intrinsic 3D structures without the conditioning DNA.
- Filter designs where the predicted (Cα RMSD < 2.0 Å) and (pLDDT > 80) indicate high design accuracy and model confidence [54].
- Binding Affinity and Specificity Assessment:
- Use Boltz-2 to rapidly predict the binding affinity (Kd) between the validated designed DBD and the target DNA sequence versus non-target sequences [55].
- Select top candidates showing high affinity for the target and low affinity for non-targets.
- Stability Analysis:
- Perform short MD simulations to assess the thermodynamic stability of the designed factor and its complex with DNA.
Protocol 2: Functional Characterization in Cellular Models
Objective: To experimentally test the reprogramming efficiency and safety of lead designed factor variants in mammalian cells.
Materials:
- Plasmids: Lentiviral or Sendai viral vectors encoding wild-type OSKM and designed variants.
- Cell Lines: Primary human fibroblasts (e.g., from HGPS patients for progeria context [6]), HEK293T cells for virus production.
- Media: Standard fibroblast culture medium, iPSC reprogramming medium.
- Assay Kits: qPCR kits for pluripotency markers, ELISA/Western blot reagents, immunocytochemistry antibodies.
Methodology:
- Virus Production and Transduction:
- Generate replication-incompetent lentiviral particles for OSKM (wild-type and variants) in HEK293T cells.
- Transduce primary fibroblasts at a low multiplicity of infection (MOI ~3-5) to avoid overexpression artifacts.
- Cyclic Induction for Partial Reprogramming:
- Efficiency Quantification:
- Quantitative PCR (qPCR): Measure expression of pluripotency markers (NANOG, REX1) and senescence-associated genes (p16, p21) at days 7, 14, and 21 post-induction.
- Immunofluorescence: Stain for TRA-1-60 and SSEA4 to quantify the emergence of pluripotent cells.
- High-Content Imaging: Use automated microscopy to track morphological shifts toward a rejuvenated cell state.
- Safety Profiling:
- RNA-Seq: Perform transcriptome analysis on reprogrammed cells to compare off-target gene activation profiles of variants versus wild-type OSKM.
- Teratoma Assay: Subcutaneously inject ~1x10^6 candidate iPSCs into immunodeficient mice. Monitor for 8-12 weeks for tumor formation; histologically analyze any masses to confirm teratoma structure [6] [9].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for AI-Engineered Factor Testing
| Reagent / Tool | Function / Application | Example & Notes |
|---|---|---|
| Doxycycline-Inducible System | Enables precise temporal control of factor expression in vitro and in vivo. | Tet-O-OSKM transgenic mouse models (e.g., 4Fj, 4Fk) [9]. |
| Small Molecule Reprogramming Cocktails | Can replace some transcription factors or enhance reprogramming efficiency/safety. | Combinations of chemicals that modulate signaling pathways (e.g., TGF-β, GSK3β). |
| Progeria Cell Models | Provides a validated system for testing rejuvenation efficacy. | HGPS patient-derived fibroblasts [6]. |
| Single-Cell Multi-Omics Assays | Allows deep characterization of heterogeneous reprogramming populations. | 10x Genomics scRNA-seq + scATAC-seq to map epigenetic and transcriptional changes. |
| Biolayer Interferometry (BLI) | Measures binding kinetics (Kon, Koff, Kd) of engineered factors to DNA. | Octet RED96 system for high-throughput analysis. |
| Genome-Wide Profiling | Maps binding sites of engineered factors across the genome. | ChIP-seq with epitope-tagged factors (e.g., HA-tag) [56]. |
AI-driven protein engineering marks a transformative advancement in the field of cellular reprogramming. By enabling the de novo design of Yamanaka factor variants with enhanced efficacy and superior safety profiles, this technology directly addresses the critical barriers to clinical translation. The integrated computational and experimental frameworks outlined in this Application Note provide a roadmap for researchers to develop next-generation reprogramming factors. These high-precision tools will be instrumental in realizing the therapeutic potential of in vivo reprogramming for regenerative medicine and the treatment of age-related diseases.
Addressing Tissue-Specific Toxicity and Inflammatory Responses
The therapeutic application of Yamanaka factors (OCT4, SOX2, KLF4, c-MYC) for in vivo reprogramming represents a paradigm shift in regenerative medicine. However, a significant barrier to clinical translation is the induction of tissue-specific toxicities and unintended inflammatory responses [6]. These adverse effects stem from several factors: the oncogenic potential of reprogramming factors like c-MYC, the generation of teratomas from poorly controlled pluripotent cells, and the immune system's recognition of reprogrammed cells [6]. This document provides a structured experimental framework to quantify these risks and outlines detailed protocols for evaluating and mitigating tissue-specific toxicities within a preclinical research setting. The focus is on aligning delivery system design with organ-specific vulnerability profiles to enable safer in vivo reprogramming.
Quantitative Profiling of Organ-Specific Toxicities
A critical first step is understanding the common toxicity profiles associated with chemotherapeutic and gene delivery agents, which provide a valuable proxy for the potential toxicities of in vivo reprogramming. The table below summarizes major organ-specific toxicities, their mechanisms, and corresponding formulation strategies that can be adapted for reprogramming factor delivery [57].
Table 1: Organ-Specific Toxicity Mechanisms and Mitigation Strategies for Delivery Systems
| Organ System | Key Toxicity Mechanisms | Proposed Mitigation Strategy for Reprogramming |
|---|---|---|
| Cardiac | Generation of reactive oxygen species (ROS), induction of DNA damage, apoptosis of cardiomyocytes [57]. | Use of ROS-responsive nanoparticles; cardiac-specific targeting ligands; continuous infusion pumps for systemic delivery. |
| Hepatic | Oxidative stress, glutathione depletion, bile duct damage, accumulation in liver macrophages (Kupffer cells) [57]. | Design of nanoparticles with neutral surface charge and PEGylation to reduce macrophage uptake; liver-avoidant formulations. |
| Renal | Glomerular damage, endothelial cell apoptosis, accumulation in renal tubules due to small size [57]. | Precise control of nanoparticle hydrodynamic diameter (>10 nm) to prevent renal filtration. |
| Neurological | Damage to peripheral nerve axons, ROS-induced damage to Schwann cells [57]. | Formulations engineered for low peripheral nervous system penetration; encapsulation to reduce free factor circulation. |
| Hematological | Disruption of hematopoietic stem cells (HSCs), DNA crosslinking, oxidative stress impairing HSC maintenance signals [57]. | Transient, pulsed delivery of factors; myeloid-specific promoter systems to restrict off-target effects. |
Experimental Protocols for Toxicity Assessment
This section provides detailed methodologies for profiling and mitigating the toxicity of Yamanaka factor delivery in vivo.
Protocol: In Vitro Cytotoxicity and Pro-inflammatory Cytokine Profiling
This protocol assesses the baseline toxicity and immunogenicity of delivery systems in different cell types before moving to in vivo models.
- Background: Polymeric nanoparticles (PNPs) are promising carriers for factor delivery. Their surface properties and composition directly influence cell viability and can trigger unwanted immune activation [58]. This assay quantifies these effects.
- Materials:
- Biological Materials: Primary human hepatocytes, cardiomyocytes (e.g., iCell Cardiomyocytes), and human umbilical vein endothelial cells (HUVECs).
- Reagents: Culture media for each cell type; Empty PNPs (PLGA-PEG) and Yamanaka factor-loaded PNPs; IL-6, TNF-α, and IL-1β ELISA kits; CellTiter-Glo Luminescent Cell Viability Assay kit.
- Equipment: Tissue culture hood, CO2 incubator, multi-well microplate reader (capable of luminescence and absorbance).
- Procedure:
- Seed cells in 96-well plates at a density of 1 x 104 cells per well and culture for 24 hours.
- CRITICAL STEP: Ensure cells are in the logarithmic growth phase at the time of treatment.
- Treat cells with a concentration series of empty and loaded PNPs (e.g., 0.1, 1, 10, 100 µg/mL). Include a media-only control.
- Incubate for 48 hours.
- PAUSE POINT: After 24 hours, carefully collect 100 µL of supernatant from each well and store at -80°C for subsequent cytokine analysis.
- After 48 hours, add CellTiter-Glo reagent to each well to measure cell viability via ATP quantification.
- Perform ELISA on the thawed supernatants to quantify IL-6, TNF-α, and IL-1β levels according to the manufacturer's instructions.
- Troubleshooting:
- Problem: High background in viability assay.
- Solution: Ensure nanoparticles are sterile-filtered and free of aggregates. Include a vehicle control to account for any effects from the suspension buffer.
- Expected Results: Dose-dependent decrease in viability and a concomitant increase in pro-inflammatory cytokines are expected. A safe formulation will show minimal cytotoxicity and cytokine secretion at therapeutic concentrations.
Protocol: In Vivo Profiling of Tissue-Specific Toxicity
This protocol provides a workflow for a comprehensive in vivo toxicity study following systemic administration of the delivery system.
- Background: In vivo models are essential for evaluating organ-specific accumulation, histopathological changes, and systemic inflammatory responses that cannot be captured in vitro [57].
- Materials:
- Biological Materials: C57BL/6 mice (8-10 weeks old).
- Reagents: Yamanaka factor-loaded PNPs; Isoflurane; Paraformaldehyde (4%); Hematoxylin and Eosin (H&E) stain; Antibodies for immunohistochemistry (e.g., anti-F4/80 for macrophages, anti-Ly6G for neutrophils).
- Equipment: IVIS Spectrum imaging system; Automatic hematology analyzer; Clinical chemistry analyzer; Microtome.
- Procedure:
- Randomize mice into control (PBS) and treatment (Yamanaka factor-PNPs) groups (n=6-8).
- Administer a single intravenous dose via the tail vein.
- PAUSE POINT: At 24 hours post-injection, collect blood via retro-orbital bleeding under anesthesia for complete blood count (CBC) and plasma cytokine analysis.
- At 7 days post-injection, euthanize mice and perfuse with PBS followed by 4% PFA.
- Harvest major organs (heart, liver, spleen, lungs, kidneys) and divide each: one half for histology (fixed in PFA), the other for molecular analysis (snap-frozen).
- Process fixed tissues, embed in paraffin, section, and stain with H&E. Analyze slides for signs of necrosis, inflammation, and tissue architecture disruption.
- Perform immunohistochemistry on sections to identify and quantify immune cell infiltration.
- General Notes: This protocol provides a snapshot of acute toxicity. For reprogramming studies, multiple doses may be required, necessitating longer-term studies with additional time points.
- Expected Results: The heart and liver are expected to be primary sites of toxicity. Histology may reveal inflammatory infiltrates and apoptotic cells in these organs. Hematology may show transient changes in white blood cell counts.
Pathway and Workflow Visualization
The following diagrams map the key signaling pathways involved in toxicity and the logical workflow for its assessment.
Toxicity Mechanism and Assessment Pathway
Experimental Workflow for Toxicity Evaluation
The Scientist's Toolkit: Research Reagent Solutions
The table below lists key materials and their functions for conducting the described toxicity assessments.
Table 2: Essential Reagents for Toxicity and Inflammation Assessment
| Item Name | Function/Application | Specifications & Notes |
|---|---|---|
| PLGA-PEG Nanoparticles | Biodegradable core-shell delivery vehicle for Yamanaka factors. | Tunable degradation rate via LA:GA ratio; PEG coating extends circulation time [58]. |
| CellTiter-Glo 3D | Luminescent assay for quantifying cell viability based on ATP content. | Ideal for 2D and 3D cell cultures; highly sensitive. |
| Pro-inflammatory ELISA Kits | Quantification of secreted cytokines (IL-6, TNF-α, IL-1β) from cell supernatants or plasma. | High specificity; provides picogram-level sensitivity for immune monitoring. |
| Anti-F4/80 Antibody | Immunohistochemistry marker for identifying tissue-infiltrating macrophages. | Crucial for quantifying innate immune response in harvested organs. |
| ROS-Sensitive Probe (e.g., DCFDA) | Flow cytometry or fluorescence microscopy to detect intracellular ROS. | Directly measures oxidative stress, a key mechanism of cardiotoxicity [57]. |
| IVIS Spectrum Imaging | Non-invasive, real-time biodistribution and persistence tracking of fluorescently-labeled PNPs. | Enables longitudinal analysis in live animals, reducing subject numbers. |
GMP-Compliant Workflows for Clinical Translation
The transition of induced pluripotent stem cell (iPSC) technologies from research to clinical application represents a frontier in regenerative medicine. The discovery that somatic cells can be reprogrammed using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) has opened unprecedented possibilities for treating degenerative diseases and aging itself [59]. However, the clinical translation of these technologies, particularly for in vivo reprogramming approaches, demands rigorous Good Manufacturing Practice (GMP)-compliant workflows to ensure patient safety, product consistency, and regulatory approval [60]. This application note details the establishment of GMP-compliant platforms specifically framed within the context of Yamanaka factor delivery methods for in vivo reprogramming research, providing researchers with practical protocols and quality control frameworks for clinical translation.
GMP Platform Establishment for iPSC Lines
The foundation of clinically applicable reprogramming therapies begins with the generation of GMP-compliant human iPSC (hiPSC) lines. Adapted from research protocols, a comprehensive GMP platform must address the entire workflow from donor selection to cell banking [60].
Key Platform Components
- Donor Recruitment and Screening: Rigorous donor eligibility assessment, informed consent, and comprehensive infectious disease testing are mandatory starting points.
- GMP-Compliant Reprogramming: Use of integration-free reprogramming methods to generate hiPSCs without genomic alterations, crucial for long-term safety.
- Master Cell Bank (MCB) Establishment: Creation of extensively characterized cell banks demonstrating safety, identity, purity, and potency.
- Stability Testing: Assessment of MCB stability under defined storage conditions to ensure product consistency throughout clinical use.
Quality Control Metrics for Released MCBs
Table 1: Essential Quality Control Tests for GMP-Compliant hiPSC Master Cell Banks
| Test Category | Specific Parameter | Release Criteria | Validation Method |
|---|---|---|---|
| Safety | Sterility | No microbial growth | Sterility testing |
| Mycoplasma | Negative | PCR/culture method | |
| Endotoxin | <5.0 EU/mL | LAL assay | |
| Adventitious Agents | Negative | In vitro/vivo assays | |
| Cell Identity | Pluripotency Markers | >75% expression | Flow cytometry/ICC |
| STR Profile | Identical to donor | DNA profiling | |
| Karyotype | Normal in >20 metaphases | K-band analysis | |
| Purity | Residual Vector | Negative | PCR |
| Plasmid Integration | No integration | Southern blot | |
| Potency | Germ Layer Differentiation | Expression of ≥2 markers per germ layer | Directed differentiation |
This quality framework ensures that hiPSC lines meet the stringent requirements for differentiation into various clinical products, including pancreatic islets (endoderm), cardiomyocytes (mesoderm), and neurons (ectoderm) [60].
Experimental Protocols for GMP-Compliant hiPSC Generation
Donor Material Procurement and Processing
Objective: To obtain and process starting donor material under GMP conditions for hiPSC generation. Materials: GMP-grade reagents, closed-system processing equipment, validated testing kits. Procedure:
- Recruit eligible donors following ethical approval and obtain informed consent
- Collect donor material (e.g., peripheral blood, skin biopsy) using sterile, GMP-compliant procedures
- Process samples within closed-system platforms (e.g., CliniMACS Prodigy system) to minimize contamination risk [61]
- Isolate target cells (e.g., CD34+ cells, fibroblasts) using GMP-grade separation methods
- Aliquot samples for mandatory quality control and adventitious agent testing
- Cryopreserve cells using controlled-rate freezing in validated media
Integration-Free Reprogramming
Objective: To generate hiPSCs from somatic cells without genomic integration of reprogramming factors. Materials: GMP-grade vectors (episomal, Sendai virus, mRNA), GMP-grade culture media and supplements, qualified cell culture equipment. Procedure:
- Thaw and expand isolated somatic cells in GMP-grade media for 2-3 passages
- Transduce/transfect cells with non-integrating vectors expressing OSKM factors at optimized MOI
- Culture cells in GMP-grade maintenance media with daily monitoring for morphological changes
- Harvest emerging hiPSC colonies 3-4 weeks post-transduction based on characteristic ESC-like morphology
- Mechanically pick and expand individual colonies in GMP-qualified matrix-coated plates
- Validate pluripotency through flow cytometry for markers (OCT4, SOX2, NANOG, SSEA-4, TRA-1-60)
- Perform genomic integration analysis to confirm vector-free status
Point-of-Care Manufacturing for Cell Therapies
Decentralized manufacturing models offer practical solutions for GMP-compliant production of cell and extracellular vesicle therapies, particularly relevant for autologous in vivo reprogramming approaches [62].
Isolator-Based POC Manufacturing
Objective: To implement isolator-based systems for GMP-compliant cell manufacturing at clinical sites. Key Advantages:
- Modular, sterile environments that reduce contamination risks and lower facility requirements
- Automation compatibility supporting both autologous and selected allogeneic product manufacturing
- Integration with real-time QC testing enabling rapid product release
- Closed-system processing maintaining aseptic conditions throughout manufacturing
Implementation Protocol:
- Install isolator systems at clinical sites with appropriate environmental monitoring
- Implement closed-system bioreactors for automated cell expansion and reprogramming
- Establish digitalized QC workflows for rapid product assessment
- Validate entire process through three consecutive successful production runs
- Train specialized personnel in isolator operation and maintenance procedures
Table 2: Comparison of Centralized vs. Point-of-Care Manufacturing Models
| Characteristic | Centralized Model | POC Model |
|---|---|---|
| Facility Requirements | High (Grade B/C cleanrooms) | Reduced (Isolator technology) |
| Production Timeline | Extended (weeks) | Condensed (days) |
| Logistical Complexity | High (cryopreservation, shipping) | Low (immediate administration) |
| Product Personalization | Challenging | Facilitated |
| Regulatory Oversight | Established | Evolving framework |
| Cost Structure | High capital investment | Distributed operational costs |
Yamanaka Factor Delivery for In Vivo Reprogramming
In vivo reprogramming using Yamanaka factors holds promise for tissue regeneration and rejuvenation but presents unique manufacturing and safety challenges [8] [10].
In Vivo Reprogramming Mechanisms
The Yamanaka factors (OSKM) orchestrate a complex reprogramming process through specific molecular mechanisms:
- OCT4 and SOX2 upregulate embryonic genes while inhibiting differentiation-associated genes
- KLF4 activates pluripotency genes like NANOG and facilitates mesenchymal-to-epithelial transition
- c-MYC modifies chromatin structure to increase accessibility for other reprogramming factors
Transient expression of these factors in vivo has demonstrated potential to restore youthful epigenetic patterns and promote functional regeneration across multiple tissues, including retina, skeletal muscle, heart, liver, brain, and intestine [10]. However, precise spatiotemporal control is essential to avoid teratoma formation, organ failure, and loss of cellular identity.
Diagram: Molecular Mechanisms of OSKM-Mediated Reprogramming
Safety Considerations for In Vivo Applications
Critical Safety Challenges:
- Teratoma formation from incomplete or uncontrolled reprogramming
- Tissue dysfunction due to loss of cellular identity in critical organs
- Oncogenic potential associated with prolonged c-MYC expression
- Inflammatory responses to delivery vectors or reprogramming process
Risk Mitigation Strategies:
- Cyclic Induction: Transient, pulsed expression of reprogramming factors rather than continuous expression [10]
- Tissue-Specific Targeting: Use of tissue-specific promoters or delivery systems to limit off-target effects
- Factor Modification: Development of modified factors with reduced oncogenic potential
- Small Molecule Alternatives: Exploration of small molecule cocktails that can replace or supplement transcription factors [6]
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for GMP-Compliant Reprogramming Workflows
| Reagent Category | Specific Product | Function | GMP Compliance |
|---|---|---|---|
| Reprogramming Vectors | Sendai viral vectors OSKM | Delivery of reprogramming factors | GMP-grade, non-integrating |
| Episomal plasmids OSKM | Non-viral factor delivery | GMP-grade, integration-free | |
| Cell Culture Media | MSC-Brew GMP Medium | Xeno-free MSC expansion | USP <1043> compliant |
| E8 flex Medium | Feeder-free hiPSC culture | GMP-grade formulation | |
| Culture Surfaces | Recombinant vitronectin | Defined substrate for pluripotency | GMP-grade, xeno-free |
| Laminin-521 | Defined substrate for expansion | GMP-grade, xeno-free | |
| Characterization Tools | MSC phenotyping kit | Standardized cell surface marker analysis | ISCT standard markers |
| Pluripotency marker panel | Confirmation of pluripotent state | Flow cytometry validated |
Signaling Pathways in Reprogramming
The molecular pathway of Yamanaka factor-mediated reprogramming involves a coordinated sequence of events that can be visualized as follows:
Diagram: Reprogramming Pathway and Cell Fate Decisions
GMP-compliant workflows are essential for the clinical translation of Yamanaka factor-based reprogramming therapies. The integration of robust manufacturing platforms, stringent quality control systems, and innovative point-of-care solutions addresses the critical challenges of safety, consistency, and scalability. As research advances in partial reprogramming and in vivo delivery methods, these GMP frameworks provide the necessary foundation for translating revolutionary reprogramming technologies into clinically viable treatments for aging and degenerative diseases. Future developments will likely focus on enhancing the precision of spatiotemporal control, optimizing delivery vectors, and establishing standardized safety assessments specific to in vivo reprogramming applications.
Assessing Efficacy: Biomarkers, Functional Outcomes, and Comparative Analysis
The advent of in vivo reprogramming, particularly using the Yamanaka factors (Oct4, Sox2, Klf4, c-Myc, collectively OSKM), has revolutionized regenerative medicine by demonstrating that aged somatic cells can be returned to a more youthful state without passing through pluripotency [4]. This approach, known as partial reprogramming, has shown remarkable potential in ameliorating age-related physiological decline and extending healthspan in animal models [4] [63]. However, the field urgently requires robust, quantitative methods to validate and measure the extent of rejuvenation induced by these interventions.
Molecular aging clocks have emerged as powerful tools to quantify biological age and assess the efficacy of rejuvenation strategies [64] [65]. These computational models estimate biological age from molecular biomarkers and often outperform chronological age in predicting health outcomes, disease risk, and mortality [65]. The most advanced aging clocks leverage various molecular layers, including epigenetic, transcriptomic, proteomic, and metabolomic signatures, providing a multi-dimensional view of the aging process and its reversal through reprogramming interventions [64] [65].
This Application Note provides a comprehensive framework for quantifying reprogramming-induced rejuvenation, with a specific focus on applications within Yamanaka factor research. We detail experimental protocols, analytical pipelines, and practical considerations for implementing these biomarkers in both basic research and drug development settings.
Types of Molecular Aging Clocks and Their Applications
Table 1: Comparison of Major Aging Clock Types
| Clock Type | Molecular Basis | Key Advantages | Limitations | Reprogramming Applications |
|---|---|---|---|---|
| Epigenetic Clocks | DNA methylation patterns at specific CpG sites [64] | High accuracy for chronological age; Well-validated across tissues [64] | May not directly reflect functional aging; Population-specific biases [65] | Gold standard for assessing OSKM-mediated rejuvenation [4] |
| Transcriptomic Clocks | Genome-wide gene expression patterns [65] | Captures active regulatory pathways; Functional insights | High sample variability; Complex normalization | Evaluating functional restoration in aged tissues [65] |
| Proteomic Clocks | Protein abundance and post-translational modifications [64] [65] | Direct reflection of functional state; Clinical utility | Limited coverage; Technical variability | Assessing tissue-level functional rejuvenation [64] |
| Metabolomic Clocks | Metabolite concentrations in biofluids [65] | Real-time metabolic snapshot; Non-invasive | Influenced by transient factors (diet, medication) [65] | Monitoring rapid responses to reprogramming factors [65] |
Each clock type provides unique insights into the rejuvenation process. Epigenetic clocks are particularly valuable in reprogramming research because they directly measure the molecular layers that Yamanaka factors target. During reprogramming, OSKM factors cooperate with epigenetic regulators like Polycomb repressive complex (PRC2) to remodel chromatin states and DNA methylation patterns [66]. The reversal of epigenetic aging signatures through partial reprogramming has been demonstrated to restore youthful function in multiple tissues [4].
Table 2: Selection Guide for Aging Clocks in Reprogramming Research
| Research Question | Recommended Clock Types | Sample Types | Key Readouts |
|---|---|---|---|
| Initial validation of rejuvenation | Multi-omic panel (Epigenetic + Proteomic) [4] [64] | Target tissue + blood | ΔBiological age; Pathway-specific changes |
| Longitudinal monitoring | Metabolomic + Clinical biochemistry [64] [65] | Blood, serum | Dynamic response; Intervention efficacy |
| Safety assessment | Epigenetic + Transcriptomic [4] [6] | Multiple tissues | Teratoma risk; Off-target effects |
| Mechanistic studies | Multi-omic integration [64] | Target tissue + primary cells | Pathway activation; Cellular identity |
Experimental Protocols for Biomarker Assessment
Sample Collection and Processing for Multi-Omic Analysis
Materials:
- DNA/RNA shield collection tubes
- Multiplex immunoassay platforms (e.g., Olink, SomaScan)
- NMR spectroscopy or LC-MS systems for metabolomics [65]
- DNA methylation array platforms (e.g., Illumina Epic Array)
Procedure:
- Pre-collection handling: Standardize collection timepoints to control for circadian effects. For in vivo reprogramming studies, establish baseline measurements prior to OSKM induction.
- Sample collection:
- Tissue biopsies: Snap-freeze in liquid nitrogen within 2 minutes of collection
- Blood collection: Draw into PAXgene Blood DNA tubes (epigenetics), Tempus Blood RNA tubes (transcriptomics), and EDTA plasma tubes (proteomics/metabolomics)
- Process plasma/serum samples within 30 minutes of collection; aliquot and store at -80°C
- Quality control:
- Assess DNA/RNA integrity numbers (DIN/RIN) >7.0
- Confirm plasma/serum absence of hemolysis
- Document all sample handling in laboratory information management system (LIMS)
DNA Methylation Analysis for Epigenetic Clock Assessment
Materials:
- EZ-96 DNA Methylation Kit (Zymo Research)
- Illumina Infinium HD Methylation Assay
- Epic BeadChip arrays
- Bioinformatics pipelines (e.g., SeSAMe, ENmix)
Procedure:
- DNA extraction and bisulfite conversion:
- Extract genomic DNA using column-based methods
- Treat 500ng DNA with bisulfite using EZ-96 DNA Methylation Kit
- Verify conversion efficiency with control probes
- Methylation array processing:
- Hybridize converted DNA to Illumina Epic BeadChips per manufacturer's protocol
- Scan arrays using iScan or NextSeq scanner
- Data preprocessing:
- Process intensity data with SeSAMe pipeline to minimize technical artifacts
- Normalize data using functional normalization
- Annotate probes to genomic features
- Exclude cross-reactive and polymorphic probes
- Epigenetic clock calculation:
- Implement Horvath's pan-tissue clock or Skin&Blood clock for tissue-specific applications
- Calculate biological age using published coefficients
- Compute ΔAge (Biological age - Chronological age) as primary rejuvenation metric
NMR-Based Metabolomic Profiling Protocol
Materials:
- Bruker 600 MHz NMR spectrometer with SampleJet
- Phosphate buffer (pH 7.4) in D₂O
- 3 mm NMR tubes
- Standard reference compounds for quantification
Procedure:
- Sample preparation:
- Thaw plasma/serum samples on ice
- Mix 300 μL plasma with 300 μL phosphate buffer
- Centrifuge at 10,000 × g for 10 minutes at 4°C
- Transfer 550 μL supernatant to 3 mm NMR tube
- NMR acquisition:
- Acquire ¹H-NMR spectra at 300K using NOESYGPPR1D pulse sequence
- Set acquisition time to 3.5 seconds, relaxation delay to 4 seconds
- Collect 64 transients for adequate signal-to-noise ratio
- Spectral processing:
- Apply exponential line broadening of 0.3 Hz before Fourier transformation
- Reference spectra to glucose α-anomer signal (δ 5.24)
- Perform baseline correction with asymmetric least squares algorithm
- Metabolite quantification:
- Use Chenomx NMR Suite for targeted profiling
- Quantify 50-100 metabolites across key pathways
- Normalize data using probabilistic quotient normalization
- Metabolomic age calculation:
- Apply pre-trained machine learning models (e.g., ensemble stacking)
- Calculate metabolic age deviation from chronological age
Integration with Yamanaka Factor Delivery Studies
Temporal Biomarker Assessment in Reprogramming Protocols
The timing of biomarker assessment is critical when evaluating Yamanaka factor-induced rejuvenation. Partial reprogramming protocols typically use cyclic induction (e.g., 2-day OSKM expression followed by 5-day withdrawal) to avoid complete dedifferentiation [4]. Biomarker sampling should align with these cycles to capture both immediate and persistent effects.
Recommended sampling scheme:
- Baseline: Pre-induction measurements
- Early phase: 24-48 hours after OSKM induction initiation
- Withdrawal phase: End of each rest period
- Cumulative effect: After 3-5 complete cycles
Longitudinal sampling enables distinction between transient responses and stable rejuvenation, with metabolomic clocks capturing rapid changes and epigenetic clocks reflecting more permanent restructuring [4] [65].
Tissue-Specific Considerations for In Vivo Applications
Different tissues show variable responsiveness to Yamanaka factor reprogramming, necessitating tailored biomarker approaches:
Central Nervous System Applications:
- Reprogramming factors induce neural stem cell generation and cognitive enhancement [15] [67]
- Primary biomarkers: Epigenetic clocks trained on neural tissue, inflammation markers (GlycA, GlycB via NMR) [65]
- Secondary validation: Behavioral tests, electrophysiology [15]
Cardiovascular System Applications:
- Partial reprogramming improves vascular function and reduces age-related pathology [6]
- Primary biomarkers: Proteomic clocks (reflecting inflammatory and coagulation pathways), metabolomic clocks (lipoprotein profiles) [64] [65]
Musculoskeletal System Applications:
- Enhanced regeneration capacity in aged muscle and bone [4]
- Primary biomarkers: Epigenetic clocks, collagen turnover metabolites via NMR [65]
Research Reagent Solutions
Table 3: Essential Research Reagents for Reprogramming and Rejuvenation Studies
| Reagent Category | Specific Products | Application | Key Considerations |
|---|---|---|---|
| Reprogramming Factor Delivery | Doxycycline-inducible lentiviral OSKM vectors [4] [68] | Controlled factor expression in vivo | Titrate doxycycline dose to achieve partial reprogramming |
| Epigenetic Clock Tools | Illumina Epic Methylation BeadChip [64] | Genome-wide DNA methylation profiling | Use consistent preprocessing pipelines for cross-study comparisons |
| Metabolomic Profiling | Bruker IVDr NMR platform with B.I.Quant-PS [65] | Quantitative plasma/serum metabolomics | Standardize sample collection to minimize pre-analytical variability |
| Single-Cell Multi-omics | 10x Genomics Multiome (ATAC + Gene Expression) | Cell-type specific rejuvenation assessment | Critical for heterogeneous tissues; identifies target cell populations |
| Aging Clock Algorithms | Horvath's pan-tissue clock, PhenoAge, DunedinPACE [64] [65] | Biological age calculation | Select clocks validated for specific tissues and species |
Data Analysis and Interpretation Framework
Multi-Omic Data Integration
Integrating data from multiple molecular layers provides the most comprehensive assessment of rejuvenation:
Statistical Integration Approach:
- Individual clock analysis: Calculate ΔAge for each omics layer separately
- Cross-omic correlation: Assess consistency of rejuvenation signals across platforms
- Composite rejuvenation score: Combine z-scores from significant biomarkers
- Pathway enrichment: Identify biological processes most affected by reprogramming
Interpretation Guidelines:
- Significant rejuvenation: Consistent ΔAge reduction across ≥2 omics layers with FDR <0.05
- Partial rejuvenation: Significant effect in one layer with trend in another
- Failed reprogramming: No ΔAge reduction or increase (potential adverse effects)
Safety and Efficacy Assessment
Biomarker analysis should include specific assessments of potential risks:
Teratoma Risk Biomarkers:
- Pluripotency gene activation signatures (NANOG, SSEA1) [68] [6]
- DNA methylation patterns at pluripotency regulator loci
- Rapid biological age reduction followed by increase may indicate dysregulated reprogramming
Tissue Function Biomarkers:
- Tissue-specific protein markers (e.g., albumin for liver, creatinine for kidney)
- Metabolomic profiles indicating normal metabolic function
- Inflammatory markers (CRP, GlycA) to detect adverse immune responses
The quantification of rejuvenation through epigenetic clocks and multi-omic biomarkers represents a critical advancement in the field of in vivo reprogramming. As Yamanaka factor delivery methods become increasingly refined for therapeutic applications, these biomarker platforms will be essential for validating efficacy, optimizing protocols, and ensuring safety. The integration of multiple molecular layers provides a comprehensive view of the rejuvenation process, from epigenetic restructuring to functional metabolic improvements. Standardized implementation of these protocols, as detailed in this Application Note, will accelerate the translation of reprogramming therapies from bench to bedside.
This application note details the functional validation of in vivo reprogramming via Yamanaka factors (OSKM: Oct4, Sox2, Klf4, c-Myc) for tissue regeneration and lifespan extension in murine models. The data and protocols herein are designed for researchers developing targeted reprogramming therapies, providing quantitative outcomes and standardized methodologies for assessing efficacy and safety.
Quantitative Data on Functional Outcomes
The following tables summarize key quantitative data from pivotal in vivo studies, demonstrating the impact of partial reprogramming on tissue regeneration, cognitive function, and overall lifespan.
Table 1: Tissue Regeneration and Functional Improvement in Mouse Models
| Tissue/Function | Intervention | Model | Key Outcomes | Citation |
|---|---|---|---|---|
| Systemic (Multiple Organs) | Cyclic OSKM induction | Progeric mice | 50% increase in mean survival time; improved wound healing, reduced fibrosis in skin and muscle | [8] [69] |
| Retina / Vision | OSK (c-Myc excluded) | Aged mice & glaucoma model | Restoration of vision; reversal of age-related vision impairment and optic nerve damage | [69] |
| Brain / Cognition | Neuron-restricted cyclic OSKM | Aged mice | Enhanced memory performance; increased neuronal activation; reversal of aging-related epigenetic markers | [70] |
| Pancreas & Muscle | Cyclic OSKM induction | Normally aging mice (12-month-old) | Increased proliferation of pancreatic beta cells; increased satellite cells in skeletal muscle | [69] |
| Vascular System | Single-short OSK in endothelial cells | Hypertensive mice | Decreased blood pressure; reversal of hypertension-induced vascular damage | [71] |
Table 2: Lifespan Extension from the Rodent Aging Interventions Database (RAID) and Related Studies
| Intervention | Subject | Mean/Median Lifespan Extension (vs. Control) | Maximum Lifespan Extension (vs. Control) | Citation |
|---|---|---|---|---|
| VEGF Overexpression | Male Mice | +360 days | +345 days | [72] |
| VEGF Overexpression | Female Mice | +270 days | +300 days | [72] |
| Rapamycin + Acarbose | Male Mice | +263 days | +218 days | [72] |
| Cyclic OSKM | Progeric Mice | +50% of mean survival | Not specified | [69] |
Detailed Experimental Protocols
Protocol: Systemic Partial Reprogramming in Murine Models
This protocol, adapted from landmark studies, describes a cyclic induction regimen for systemic partial reprogramming to achieve rejuvenation without teratoma formation [8] [69].
- Objective: To reverse age-related functional decline and extend healthspan in a progeric mouse model via transient, body-wide expression of Yamanaka factors.
- Materials:
- Animals: Progeric mice (e.g., LmnaG609G/G609G) or wild-type aged mice engineered with a doxycycline-inducible OSKM transgene.
- Induction Agent: Doxycycline hyclate (2 mg/mL) in drinking water, supplemented with 1% sucrose to mask taste. Protect solution from light.
- Control: Age-matched transgenic mice receiving standard drinking water.
- Procedure:
- Animal Age: Begin treatment when progeric mice show early signs of aging, or in wild-type mice at middle age (e.g., 12 months).
- Cyclic Induction Regimen:
- Administer doxycycline water for 48 hours.
- Replace with standard water for a 5-day rest period.
- This constitutes one 7-day cycle.
- Treatment Duration: Continue cycles for a pre-defined period (e.g., 6-12 weeks) or for the duration of the study.
- Functional Validation (Endpoint Analysis):
- Lifespan Monitoring: Record survival daily.
- Tissue Regeneration Assay: At a defined timepoint (e.g., after 6 weeks), perform a wound healing assay by creating a standard dorsal skin punch biopsy and measuring the rate of wound closure.
- Histopathology: Harvest and process tissues (skin, kidney, spleen, liver) for H&E staining to assess fibrosis reduction and general tissue architecture.
- Epigenetic Analysis: Isolve DNA from tissues (e.g., liver, blood) for bulk or single-cell DNA methylation analysis using established epigenetic clocks.
- Key Considerations: Continuous, unregulated OSKM expression leads to teratoma formation and organ failure. The cyclic, transient nature of this protocol is critical for its safety, allowing for epigenetic reset without loss of cellular identity [8] [73].
Protocol: Neuron-Restricted Partial Reprogramming for Cognitive Enhancement
This protocol details a method for achieving cell-type-specific reprogramming to reverse brain aging, minimizing risks associated with systemic factor delivery [70].
- Objective: To rejuvenate a specific neuronal population and reverse age-associated cognitive decline.
- Materials:
- Animals: Transgenic mice with Yamanaka factors (OSKM) under a neuron-specific promoter (e.g., α-CaMKII) and a doxycycline-inducible system (Tet-On).
- Induction Agent: Doxycycline diet (625 mg/kg) or water.
- Procedure:
- Animal Age: Use aged mice (e.g., 18-20 months old).
- Induction Regimen: Administer doxycycline chow for a minimum of 4 months to induce cyclic, low-level expression of OSKM specifically in α-CaMKII-positive excitatory neurons.
- Cognitive and Molecular Analysis:
- Behavioral Testing: Assess spatial learning and memory using the Morris Water Maze or contextual fear conditioning.
- Epigenetic Analysis: Perform immunofluorescence or Western Blot on cortical and hippocampal lysates for age-related epigenetic markers (e.g., H3K9me3, H4K16ac).
- Synaptic Plasticity: Analyze changes in the extracellular matrix and synaptic density via immunohistochemistry (e.g., WFA staining for perineuronal nets, PSD-95).
- Key Considerations: Restricting expression to post-mitotic neurons virtually eliminates the risk of teratoma formation. Cyclic induction is superior to continuous expression for achieving functional cognitive benefits [70].
Signaling Pathways and Experimental Workflows
The following diagrams, generated using Graphviz DOT language, illustrate the core experimental workflow and the underlying signaling logic of partial reprogramming.
Diagram 1: In Vivo Reprogramming Workflow
Diagram 2: Molecular Signaling in Partial Reprogramming
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for In Vivo Reprogramming Research
| Reagent / Tool | Function | Example Use Case | Key Consideration |
|---|---|---|---|
| Doxycycline-Inducible (Tet-On) System | Enables precise temporal control over OSKM expression. | Systemic or tissue-specific cyclic induction protocols. | Prevents continuous expression, mitigating cancer risk. [8] [70] |
| Tissue-Specific Promoters (e.g., α-CaMKII) | Restricts OSKM expression to target cell types (e.g., neurons, hepatocytes). | Studying cell-type-specific rejuvenation without systemic effects. | Critical for safety and for deconvoluting tissue-specific mechanisms. [73] [70] |
| Non-Viral Delivery Vectors (e.g., Plasmid DNA) | Delivers OSKM transgenes without viral integration. | Hydrodynamic tail vein injection for liver-specific reprogramming. | Safer profile but often lower efficiency than viral methods. [73] |
| Truncated Factor Set (OSK) | Uses Oct4, Sox2, Klf4 while omitting the oncogene c-Myc. | Ocular neuro-regeneration to improve safety profile. | May reduce efficacy in some tissues but significantly lowers tumorigenic potential. [69] |
| Epigenetic Clock Assays | Quantifies biological age reversal via DNA methylation patterns. | Validating epigenetic rejuvenation in tissue samples post-treatment. | Key biomarker for confirming the mechanism of action. [8] [69] |
Head-to-Head Comparison of Delivery Delivery Modalities: Efficiency vs. Safety
The clinical translation of in vivo reprogramming, a revolutionary strategy for tissue regeneration and rejuvenation, is critically dependent on the safe and efficient delivery of reprogramming factors such as the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC/OSKM). The ideal delivery system must achieve high transduction efficiency in target cells while minimizing risks such as insertional mutagenesis, immunogenicity, and teratoma formation. This document provides a structured, comparative analysis of current delivery modalities—viral, non-viral, and chemical reprogramming—framed within the context of in vivo applications. It includes detailed protocols for evaluating these systems and a decision-making framework to guide researchers in selecting the most appropriate delivery strategy for their preclinical in vivo reprogramming studies.
In vivo reprogramming, which involves the direct conversion of somatic cells to a pluripotent or rejuvenated state within a living organism, holds immense promise for regenerative medicine and the treatment of age-related diseases [63]. The cornerstone of this technology is the delivery of specific transcription factors that orchestrate this cellular reprogramming. However, the path from concept to clinic is fraught with challenges, primarily centered on delivery. The safety concerns are paramount; for instance, the use of the original Yamanaka factor c-Myc, an oncogene, poses significant tumorigenic risks, and the integration of viral vectors into the host genome can lead to insertional mutagenesis [7] [6]. Conversely, achieving sufficient reprogramming efficiency in a complex in vivo environment is a major hurdle. The delivery vehicle must successfully navigate biological barriers, target the desired cell population, and express factors at levels and durations that are both effective and safe. This application note directly addresses this efficiency-safety trade-off by providing a head-to-head comparison of available modalities, enabling researchers to make informed decisions for their experimental designs.
Comparative Analysis of Delivery Modalities
The following tables summarize the key characteristics, advantages, and disadvantages of the primary delivery modalities used for in vivo reprogramming.
Table 1: High-Level Comparison of Delivery Modality Platforms
| Modality | Key Sub-Types | Mechanism of Delivery | Integration into Genome | Best Use Case in In Vivo Research |
|---|---|---|---|---|
| Viral Vectors | Retrovirus, Lentivirus (LV) | Infects cells; delivers genetic cargo. | Yes (Retro/Lenti) | Stable, long-term expression in dividing cells (LV) [74]. |
| Adeno-associated Virus (AAV) | Infects cells; delivers genetic cargo. | No (episomal) | High-efficiency transduction in vivo; long-term expression in non-dividing cells [74] [75]. | |
| Adenovirus (AdV) | Infects cells; delivers genetic cargo. | No (episomal) | High transduction efficiency; large cargo capacity (up to 36kb) [74]. | |
| Non-Viral Delivery | Lipid Nanoparticles (LNPs) | Encapsulates and delivers cargo (mRNA, protein). | No | Transient, high-level expression; suitable for redosing [74] [76]. |
| Engineered Exosomes/EVs | Natural vesicle-mediated delivery. | No | Low immunogenicity; potential for inherent tissue targeting [75] [77]. | |
| Cell-Penetrating Peptides (CPPs) | Direct translocation or endocytosis of cargo. | No | Delivery of protein-based factors (e.g., recombinant OSKM) [74]. | |
| Chemical Reprogramming | Small Molecule Cocktails | Activates endogenous signaling pathways. | Not Applicable | Enhanced safety profile by avoiding exogenous genetic material [7]. |
Table 2: Quantitative and Qualitative Assessment of Key Parameters
| Modality | Typical Payload | Reprogramming Efficiency | Safety & Immunogenicity Profile | Tumorigenicity Risk | Key Limitations |
|---|---|---|---|---|---|
| Lentivirus (LV) | DNA (~8kb) | High | Moderate; integration risk, immunogenic concerns [74]. | High (integration) | Limited packaging capacity; complex production [74]. |
| AAV | DNA (<4.7kb) | Moderate to High | Favorable; mild immune response, non-integrating [74] [75]. | Low | Severely limited payload capacity; pre-existing immunity in populations [74] [75]. |
| Adenovirus (AdV) | DNA (~36kb) | Very High | Poor; can trigger strong inflammatory immune responses [74]. | Low | High immunogenicity; potential for host cell damage [74]. |
| Lipid Nanoparticles (LNPs) | mRNA, RNP | High (transient) | Favorable; low immunogenicity, allows redosing [74] [76]. | Low (transient) | Primarily targets liver; endosomal escape inefficiency [74] [77]. |
| Chemical Reprogramming | N/A | Low to Moderate | Highly Favorable; no viral components or genetic integration [7]. | Very Low | Complex optimization of cocktail; often lower efficiency [7] [77]. |
Detailed Experimental Protocols
This section outlines a standardized workflow for producing and evaluating an AAV-based delivery system for the Yamanaka factors in a mouse model, a common starting point for in vivo reprogramming studies. The protocol can be adapted for other modalities by substituting the relevant production and formulation steps.
Protocol 1: Production and Purification of AAV-OSKM for In Vivo Delivery
Principle: Generate high-titer, serotype-specific AAV vectors encoding the OSKM factors to achieve efficient in vivo transduction with a favorable safety profile [74] [75].
Materials:
- Plasmids: AAV transfer plasmid(s) with OSKM expression cassettes (note: due to AAV's cargo limit, split across multiple AAVs may be necessary), pAAV Helper Plasmid, pRC Plasmid (for serotype).
- Cell Line: HEK293T cells [74].
- Key Reagents: Polyethylenimine (PEI), Benzonase, Iodixanol, PBS-MK buffer.
- Equipment: Ultracentrifuge, 0.22 µm filter.
Procedure:
- Vector Design and Packaging: Clone the cDNAs for human OCT4, SOX2, KLF4, and a safer alternative to c-MYC (e.g., L-MYC) into AAV transfer plasmids. Use minimal promoters and synthetic polyA signals to fit within packaging constraints [7].
- Cell Transfection: Cultivate HEK293T cells in a cell factory. Co-transfect the cells with the AAV transfer, helper, and pRC plasmids using PEI as the transfection reagent.
- Harvest and Lysis: 72 hours post-transfection, harvest the cells and media. Subject the crude lysate to multiple freeze-thaw cycles. Treat the lysate with Benzonase to digest unprotected nucleic acids.
- Purification: Purify the AAV vectors via iodixanol density gradient ultracentrifugation. Concentrate and buffer-exchange the purified virus into PBS-MK using an Amicon centrifugal filter unit.
- Titration and QC: Determine the genomic titer (vg/mL) of the final preparation using quantitative PCR (qPCR). Confirm the absence of replication-competent AAV and assess sterility.
Protocol 2: In Vivo Evaluation of Reprogramming Efficiency and Safety in a Mouse Model
Principle: Systemically administer AAV-OSKM to adult mice and quantitatively assess the extent of reprogramming (efficiency) and the absence of teratomas (safety).
Materials:
- Animals: Adult immunocompromised mice (e.g., NSG) to mitigate immune response against AAV and human transgenes.
- Test Article: Purified AAV-OSKM from Protocol 1.
- Key Reagents: Antibodies for FACS (anti-TRA-1-60, anti-SSEA4), RNA extraction kit, primers for pluripotency markers (Nanog, Rex1).
- Equipment: IVIS Imaging System, Flow Cytometer, RT-qPCR machine, Histology equipment.
Procedure:
- Dosing and Administration: Randomize mice into experimental (AAV-OSKM) and control (AAV-GFP) groups. Adminish the vectors via intravenous (IV) tail vein injection. Note: Dose escalation is recommended to establish a therapeutic window.
- In Vivo Imaging: If using a bioluminescent reporter (e.g., luciferase under a pluripotency promoter), monitor the activation of the pluripotency network weekly using an IVIS imager.
- Endpoint Analysis (4-6 weeks post-injection):
- Efficiency Metric 1 (Flow Cytometry): Harvest target organs (e.g., liver, spleen). Create a single-cell suspension and stain for surface pluripotency markers (TRA-1-60, SSEA4). Analyze via FACS to determine the percentage of reprogrammed cells.
- Efficiency Metric 2 (RT-qPCR): Isolate total RNA from tissues. Perform RT-qPCR to quantify the expression of endogenous pluripotency genes (Nanog, Rex1). Normalize to housekeeping genes.
- Safety Metric 1 (Histopathology): Fix organs in formalin, embed in paraffin, section, and stain with H&E. Examine thoroughly for any evidence of teratoma formation or abnormal tissue growth.
- Safety Metric 2 (Blood Chemistry): Collect blood serum and analyze for markers of organ damage (e.g., ALT, AST for liver function) to assess acute toxicity.
Visualization of Delivery Modality Selection and Evaluation Workflow
The following diagram illustrates the critical decision points and experimental workflow for selecting and evaluating a delivery modality for in vivo reprogramming, based on the core research objectives.
Table 3: Essential Research Reagent Solutions for In Vivo Reprogramming Delivery Studies
| Item / Resource | Function / Description | Example Application in Protocol |
|---|---|---|
| AAV Serotype Library | Different serotypes (e.g., AAV8, AAV9, AAV-DJ) exhibit distinct tissue tropisms, enabling targeted delivery to organs like liver, muscle, or CNS. | Screening for optimal transduction of a specific target organ (e.g., liver for AAV8) before committing to a large-scale in vivo study [74] [75]. |
| LNP Formulation Kits | Ionizable lipids and pre-formed kits for encapsulating mRNA or saRNA encoding Yamanaka factors. | Generating nanoparticles for transient, non-integrating reprogramming, as demonstrated in recent CRISPR clinical trials [74] [76]. |
| Small Molecule Reprogramming Cocktails | Combinations of chemicals (e.g., VPA, RepSox, 8-Br-cAMP) that replace or enhance the activity of transcription factors. | Implementing a non-genetic integration-free reprogramming strategy to improve safety, albeit often at lower efficiency [7]. |
| Pluripotency Reporter Cell Line | Cells with a fluorescent or luminescent reporter (e.g., GFP under Nanog promoter) to track reprogramming in real-time. | Quantifying reprogramming efficiency in vitro before moving to in vivo models, as part of the vector validation process [6]. |
| In Vivo Imaging System (IVIS) | Non-invasive optical imaging technology to track bioluminescent reporters in live animals over time. | Monitoring the activation and duration of the pluripotency program in Protocol 2, allowing for longitudinal assessment in the same animal. |
The pursuit of safe and efficient in vivo reprogramming demands a meticulous, context-dependent selection of delivery modalities. No single platform currently holds all the ideal properties; the choice involves a calculated trade-off. Viral vectors, particularly AAVs, offer high efficiency and are invaluable for proof-of-concept studies, but their immunogenicity and cargo limits are significant drawbacks. Non-viral methods, especially LNPs delivering mRNA, are rapidly gaining traction due to their transient nature, reduced safety concerns, and the emerging potential for redosing, as seen in recent clinical breakthroughs [76]. The ultimate in safety may come from chemical reprogramming, though its efficiency requires further optimization.
Future advancements will likely hinge on the development of next-generation delivery systems. This includes engineered AAV capsids with enhanced tropism and larger cargo capacities, as well as LNPs that can target organs beyond the liver. Furthermore, the concept of partial reprogramming—achieving cellular rejuvenation without full reversion to pluripotency—may relax the requirements for prolonged factor expression, making safer, transient delivery systems like LNPs increasingly sufficient [6] [63]. By carefully weighing the parameters outlined in this application note, researchers can strategically navigate the current landscape and contribute to the accelerated and responsible translation of in vivo reprogramming therapies.
Validating Pluripotency and Genomic Stability in Derived Cell Lines
The derivation of cell lines via induced pluripotency represents a cornerstone of modern regenerative medicine and disease modeling research. Within the context of a broader thesis on Yamanaka factor (OCT4, SOX2, KLF4, c-MYC/OSKM) delivery methods for in vivo reprogramming, rigorous validation of the resulting cells is paramount [7]. The transition from a somatic to a pluripotent state, whether for generating fully pluripotent induced pluripotent stem cells (iPSCs) or for achieving a partially reprogrammed state in vivo, must be confirmed through a multifaceted characterization approach [63]. This protocol details the essential assays for validating two fundamental properties of any derived cell line: pluripotency, the capacity to differentiate into all germ layers, and genomic stability, the maintenance of a normal genetic complement free of deleterious mutations [78]. The reliability of downstream applications, from drug screening to cell therapy, is contingent upon this initial rigorous validation, which ensures that cellular models are both accurate and reproducible [79] [80].
Validating Pluripotency: Markers and Functional Assays
Confirmation of pluripotency involves assessing the expression of key molecular markers and demonstrating functional differentiation potential. A combination of these methods is necessary to provide conclusive evidence.
Molecular Marker Analysis
Pluripotent stem cells (PSCs) express a characteristic set of surface and intracellular markers. The International Stem Cell Initiative (ISCI) has identified a core set including the surface antigens SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81, and the transcription factors NANOG, OCT4, and SOX2 [78]. SSEA-1, by contrast, is typically expressed only upon differentiation [78].
- Flow Cytometry: This is a mandatory, quantitative technique for assessing the percentage of cells in a population that express pluripotency markers. It allows for simultaneous analysis of multiple markers and provides robust statistical data on population homogeneity [78].
- Immunocytochemistry (ICC): This method provides visual confirmation of marker expression within the characteristic morphology of undifferentiated colonies. It is crucial for demonstrating that the markers are localized to the nucleus (e.g., OCT4, NANOG) or cell surface (e.g., TRA-1-60) of cells within compact colonies with a high nucleus-to-cytoplasm ratio [81].
- Quantitative RT-PCR (qRT-PCR): This sensitive technique quantifies the expression levels of genes associated with pluripotency. It can detect the upregulation of endogenous pluripotency genes and the silencing of any exogenous reprogramming transgenes, which is a key indicator of successful reprogramming [78].
Table 1: Key Pluripotency Markers and Assessment Methods
| Marker Type | Marker Examples | Assessment Method | Key Interpretation |
|---|---|---|---|
| Surface Antigens | SSEA-3, SSEA-4, TRA-1-60, TRA-1-81 | Flow Cytometry, ICC | High expression (>80%) indicates an undifferentiated state. |
| Transcription Factors | NANOG, OCT4, SOX2 | ICC, qRT-PCR, Flow Cytometry (for intracellular staining) | Nuclear localization and high gene expression confirm pluripotency network activity. |
| Enzymatic Activity | Alkaline Phosphatase (AP) | AP Staining | High AP activity is a characteristic of undifferentiated PSCs. |
Functional Differentiation Assays
The definitive proof of pluripotency is the demonstration of a cell's ability to differentiate into derivatives of all three primary germ layers: ectoderm, mesoderm, and endoderm.
- In Vitro Embryoid Body (EB) Formation: This is a standard and mandatory assay. When PSCs are cultured in low-attachment conditions, they form three-dimensional aggregates called EBs, which spontaneously differentiate. The resulting EBs can be analyzed via qRT-PCR or ICC for markers of the three germ layers, such as:
- In Vivo Teratoma Formation: This is the gold-standard functional assay for pluripotency. Cells are injected into immunocompromised mice, where they form teratomas. Histological analysis of these tumors must reveal well-differentiated tissues derived from all three germ layers, such as cartilage (mesoderm), neural rosettes (ectoderm), and gut-like epithelium (endoderm) [78] [81]. Due to its complexity and ethical considerations, it is often used as a confirmatory assay.
The following diagram illustrates the core workflow for validating pluripotency, integrating both molecular and functional analyses:
Diagram 1: Workflow for comprehensive pluripotency validation.
Assessing Genomic Stability
Genomic instability is a major concern in derived cell lines, particularly in iPSCs, where the reprogramming process and subsequent culture can induce mutations. These abnormalities can alter cellular behavior, compromise differentiation potential, and pose a significant tumorigenic risk [79] [81]. Therefore, comprehensive genetic assessment is a critical release criterion.
Karyotypic Analysis and Copy Number Variations
The initial screening often focuses on detecting large-scale chromosomal abnormalities.
- Karyotyping (G-banding): This traditional cytogenetic method provides a genome-wide view of the chromosome number (ploidy) and large structural variations (translocations, deletions, inversions). It is a standard, low-resolution method for confirming a normal diploid karyotype [78].
- Chromosomal Microarray (CMA) and qPCR Assays: These techniques offer higher resolution for detecting submicroscopic copy number variations (CNVs), such as deletions or duplications. Targeted RT-qPCR assays for common karyotypic abnormalities in human iPSCs (e.g., gains of chromosomes 1, 12, 17, 20) have been shown to be a practical and effective strategy. Cultures derived from genomically stable iPSCs exhibit significantly reduced variance and improved differentiation efficiency [79] [80].
The Role of Next-Generation Sequencing (NGS)
Next-Generation Sequencing has emerged as the most powerful and comprehensive tool for genetic stability testing, capable of identifying point mutations, small insertions/deletions (indels), and CNVs with base-pair precision [82] [83].
- Whole Genome Sequencing (WGS): WGS provides a base-by-base view of the entire genome, enabling the detection of single nucleotide variations (SNVs), CNVs, and structural variants without prior knowledge of their location. It is ideal for comprehensive characterization of master cell banks [82].
- Targeted NGS Approaches: For more focused and cost-effective monitoring, techniques like Targeted Locus Amplification (TLA) or Cas9-enrichment can be coupled with NGS to deeply sequence the integration sites of exogenous reprogramming transgenes and their flanking regions. This is crucial for verifying the integrity of the inserted sequence and identifying potential structural variants or recombination events [83].
- Monitoring Genetic Drift: NGS can be used to track the emergence of genetic variants over time by sequencing cells at early and late passages. Studies have shown that the choice of reprogramming method significantly impacts genomic instability; for instance, Sendai virus (SV)-derived iPSCs have been observed to accumulate a higher frequency of CNVs and SNVs compared to those generated with integration-free episomal vectors (Epi) [81].
Table 2: Methods for Assessing Genomic Stability
| Method | Target Abnormality | Resolution | Key Advantage |
|---|---|---|---|
| Karyotyping | Aneuploidy, Large Translocations | ~5-10 Mb | Low-cost, genome-wide standard screen. |
| qPCR Assay | Common CNVs (e.g., Chr 1, 12, 17, 20) | Gene-specific | Practical, accessible, and quantitative [79]. |
| Chromosomal Microarray | CNVs, Loss of Heterozygosity | 10-100 kb | High-resolution, genome-wide CNV detection. |
| Next-Generation Sequencing | SNVs, Indels, CNVs, Structural Variants | Single Base Pair | Most comprehensive; detects all variant types [82] [83]. |
The following diagram outlines a strategic workflow for genomic stability assessment, from initial screening to deep investigation:
Diagram 2: Decision workflow for genomic stability assessment, highlighting the influence of reprogramming method on risk.
The Scientist's Toolkit: Essential Reagents and Materials
Successful validation requires a suite of reliable reagents and tools. The following table details key solutions for the experiments described in this protocol.
Table 3: Research Reagent Solutions for Pluripotency and Genomic Stability Validation
| Reagent / Kit | Primary Function | Application Note |
|---|---|---|
| mTeSR1 / Essential 8 Medium | Defined, xeno-free culture medium for maintaining PSCs. | Promotes consistent, high-quality growth of pluripotent cells, reducing spontaneous differentiation. |
| STEMdiff Trilineage Differentiation Kit | Directed differentiation of PSCs into ectoderm, mesoderm, and endoderm. | Standardized, kit-based approach for robust and reproducible in vitro functional validation of pluripotency. |
| CytoTune-iPS Sendai Reprogramming Kit | Non-integrating viral vectors for delivering OSKM factors. | Common method for generating iPSCs; note associated higher genomic instability risk requires careful monitoring [81]. |
| Episomal iPSC Reprogramming Vectors | Non-viral, integration-free plasmid vectors for reprogramming. | Safer alternative to viral methods; often includes additional factors like L-Myc and shp53 to enhance efficiency [81]. |
| Flow Cytometry Antibody Panels (e.g., anti-OCT4, SSEA-4, TRA-1-60) | Quantitative analysis of pluripotency marker expression. | Multiplexed panels allow for simultaneous assessment of multiple markers, providing a purity score for the population. |
| Karyostat+ / CNV qPCR Assay | Targeted qPCR assay for common CNVs in human PSCs. | A rapid, accessible, and cost-effective method for routine genomic stability screening [79]. |
| Genedata Selector | Bioinformatics platform for NGS data analysis. | Automates the analysis of genetic stability data from NGS, streamlining variant calling and report generation for regulatory compliance [82]. |
Integrated Application Note: Variability in iPSC-Derived Motor Neurons
Context: A research group is using an iPSC-derived motor neuron (MN) model to study amyotrophic lateral sclerosis (ALS). Despite using a standardized small-molecule differentiation protocol, they observe high batch-to-batch variability in MN purity and morphology, hindering their drug discovery efforts [79] [80].
Investigation & Solution:
- Quantify Variability: The group first analyzes their historical differentiation data using coefficient of variance and linear modeling. They find that non-genetic factors ("Induction Set" and "Operator") are the predominant sources of variability (explaining 30-70% of variance), outweighing the contribution from "Cell Line" genetics (2-30%) [79] [80].
- Hypothesize Genetic Cause: Suspecting that underlying genomic instability in the iPSC lines is amplifying technical variability, they implement a targeted RT-qPCR assay to screen for common karyotypic abnormalities.
- Findings: They identify that differentiations from genomically stable iPSCs (no detectable abnormalities) show a decreased coefficient of variance and significantly improved MN marker expression profiles (e.g., ISL1+MAP2) compared to those from abnormal lines [79] [80]. Morphological abnormalities, including a complete lack of neuronal organization or dense, non-neuronal clusters, are strongly associated with confirmed chromosomal deletions [80].
Conclusion and Protocol Integration: This case underscores that even with optimized Yamanaka factor delivery, the genomic integrity of the starting cell material is a critical determinant of experimental reproducibility. It validates the protocols described herein, demonstrating that routine genomic assessment is not just a quality control step but a practical strategy to enhance model reliability in downstream applications like preclinical drug discovery [79] [80] [7].
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) represented a transformative breakthrough in regenerative biology [7]. However, the clinical translation of this technology has faced significant challenges, including low reprogramming efficiency (typically <0.1% of cells convert), extended reprogramming timelines (3 weeks or more), and potential safety concerns regarding oncogenic transformation, particularly associated with c-MYC [84] [4]. Recent advances in artificial intelligence (AI) and machine learning are now overcoming these limitations through the rational design of enhanced reprogramming factors with superior functionality and rejuvenation potential [84] [85].
This case study examines a groundbreaking collaboration between OpenAI and Retro Biosciences that leveraged a specialized AI model to engineer novel variants of Yamanaka factors, achieving unprecedented gains in reprogramming efficiency and cellular rejuvenation [84]. We present comprehensive quantitative data, detailed experimental protocols, and analytical frameworks to guide the application of these AI-enhanced factors in in vivo reprogramming research.
AI-Driven Protein Engineering Platform
GPT-4b Micro Model Development
The core innovation enabling this breakthrough was the development of GPT-4b micro, a domain-specific AI model engineered for biological applications [84]. The model architecture and training methodology included:
- Model Initialization: GPT-4b micro was initialized from a scaled-down version of GPT-4o to leverage existing knowledge of biological systems while maintaining computational efficiency for specialized tasks [84].
- Specialized Training Data: The model was trained on a multimodal dataset comprising:
- Protein sequences from diverse biological contexts
- Biological text from scientific literature
- Tokenized 3D protein structure data (omitted by most protein language models) [84]
- Contextual Enrichment: A substantial portion of the training data was enriched with additional contextual information, including textual descriptions of protein function, co-evolutionary homologous sequences, and protein interaction networks [84].
- Extended Context Processing: The training approach substantially increased the effective context length, enabling processing of prompts up to 64,000 tokens during inference—unprecedented for protein sequence models [84].
Table 1: GPT-4b Micro Model Architecture and Training Specifications
| Component | Specification | Functional Advantage |
|---|---|---|
| Base Architecture | Scaled-down GPT-4o | Leverages existing knowledge with computational efficiency |
| Training Data Types | Protein sequences, biological text, tokenized 3D structures | Comprehensive understanding of structure-function relationships |
| Context Window | 64,000 tokens | Enables analysis of complex, multi-protein systems |
| Key Innovation | Contextual data enrichment with evolutionary and functional information | Enhanced capacity for generating biologically plausible variants |
Transfer Learning Approach for Cell Reprogramming
Complementary to the OpenAI approach, recent computational advances have demonstrated the effectiveness of transfer learning methodologies for predicting optimal cellular reprogramming interventions [86]. This framework involves:
- Pre-training Phase: A machine learning model is pre-trained on broad-based transcriptomic data from unperturbed cell types to learn the fundamental relationships in gene regulatory networks [86].
- Application-Specific Fine-tuning: The pre-trained model incorporates transcriptomic responses to genetic perturbations (knockdowns or overexpressions) to predict optimal interventions for specific reprogramming objectives [86].
- Pertubation Optimization: The approach identifies minimal combinatorial perturbations that maximize the probability of transitioning a cell from an initial state to a target cellular identity [86].
AI-Enhanced Yamanaka Factor Engineering
RetroSOX and RetroKLF Design
The OpenAI-Retro Biosciences team focused initial engineering efforts on SOX2 and KLF4, applying GPT-4b micro to generate novel variants with enhanced reprogramming functionality [84]. Key aspects of the engineering strategy included:
- Diverse Variant Generation: The AI model was prompted to propose a diverse set of "RetroSOX" and "RetroKLF" sequences that substantially diverged from wild-type proteins while maintaining core functionality [84].
- Functional Prioritization: Generated variants were prioritized based on predicted improvements in DNA binding affinity, protein-protein interactions, and transcriptional activation capacity [84].
The AI-driven engineering yielded remarkable results, with RetroSOX variants differing by more than 100 amino acids on average from wild-type SOX2 while maintaining or enhancing functionality [84].
Quantitative Performance Metrics
The AI-enhanced factors demonstrated exceptional performance in systematic screening assays, significantly outperforming both wild-type factors and manually engineered variants [84].
Table 2: Performance Comparison of AI-Enhanced vs. Wild-Type Yamanaka Factors
| Parameter | Wild-Type Factors | AI-Enhanced Factors | Fold Improvement |
|---|---|---|---|
| Reprogramming Marker Expression | Baseline | >50x increase in pluripotency markers | 50x [84] |
| SOX2 Variant Hit Rate | <10% (typical screens) | >30% outperformed wild-type | 3x [84] |
| KLF4 Variant Hit Rate | 1/19 (5.3%, prior attempts) | Nearly 50% superior to manual designs | 9x [84] |
| Time to Late Marker Expression | ~21 days | Several days sooner | ~30% reduction [84] |
| Endogenous Pluripotency Activation | Variable, donor-dependent | >85% in MSCs from older donors | Highly significant [84] |
Experimental Validation & Rejuvenation Assessment
In Vitro Reprogramming Protocols
Fibroblast Reprogramming Workflow
- Cell Culture Preparation:
- Factor Delivery:
- Media Transition:
- Colony Characterization:
Mesenchymal Stromal Cell (MSC) Reprogramming
- Donor-Specific Adaptation:
- mRNA-Based Reprogramming:
- Pluripotency Validation:
- Assess expression of endogenous pluripotency genes (OCT4, NANOG, SOX2, TRA-1-60) via immunostaining at day 12 [84].
- Confirm trilineage differentiation potential (ectoderm, mesoderm, endoderm) using established protocols [84].
- Perform karyotype analysis to verify genomic stability after multiple passages [84].
Rejuvenation Assessment Protocols
DNA Damage Repair Assay
The enhanced rejuvenation potential of AI-engineered factors was quantified through DNA damage response assays [84]:
- Induction of DNA Damage:
- Subject reprogrammed cells to gamma-irradiation (5 Gy) or chemical DNA damaging agents (etoposide 20 μM for 4 hours) [84].
- Immunofluorescence Staining:
- Fix cells with 4% paraformaldehyde for 15 minutes at room temperature.
- Permeabilize with 0.5% Triton X-100 for 10 minutes.
- Block with 5% BSA in PBS for 1 hour.
- Incubate with primary antibody against γ-H2AX (1:1000 dilution) overnight at 4°C [84].
- Incubate with fluorescent secondary antibody (1:2000 dilution) for 1 hour at room temperature.
- Mount with DAPI-containing mounting medium.
- Quantification and Analysis:
- Image cells using high-content fluorescence microscopy.
- Quantify γ-H2AX foci per nucleus using image analysis software (e.g., ImageJ).
- Compare DNA damage levels between cells reprogrammed with wild-type OSKM versus AI-enhanced factors [84].
Epigenetic Age Assessment
- DNA Methylation Analysis:
- Extract genomic DNA from reprogrammed cells using standard phenol-chloroform protocol.
- Perform bisulfite conversion using EZ DNA Methylation kit.
- Analyze methylation status using Illumina EPIC arrays or targeted bisulfite sequencing.
- Apply established epigenetic clocks (Horvath's pan-tissue clock, etc.) to estimate biological age [4].
- Transcriptomic Age Assessment:
- Extract total RNA using TRIzol method.
- Prepare RNA-seq libraries using poly-A selection.
- Sequence on Illumina platform (minimum 30 million reads per sample).
- Apply transcriptomic aging clocks to assess rejuvenation at gene expression level [4].
Research Reagent Solutions
Table 3: Essential Research Reagents for AI-Enhanced Factor Reprogramming
| Reagent/Category | Specific Examples | Function & Application |
|---|---|---|
| AI-Enhanced Factors | RetroSOX, RetroKLF variants | Core reprogramming factors with enhanced efficiency and rejuvenation potential [84] |
| Delivery Vectors | Lentivirus, mRNA-LNPs, Adenovirus, Sendai virus | Enable safe and efficient factor delivery; mRNA preferred for transient expression [84] [7] |
| Reprogramming Media | DMEM/F12, KnockOut SR, bFGF, NEAA, β-mercaptoethanol | Support pluripotent state acquisition and maintenance [7] |
| Pluripotency Markers | Anti-SSEA-4, Anti-TRA-1-60, Anti-NANOG, AP Staining | Validate successful reprogramming at different stages [84] |
| DNA Damage Assays | Anti-γ-H2AX, Etoposide, Gamma-irradiator | Quantify cellular rejuvenation through DNA repair capacity [84] |
| Epigenetic Clocks | Illumina EPIC arrays, Bisulfite conversion kits | Assess biological age reversal at epigenetic level [4] |
Signaling Pathway Framework for Enhanced Reprogramming
The AI-enhanced factors appear to operate through optimized engagement of core reprogramming pathways, with particular efficiency in activating the pluripotency network and enhancing DNA damage response mechanisms.
Application Notes for In Vivo Reprogramming Research
Delivery System Considerations for In Vivo Applications
The translation of AI-enhanced factors to in vivo settings requires careful selection of delivery modalities to maximize safety and efficacy:
Viral Vector Systems:
- Adeno-Associated Viruses (AAVs): AAV9 demonstrates broad tissue tropism and has been successfully used for in vivo reprogramming with OSK factors (excluding c-MYC) in mouse models, showing extended lifespan and reduced frailty [4].
- Lentiviral Vectors: Suitable for stable integration but raise safety concerns for clinical applications; can be used with inducible systems (tet-on) for temporal control [15].
- Sendai Virus: Non-integrating RNA virus that provides high transduction efficiency and transient expression, reducing tumorigenesis risk [7].
Non-Viral Delivery Methods:
- mRNA-LNP Formulations: Lipid nanoparticles encapsulating modified mRNA offer transient expression with minimal immunogenicity, ideal for controlled partial reprogramming [84] [4].
- Chemical Reprogramming: Small molecule cocktails (e.g., 7c cocktail) can induce partial reprogramming without genetic integration, though efficiency may be lower than factor-based approaches [4].
Partial vs. Complete Reprogramming Strategies
For in vivo rejuvenation applications, partial reprogramming approaches have demonstrated significant advantages:
- Cyclic Induction Protocols: Short, repetitive cycles of factor expression (e.g., 2-day induction followed by 5-day rest periods) have shown efficacy in reducing epigenetic age while maintaining cellular identity [4].
- Tissue-Specific Optimization: Different tissues exhibit varying responsiveness to reprogramming factors; protocols must be optimized for target tissue (CNS, liver, muscle, etc.) [15] [4] [87].
- Dosage Titration: Factor expression levels must be carefully calibrated to achieve rejuvenation without inducing full pluripotency or teratoma formation [4].
Safety and Efficacy Assessment
- Teratoma Risk Mitigation: Exclusion of c-MYC from reprogramming cocktails significantly reduces tumorigenic potential while maintaining rejuvenation efficacy [4].
- Immunological Compatibility: Assess host immune responses to delivery vectors and expressed factors, particularly for chronic or repeated administration regimens.
- Functional Outcome Measures: Beyond molecular rejuvenation markers, prioritize assessment of tissue function restoration (e.g., liver regeneration, cognitive improvement, motor function) [15] [87].
The integration of AI-driven protein engineering with cellular reprogramming technologies represents a paradigm shift in regenerative biology and rejuvenation research. The documented performance of RetroSOX and RetroKLF variants—achieving >50x increases in pluripotency marker expression, significantly higher hit rates than traditional approaches, and enhanced DNA damage repair—demonstrates the transformative potential of this methodology [84].
These AI-enhanced factors offer particular promise for in vivo reprogramming applications, where efficiency, safety, and temporal control are paramount considerations. The experimental protocols and analytical frameworks presented herein provide researchers with robust methodologies for implementing these advanced tools in diverse reprogramming contexts.
As the field progresses, the combination of AI-optimized reprogramming factors with sophisticated delivery systems and precise temporal control mechanisms will likely enable unprecedented capabilities for tissue rejuvenation and age-related disease intervention. Future research directions should focus on further optimizing factor combinations, developing tissue-specific delivery platforms, and establishing comprehensive safety profiles for clinical translation.
Conclusion
The strategic development of in vivo Yamanaka factor delivery methods marks a frontier in regenerative medicine, with the potential to directly intervene in the aging process and age-related diseases. The convergence of refined delivery systems, such as AAVs and mRNA, with safety-focused protocols like cyclic induction and AI-engineered factors, is paving a tangible path toward clinical application. Future progress hinges on achieving precise spatiotemporal control to ensure tissue-specific efficacy while eliminating oncogenic risk. The emerging success of chemical reprogramming and non-integrating methods promises a safer, more controllable therapeutic paradigm. For researchers and drug developers, the priority must be the establishment of robust GMP-compliant manufacturing and validation workflows that can transform these potent laboratory findings into viable, life-extending human therapies.
References
- 1. Mechanisms, pathways and strategies for rejuvenation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11058000/]
- 2. The occurrence and development of induced pluripotent ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2024.1389558/full]
- 3. Cellular reprogramming and epigenetic rejuvenation [https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-021-01158-7]
- 4. The long and winding road of reprogramming-induced ... [https://www.nature.com/articles/s41467-024-46020-5]
- 5. In vitro reconstitution of epigenetic reprogramming in the ... [https://www.nature.com/articles/s41586-024-07526-6]
- 6. Current advances and future prospects of cell ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1546423/full]
- 7. Advances in Induced Pluripotent Stem Cell Reprogramming ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12577567/]
- 8. A Review of the Present State of Epigenetic ... [https://www.fightaging.org/archives/2025/10/a-review-of-the-present-state-of-epigenetic-reprogramming-to-treat-aging/]
- 9. Organ‐Specific Dedifferentiation and Epigenetic Remodeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12610414/]
- 10. Organ-Specific Dedifferentiation and Epigenetic ... [https://pubmed.ncbi.nlm.nih.gov/41114535/]
- 11. Hallmarks of aging: An expanding universe [https://www.sciencedirect.com/science/article/pii/S0092867422013770]
- 12. Molecular mechanisms of aging and anti-aging strategies [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-024-01663-1]
- 13. Chemically induced reprogramming to reverse cellular aging [https://pmc.ncbi.nlm.nih.gov/articles/PMC10373966/]
- 14. Cellular population dynamics shape the route to human ... [https://www.nature.com/articles/s41467-023-37270-w]
- 15. In Vivo Reprogramming Using Yamanaka Factors in the CNS [https://pmc.ncbi.nlm.nih.gov/articles/PMC10886652/]
- 16. Human iPSC Reprogramming Success: The Impact of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11756937/]
- 17. In Vivo Base Editing Rescues Hutchinson-Gilford Progeria ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7872200/]
- 18. In vivo stress reporters as early biomarkers of the cellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9639039/]
- 19. Experimental mouse model reveals potential progeria ... [https://www.drugtargetreview.com/news/98981/experimental-mouse-model-reveals-potential-progeria-treatment-strategy/]
- 20. In Vivo Reprogramming Using Yamanaka Factors in the CNS [https://www.mdpi.com/2073-4409/13/4/343]
- 21. Viral Vectors Compared: AAV vs. Lentivirus vs. Adenovirus [https://synapse.patsnap.com/article/viral-vectors-compared-aav-vs-lentivirus-vs-adenovirus]
- 22. AAV Vs. Lentiviral Vectors - Life in the Lab [https://www.thermofisher.com/blog/life-in-the-lab/aav-vs-lv/]
- 23. Choosing the Right Vector for Gene Therapy and Research [https://www.cd-genomics.com/biomedical-ngs/resource/aav-lentivirus-adenovirus-gene-therapy-vaccine.html]
- 24. Adeno-Associated Virus Vectors: Principles, Practices, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11861813/]
- 25. AAV vs. Lentiviral Vectors | Gene Transfer Tools Comparison [https://www.corning.com/worldwide/en/products/life-sciences/resources/stories/at-the-bench/comparing-gene-transfer-tools-adeno-associated-virus-and-lentiviral-vectors.html]
- 26. AAV vector-mediated in vivo reprogramming into pluripotency [https://www.nature.com/articles/s41467-018-05059-x]
- 27. Non-Viral Gene Delivery Systems: Current Advances and ... [https://pubmed.ncbi.nlm.nih.gov/40874945/]
- 28. Nonviral Gene Delivery: Principle, Limitations, and Recent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2782077/]
- 29. Lipid nanoparticles for mRNA delivery [https://www.nature.com/articles/s41578-021-00358-0]
- 30. Current landscape of mRNA technologies and delivery ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-024-01080-z]
- 31. Episomes and Transposases—Utilities to Maintain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9601623/]
- 32. An episomal DNA vector platform for the persistent genetic ... [https://www.sciencedirect.com/science/article/pii/S2213671121005920]
- 33. Small-molecule-mediated reprogramming: a silver lining ... [https://www.nature.com/articles/s12276-020-0383-3]
- 34. The Combination of Two Small Molecules Improves ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12123452/]
- 35. Small molecules reprogram reactive astrocytes into neuronal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11081968/]
- 36. Exploring 7c versus 2c Small Molecule Reprogramming ... [https://www.fightaging.org/archives/2025/07/exploring-7c-versus-2c-small-molecule-reprogramming-combinations-for-rejuvenation/]
- 37. Tetracycline (Tet) Inducible Expression [https://www.addgene.org/collections/tetracycline/]
- 38. The Tet-On/Tet-Off Inducible Expression System [https://bitesizebio.com/24730/gain-control-the-tet-ontet-off-inducible-expression-system/]
- 39. Tet-On 3G tetracycline-inducible expression systems [https://www.takarabio.com/products/gene-function/tet-inducible-expression-systems/tet-on-3g-systems/tet-on-3g-systems?srsltid=AfmBOoqkxdkcy_VPF6OiivdsopmVWDXzr22zjVyPl3aT9mzQ_I8MM_Yg]
- 40. Tetracycline inducible expression—Tet-One systems [https://www.takarabio.com/products/gene-function/tet-inducible-expression-systems/tet-one-systems?srsltid=AfmBOoqm6GZ801nnqvDgm4BPoVRh-ysZJHL5jnD_oNBItjFRZXbjAXvL]
- 41. Engineering a synthetic gene circuit for high-performance ... [https://www.nature.com/articles/s41467-024-47592-y]
- 42. A versatile in vivo platform for reversible control of ... [https://www.sciencedirect.com/science/article/pii/S2213671124003175]
- 43. In vivo partial cellular reprogramming enhances liver ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9807246/]
- 44. Converting Cardiac Fibroblasts to Cardiomyocytes [https://pmc.ncbi.nlm.nih.gov/articles/PMC7897695/]
- 45. Inverse genetics tracing the differentiation pathway of ... [https://www.sciencedirect.com/science/article/pii/S1063458424012627]
- 46. Direct reprogramming of mouse fibroblasts to ... [https://www.sciencedirect.com/science/article/abs/pii/S0142961213006273]
- 47. Prevention of tumor risk associated with the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7268627/]
- 48. Prevention of tumor risk associated with the reprogramming of ... [https://jeccr.biomedcentral.com/articles/10.1186/s13046-020-01584-0]
- 49. MYC as a Target for Cancer Treatment: from Undruggable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12454499/]
- 50. c-Myc is essential for vasculogenesis and angiogenesis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC187450/]
- 51. Lysine-specific demethylase 1 inhibitors prevent teratoma ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5814224/]
- 52. The Safety of Embryonic Stem Cell Therapy Relies on ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3292887/]
- 53. The Role of AI-Driven De Novo Protein Design in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467925/]
- 54. Beyond evolution: De novo designed protein toolkit ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12624534/]
- 55. 2025 Guide to AI-Driven Protein Design & Machine Learning ... [https://nanohelix.ai/blog/ai-protein-design-guide-2025]
- 56. Widespread variation in molecular interactions and ... [https://www.sciencedirect.com/science/article/abs/pii/S1097276525001972]
- 57. Aligning organ-specific toxicities with formulation strategies [https://www.sciencedirect.com/science/article/abs/pii/S0378517325009585]
- 58. Inflammation-modulating polymeric nanoparticles: design ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12271758/]
- 59. Induced Pluripotent Stem Cells: Reprogramming Platforms ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7194323/]
- 60. Good Manufacturing Practice–compliant human induced ... [https://www.sciencedirect.com/science/article/pii/S1465324924000707]
- 61. MSC Workflow for Efficient Clinical Translation [https://www.labroots.com/webinar/mesenchymal-stromal-stem-cell-msc-workflow-efficient-clinical-translation-automated-gmp-compliant-ce?srsltid=AfmBOooyDH1LhIwOQaVw66Kjq0NaqqT3y-o1nge9Pwbx7tGDDVVzmPOc]
- 62. Isolator-based point-of-care manufacturing: a practical ... [https://pubmed.ncbi.nlm.nih.gov/40969223/]
- 63. Review Research of in vivo reprogramming toward clinical ... [https://www.sciencedirect.com/science/article/pii/S2352320424002001]
- 64. From ageing clocks to human digital twins in personalising ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12371080/]
- 65. Metabolomic-based aging clocks | npj Metabolic Health ... [https://www.nature.com/articles/s44324-025-00078-x]
- 66. Epigenetic reprogramming of cell identity: lessons from ... [https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-021-01131-4]
- 67. Expansion of the neocortex and protection from ... [https://www.sciencedirect.com/science/article/pii/S1934590924003278]
- 68. Restricting epigenetic activity promotes the reprogramming ... [https://www.nature.com/articles/s41420-023-01533-8]
- 69. And Cellular Yamanaka Factors Reprogramming [https://www.lifespan.io/topic/yamanaka-factors/]
- 70. cyclic overexpression of In restricted to... vivo Yamanaka factors [https://www.nature.com/articles/s42003-024-06328-w?error=cookies_not_supported&code=d05418e5-6e4e-441a-9ca6-de9f76f2644a]
- 71. Rejuvenation Roundup September 2025 [https://www.lifespan.io/news/rejuvenation-roundup-september-2025/]
- 72. The rodent aging interventions database (RAID): a data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11984422/]
- 73. of Adult Somatic Cells to Pluripotency by... In vivo Reprogramming [https://www.jove.com/pl/t/50837/in-vivo-reprogramming-adult-somatic-cells-to-pluripotency]
- 74. CRISPR Delivery Methods: Cargo, Vehicles, and Challenges [https://www.synthego.com/blog/delivery-crispr-cas9/]
- 75. Advances in β-Thalassemia Gene Therapy: CRISPR/Cas ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12562837/]
- 76. CRISPR Clinical Trials: A 2025 Update [https://innovativegenomics.org/news/crispr-clinical-trials-2025/]
- 77. PROTAC Delivery Strategies for Overcoming Physicochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12030311/]
- 78. Methods for Pluripotent Stem Cell Characterization [https://pmc.ncbi.nlm.nih.gov/articles/PMC11815987/]
- 79. Examining iPSC derived motor neuron variability and genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12612198/]
- 80. Examining iPSC derived motor neuron variability and ... [https://www.nature.com/articles/s41598-025-23378-0]
- 81. Tracing genomic instability in induced mesenchymal ... [https://www.nature.com/articles/s12276-025-01439-8]
- 82. Genetic Stability Testing Using NGS and MAM [https://www.genedata.com/resources/learn/details/blog/cho-cell-lines-characterization-with-ngs]
- 83. Next-generation sequencing: A powerful multi-purpose tool ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12013335/]
- 84. Accelerating life sciences research [https://openai.com/index/accelerating-life-sciences-research-with-retro-biosciences/]
- 85. Potential Breakthrough in Cell Reprogramming as OpenAI ... [https://www.biopharmatrend.com/news/potential-breakthrough-in-cell-reprogramming-as-openai-and-retro-biosciences-report-50x-pluripotency-marker-expression-gains-1357/]
- 86. Cell reprogramming design by transfer learning of functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10945810/]
- 87. In vivo partial cellular reprogramming enhances liver ... [https://www.sciencedirect.com/science/article/pii/S2211124722004910]