mRNA Reprogramming: A Footprint-Free Path to Clinical-Grade iPSCs and Regenerative Therapies
This article provides a comprehensive overview of mRNA-based technology for cell fate reprogramming, a transformative approach in regenerative medicine and drug development.
mRNA Reprogramming: A Footprint-Free Path to Clinical-Grade iPSCs and Regenerative Therapies
Abstract
This article provides a comprehensive overview of mRNA-based technology for cell fate reprogramming, a transformative approach in regenerative medicine and drug development. It explores the foundational principles of how synthetic mRNA enables the production of induced pluripotent stem cells (iPSCs) and direct lineage conversion without the risks of genomic integration. The scope spans from the molecular design of mRNA, including cap structures and nucleoside modifications that enhance stability and reduce immunogenicity, to advanced delivery systems like lipid nanoparticles (LNPs) and tissue nanotransfection (TNT). The content further addresses key methodological applications, critical troubleshooting steps for optimizing efficiency and safety, and a comparative analysis with other gene-editing and silencing technologies. Aimed at researchers and drug development professionals, this review synthesizes current advancements and future directions for deploying mRNA as a powerful, versatile, and clinically relevant tool for cellular reprogramming.
The mRNA Blueprint: Decoding the Science Behind Transient and Safe Cell Reprogramming
The unprecedented success of messenger RNA (mRNA) vaccines during the COVID-19 pandemic marked a pivotal turning point for this versatile technology. While the global deployment of SARS-CoV-2 vaccines demonstrated the feasibility, safety, and efficacy of mRNA platforms on a massive scale, these applications merely scratch the surface of its potential [1]. The core principle of mRNA therapeutics—delivering genetic instructions that direct human cells to produce specific proteins—lends itself to applications far beyond infectious disease prevention [2]. This technological paradigm is now rapidly expanding into the realm of regenerative medicine, offering innovative solutions for cellular reprogramming, tissue repair, and the treatment of chronic diseases [3]. The convergence of mRNA technology with cell fate reprogramming research represents a transformative approach in biomedical science, potentially enabling direct in vivo reprogramming of cell phenotypes without genetic integration [4]. This review explores the scientific foundations, current applications, and future directions of mRNA-based therapeutics, with particular emphasis on their role in cell engineering and regenerative medicine.
Technical Foundations of mRNA Therapeutics
Structural Components and Design Optimization
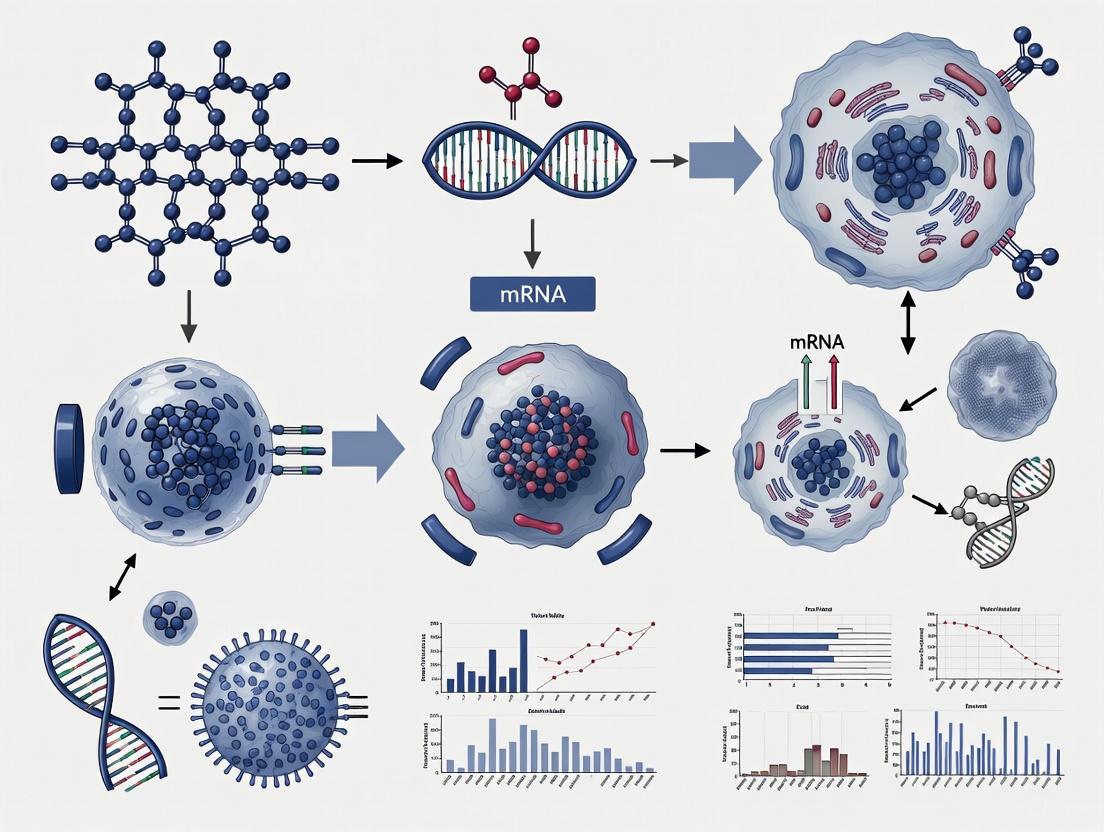
The architecture of synthetic mRNA is meticulously designed to mimic mature endogenous mRNA, comprising several critical regulatory elements that collectively influence stability, translational efficiency, and immunogenicity [1]. A standard in vitro transcribed (IVT) mRNA construct includes five essential regions: the 5' cap structure, 5' untranslated region (UTR), open reading frame (ORF) encoding the target protein, 3' UTR, and a poly(A) tail [1]. Each component serves distinct functions—the 5' cap and poly(A) tail protect against exonuclease degradation and facilitate ribosomal binding, while the UTRs contain regulatory elements that modulate translation efficiency and subcellular localization [2]. The ORF constitutes the coding sequence for the protein of interest, which can range from viral antigens for vaccines to transcription factors for cellular reprogramming [3].
Significant advances in nucleoside chemistry have been crucial for therapeutic application. The seminal work of Karikó and Weissman demonstrated that incorporating modified nucleosides such as N1-methylpseudouridine (m1Ψ) and 5-methylcytidine suppresses innate immune recognition by avoiding activation of pattern recognition receptors while simultaneously enhancing translational capacity [2] [5] [6]. This breakthrough fundamentally addressed the previously limiting issues of excessive immunogenicity and poor protein expression that hampered early mRNA therapeutic development [6].
Table 1: Key Structural Elements of Synthetic mRNA and Their Functions
| Structural Element | Composition | Primary Function | Impact on Efficacy |
|---|---|---|---|
| 5' Cap | 7-methylguanosine linked via 5'-5' triphosphate bond (Cap 0,1,2) | Ribosome recognition, initiation of translation, protection from degradation | Cap 1 structure reduces immunogenicity; Cap 2 may further evade immune detection [2] |
| 5' UTR | Nucleotide sequences upstream of ORF | Regulation of ribosome scanning and translation initiation | Optimized sequences enhance translational efficiency [1] |
| Open Reading Frame (ORF) | Protein-coding sequence with optimized codons | Encodes the therapeutic protein | Nucleoside modifications (e.g., m1Ψ) increase translation and reduce immunogenicity [5] |
| 3' UTR | Nucleotide sequences downstream of ORF | Regulation of mRNA stability and translation | Specific sequences can enhance half-life and protein yield [1] |
| Poly(A) Tail | 100-250 adenosine residues | Promotes mRNA stability and translational initiation | Optimal length crucial for protein expression; affects mRNA half-life [1] |
Advanced mRNA Synthesis and Purification Technologies
The production of therapeutic-grade mRNA relies on sophisticated in vitro transcription and purification systems. Two primary capping methodologies have emerged: post-transcriptional capping using vaccinia capping enzyme (VCE) and co-transcriptional capping using cap analogs [2]. The VCE approach, employed for Moderna's COVID-19 vaccine, enzymatically adds a Cap 0 structure to the 5' end of transcribed RNA, which can be further modified to Cap 1 by 2'-O-methyltransferase [2]. Alternatively, BioNTech/Pfizer's vaccine utilizes co-transcriptional capping with CleanCap technology, where cap analogs are incorporated during the transcription reaction [2].
A significant challenge in mRNA production has been the elimination of immunogenic byproducts, particularly double-stranded RNA (dsRNA) and 5'-uncapped mRNA. Recent innovations like the PureCap method developed by Inagaki et al. address this by employing a novel cap analog with a hydrophobic purification tag that enables chromatographic separation of fully capped mRNA from incomplete products [2]. This technology reportedly produces mRNA with translational activity more than ten times higher than conventional cap analogs and allows synthesis of the advanced Cap2 structure, which further evades immune detection [2].
Diagram 1: Synthetic mRNA structural components and their primary functions. The 5' cap facilitates ribosome binding and immune evasion, while nucleoside modifications in the ORF enhance translation and reduce immunogenicity. UTRs and the poly(A) tail collectively regulate stability and translational efficiency.
Delivery Systems: Lipid Nanoparticles and Beyond
Efficient intracellular delivery remains a critical challenge for mRNA therapeutics. Lipid nanoparticles (LNPs) have emerged as the leading delivery platform, with their composition optimized through decades of research [2] [5]. Standard LNP formulations comprise four key components: ionizable cationic lipids that complex with negatively charged mRNA, phospholipids that support lipid bilayer structure, cholesterol that enhances stability, and PEGylated lipids that improve nanoparticle pharmacokinetics [5]. These components self-assemble into approximately 100 nm particles that protect mRNA from nucleases and facilitate cellular uptake through endocytosis [5].
While current LNP systems efficiently deliver to hepatocytes and immune cells following systemic administration, their application to other tissues remains challenging. Research efforts are now focused on developing next-generation LNPs with enhanced tissue specificity through the incorporation of targeting ligands [3]. Additionally, a significant limitation of current LNP technology is the low endosomal escape efficiency—estimated at only about 2%—meaning most encapsulated mRNA never reaches the cytosol for translation [2]. Novel ionizable lipids with improved endosomolytic properties are under active investigation to address this bottleneck.
mRNA in Regenerative Medicine and Cell Fate Reprogramming
Principles of mRNA-Mediated Cellular Reprogramming
The application of mRNA technology to cell fate manipulation represents a paradigm shift in regenerative medicine. Unlike viral vector-based approaches that pose risks of genomic integration and insertional mutagenesis, mRNA offers a non-integrating alternative for transiently expressing reprogramming factors [2] [3]. This is particularly important when using oncogenic transcription factors such as Klf4 and c-Myc (Yamanaka factors), where persistent expression increases tumorigenic risk [2]. mRNA-mediated delivery provides precisely controlled, transient expression of these factors, sufficient to initiate reprogramming without permanent genetic alteration [3].
The fundamental process involves introducing synthetic mRNA encoding specific transcription factors that direct cells toward new phenotypic states. Early proof-of-concept studies demonstrated that repeated transfections of modified mRNA encoding the Yamanaka factors (Oct4, Sox2, Klf4, c-Myc) could generate induced pluripotent stem cells (iPSCs) from human fibroblasts [3]. Subsequent research has expanded this approach to direct lineage conversion, bypassing the pluripotent intermediate stage to transdifferentiate somatic cells into various target cell types, including cardiomyocytes, neurons, and hepatocytes [3].
Experimental Workflow for mRNA-Mediated Cellular Reprogramming
Diagram 2: Experimental workflow for mRNA-mediated cellular reprogramming. The process involves iterative mRNA transfections with critical quality control checkpoints at each stage to ensure successful phenotype conversion.
Applications in Specific Tissue Regeneration
Cardiovascular Regeneration
mRNA technology has shown significant promise in cardiovascular repair. Direct intramyocardial injection of VEGF-encoding mRNA has been investigated in patients undergoing coronary artery bypass grafting to promote therapeutic angiogenesis [3] [6]. In preclinical models, direct reprogramming of cardiac fibroblasts into cardiomyocyte-like cells using mRNA-encoded transcription factors (Gata4, Mef2c, Tbx5) has demonstrated potential for regenerating functional myocardium without cell transplantation [3].
Neural Regeneration
The field of neurology has witnessed remarkable advances with mRNA-based reprogramming approaches. Studies have successfully demonstrated the direct conversion of human astrocytes into functional dopamine neurons using mRNA-encoded transcription factors (Ascl1, Nurr1, Lmx1a) in Parkinson's disease models [3]. Similarly, reprogramming of fibroblasts to functional neurons has been achieved using modified mRNA cocktails, offering potential for neurodegenerative disease modeling and cell-based therapies [3].
Metabolic and Hepatic Regeneration
For metabolic disorders, mRNA technology enables in vivo production of missing or deficient enzymes. Approaches include direct reprogramming of human fibroblasts to hepatocyte-like cells using synthetic mRNA encoding hepatic transcription factors (HNF1A, HNF4A, HNF6) [3]. Clinical trials are underway for mRNA-based treatments of inherited metabolic diseases like ornithine transcarbamylase deficiency and Crigler-Najjar syndrome, where mRNA-encoded enzymes correct metabolic abnormalities in preclinical models [6].
Table 2: mRNA-Based Cellular Reprogramming Approaches in Regenerative Medicine
| Target Cell Type | Reprogramming Factors | Source Cells | Key Applications | Reference |
|---|---|---|---|---|
| iPSCs | Oct4, Sox2, Klf4, c-Myc | Human fibroblasts | Pluripotency establishment, disease modeling | [3] |
| Cardiomyocytes | Gata4, Mef2c, Tbx5 | Cardiac fibroblasts | Heart regeneration after myocardial infarction | [3] |
| Dopaminergic Neurons | Ascl1, Nurr1, Lmx1a | Human astrocytes | Parkinson's disease therapy | [3] |
| Hepatocytes | HNF1A, HNF4A, HNF6 | Human fibroblasts | Liver regeneration, metabolic disease | [3] |
| Hematopoietic Progenitors | ETV2, GATA2, c-MYB | Human pluripotent stem cells | Blood cell regeneration | [3] |
Research Reagents and Methodological Toolkit
Successful implementation of mRNA-based reprogramming protocols requires specialized reagents and methodologies. The following table summarizes essential research tools and their applications in mRNA therapeutic development.
Table 3: Essential Research Reagent Solutions for mRNA-Based Reprogramming Studies
| Reagent Category | Specific Examples | Function & Application | Technical Notes |
|---|---|---|---|
| Nucleoside Modifications | N1-methylpseudouridine, 5-methylcytidine | Reduce immunogenicity, enhance translation | Critical for in vivo applications; suppresses TLR recognition [5] |
| Capping Systems | CleanCap, VCE, PureCap | 5' cap addition for mRNA stability and translation | PureCap enables complete capping and superior purity [2] |
| In Vitro Transcription Kits | T7 polymerase-based systems | mRNA synthesis from DNA templates | Commercial systems available with optimized buffer formulations |
| Lipid Nanoparticles | Ionizable cationic lipids, PEG lipids | mRNA encapsulation and delivery | Composition affects tropism and endosomal escape efficiency [5] |
| Purification Systems | HPLC, FPLC | Removal of dsRNA contaminants and uncapped mRNA | Essential for reducing immune activation and improving yield [2] |
| Quality Control Assays | Agarose gel electrophoresis, LC-MS | mRNA purity, integrity, and modification analysis | Confirm absence of dsRNA contaminants for reduced immunogenicity |
Clinical Translation and Current Challenges
Clinical Development Landscape
The clinical application of mRNA therapeutics has expanded dramatically beyond COVID-19 vaccines. Current clinical trials encompass multiple domains, including infectious disease vaccines (influenza, RSV, HIV), cancer immunotherapies, protein replacement therapies, and regenerative medicine applications [1] [6]. Personalized cancer vaccines utilizing mRNA encoding patient-specific neoantigens have shown promising results in clinical trials, particularly when combined with immune checkpoint inhibitors [6]. In the regenerative medicine space, several mRNA-based candidates have entered clinical development, including VEGF-encoding mRNA for cardiovascular repair and mRNA-encoded enzymes for metabolic disorders [3] [6].
The manufacturing process for mRNA therapeutics has been streamlined to support clinical development, with current production timelines significantly shorter than those for traditional biologics. The entire process—from sequence design to purified mRNA—can be completed in weeks, facilitating rapid iteration and optimization [7] [1]. This agility is particularly valuable for personalized applications such as cancer neoantigen vaccines, where timely production is critical.
Technical Hurdles and Research Frontiers
Despite substantial progress, several technical challenges must be addressed to fully realize the potential of mRNA-based regenerative medicine. Precise control of protein expression dynamics remains difficult with current LNP platforms, as mRNA inherently produces transient expression requiring repeated administration for sustained effect [3]. While this transient nature is advantageous for safety, it complicates applications requiring prolonged protein expression. Emerging solutions include self-amplifying RNA (saRNA) systems derived from alphaviruses that enable longer-lasting expression from lower doses, and circular RNA (circRNA) constructs with superior stability owing to their resistance to exonuclease degradation [1].
Tissue-specific delivery represents another major hurdle. Current LNP systems predominantly target the liver following intravenous administration, limiting applications for other tissues [2] [3]. Research efforts are focused on developing selective organ targeting (SORT) LNPs through systematic modulation of lipid compositions and incorporation of targeting ligands [3]. Additionally, the inefficient endosomal escape of mRNA-LNP complexes significantly limits translational output, with typically less than 2% of internalized mRNA reaching the cytosol [2]. Novel ionizable lipids with improved endosomolytic properties are under active investigation to address this bottleneck.
Immunogenicity concerns persist even with nucleoside-modified mRNA, particularly for regenerative applications where immune activation is undesirable. While modifications reduce recognition by innate immune sensors, they do not eliminate it completely [1]. Further refinement of purification methods to remove immunogenic contaminants like dsRNA, combined with advanced cap structures and sequence engineering, may address this limitation.
mRNA technology has undergone a remarkable transformation from a pandemic response tool to a versatile platform with far-reaching implications for regenerative medicine and cellular reprogramming. The foundational breakthroughs in nucleoside chemistry, delivery systems, and manufacturing processes have positioned mRNA therapeutics as a disruptive force in biomedical science. As research advances, the convergence of mRNA technology with gene editing tools like CRISPR-Cas9 presents particularly exciting opportunities, with mRNA-encoded editors offering transient, efficient genome modification without the risks associated with viral delivery [3].
The future trajectory of mRNA-based regenerative medicine will likely focus on several key areas: development of novel delivery systems with enhanced tissue specificity and endosomal escape efficiency; creation of more sophisticated mRNA constructs with tunable expression kinetics; and integration of mRNA technology with tissue engineering approaches for complex organ regeneration. As these innovations mature, mRNA-based cell fate reprogramming stands to revolutionize therapeutic approaches for degenerative diseases, traumatic injuries, and genetic disorders, ultimately fulfilling the promise of regenerative medicine to restore form and function to damaged tissues and organs.
The discovery that somatic cells could be reprogrammed into induced pluripotent stem cells (iPSCs) using defined transcription factors marked a transformative milestone in regenerative medicine [8]. Traditional reprogramming methodologies, particularly those employing viral vectors such as retroviruses and lentiviruses, have been instrumental in advancing the field but present significant clinical translation challenges due to their inherent risk of genomic integration [8] [9]. These integrating vectors can disrupt host genomes, increase tumorigenic potential through insertional mutagenesis, and lead to persistent transgene expression that may interfere with iPSC differentiation and function [10] [9]. In response to these limitations, messenger RNA (mRNA)-based reprogramming has emerged as a powerful "footprint-free" alternative that avoids genomic integration while offering unprecedented control over reprogramming factor expression [11] [9]. This technical guide examines the scientific basis, methodological protocols, and emerging applications of mRNA technology in iPSC generation, framing this discussion within the broader context of mRNA-based cell fate reprogramming research.
The Viral Vector Challenge: Safety and Limitations
Initial iPSC generation relied heavily on viral delivery systems for introducing the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC). While effective, these systems present considerable hurdles for clinical translation:
- Genomic Integration Risks: Retroviral and lentiviral vectors integrate into the host genome, creating potential for insertional mutagenesis and oncogene activation [9]. This poses significant safety concerns for therapeutic applications.
- Persistent Transgene Expression: Integrated transgenes can remain active in iPSCs and their derivatives, potentially interfering with differentiation capacity and functional maturation of target cells [11].
- Immune Recognition: Viral components may trigger immune responses against both the delivery vector and the reprogrammed cells [12].
- Ethical and Regulatory Hurdles: The use of viral vectors involves more complex regulatory approval pathways compared to non-integrating methods [8].
Although non-integrating viral approaches such as Sendai virus have been developed, complete clearance of viral remnants from reprogrammed cells requires lengthy culture periods and rigorous validation [9].
mRNA Technology: A "Footprint-Free" Alternative
Fundamental Advantages
mRNA-based reprogramming represents a fundamentally different approach that addresses the core limitations of viral vectors:
- No Genomic Integration: As mRNA functions entirely in the cytoplasm without entering the nucleus, it presents zero risk of insertional mutagenesis [11] [9]. The reprogramming factors never interact with the host genome.
- Transient Expression Profile: mRNA has a relatively short intracellular half-life (typically 24-48 hours), allowing for precise, temporal control over reprogramming factor expression through repeated transfections [11].
- Rapid Clearance: Unlike DNA vectors or viruses, mRNA does not require a "cleanup phase" to purge residual reprogramming elements once pluripotency is established [11].
- High Reprogramming Efficiency: Modern mRNA protocols can achieve reprogramming efficiencies equaling or surpassing viral methods [11].
Key Technological Innovations
The practical implementation of mRNA reprogramming has been enabled by several critical innovations:
- Nucleotide Modification: Incorporation of modified nucleosides such as pseudouridine (ψU) and 5-methylcytidine (5mC) reduces innate immune recognition by pattern recognition receptors (TLR7, TLR8), minimizing interferon responses that otherwise inhibit reprogramming [9] [1].
- Optimized UTR Designs: Engineering 5' and 3' untranslated regions (UTRs) derived from highly stable endogenous mRNAs (e.g., α-globin, β-globin) significantly enhances mRNA stability and translational efficiency [9].
- Advanced Capping Systems: Anti-reverse cap analogs (ARCAs) ensure proper 5' cap orientation, improving ribosomal binding and translation initiation while protecting against exonuclease degradation [9].
Table 1: Comparison of iPSC Reprogramming Methods
| Method | Genomic Integration | Reprogramming Efficiency | Safety Profile | Technical Complexity | Clinical Translation Potential |
|---|---|---|---|---|---|
| Retroviral/Lentiviral | Yes | 0.01%-0.1% [9] | Low (insertional mutagenesis risk) | Moderate | Limited |
| Sendai Virus | No | 0.01%-1% [9] | Moderate (viral persistence concerns) | Moderate | Moderate |
| Episomal Plasmids | No (low frequency) | 0.04%-0.3% [9] | Moderate (potential plasmid persistence) | Moderate | Moderate |
| mRNA-Based | No | ~1-4% [11] | High (truly footprint-free) | High | High |
Experimental Protocols: Implementing mRNA Reprogramming
Core Workflow and Methodology
The following workflow outlines the optimized procedure for mRNA-based iPSC generation:
Detailed Protocol Components
mRNA Reprogramming Cocktail Formulation
Advanced mRNA cocktails have evolved beyond the canonical Yamanaka factors to enhance reprogramming kinetics and efficiency:
- Engineered Transcription Factors: The M3O variant of Oct4, incorporating an N-terminal MyoD transactivation domain, demonstrates significantly improved reprogramming efficiency compared to wild-type Oct4 [11].
- Supplemental Factors: Addition of Nanog to the core cocktail (OCT4, SOX2, KLF4, c-Myc, LIN28) dramatically improves colony formation efficiency [11].
- Stability-Optimized Constructs: The c-Myc-T58A mutation enhances protein stability by eliminating phosphorylation-dependent degradation signals [11].
Feeder-Free Culture Conditions
Modern protocols have eliminated the requirement for feeder layers through strategic culture optimization:
- Matrix Substitution: Use of synthetic substrates (e.g., Matrigel, vitronectin) replaces mouse embryonic fibroblasts, removing a source of variability and potential xeno-contamination [11].
- Density Optimization: Precise initial plating densities (e.g., 10,000 BJ fibroblasts per well in 12-well plates) maintain cell viability while preventing premature confluence that impedes transfection efficiency [11].
- Dynamic Suspension Culture: Implementation of suspension culture systems significantly enhances reprogramming efficiency for GO-PEI-RNA complex delivery [10].
mRNA Delivery and Transfection
Successful mRNA reprogramming requires optimized delivery strategies:
- Daily Transfection Protocol: Repeated transfection over 12-18 days is typically required to establish and stabilize pluripotency [11].
- Advanced Delivery Vehicles: Graphene oxide-polyethylenimine (GO-PEI) complexes demonstrate efficient mRNA delivery while protecting transcripts from RNase degradation [10].
- Dose Optimization: mRNA concentrations must balance expression efficiency against cytotoxicity, typically ranging from 0.5-1.0 µg per well in 12-well plates [11].
Table 2: Research Reagent Solutions for mRNA Reprogramming
| Reagent Category | Specific Examples | Function | Optimization Notes |
|---|---|---|---|
| Reprogramming mRNAs | M3O-Oct4, Sox2, Klf4, c-Myc-T58A, Lin28, Nanog [11] | Induce pluripotency in somatic cells | M3O-Oct4 variant provides 3x efficiency improvement vs. wild-type |
| Delivery Vehicles | GO-PEI complexes [10], lipid nanoparticles | Protect mRNA and facilitate cellular uptake | GO-PEI protects from RNase degradation; enables single-transfection |
| Culture Matrices | Matrigel, vitronectin, synthetic peptides | Replace feeder cells in xeno-free systems | Enables feeder-free reprogramming and clinical compliance |
| mRNA Modifications | Pseudouridine (ψU), 5-methylcytidine (5mC) [9] | Reduce immunogenicity, enhance stability | Critical for minimizing interferon response |
| Small Molecule Enhancers | Valproic acid, 8-Br-cAMP [13] | Improve reprogramming efficiency | 8-Br-cAMP with VPA increases efficiency 6.5-fold |
Molecular Mechanisms: How mRNA Reprogramming Reshapes Cell Identity
The process of mRNA-mediated reprogramming involves sophisticated molecular restructuring:
Key Mechanistic Insights
- Transient Factor Expression: Daily mRNA transfections create pulsatile expression of reprogramming factors that progressively reshape the epigenetic landscape without genomic integration [11].
- Metabolic Reprogramming: The reprogramming process induces a shift from oxidative phosphorylation to glycolysis, essential for establishing pluripotency [8].
- Epigenetic Restructuring: mRNA-derived factors activate TET enzymes that promote DNA demethylation at pluripotency loci like the OCT4 promoter [8].
- Endogenous Network Activation: Ultimately, exogenous factor expression is replaced by activation of endogenous pluripotency networks, establishing a self-sustaining pluripotent state [8].
Technical Challenges and Optimization Strategies
Despite its advantages, mRNA reprogramming presents specific technical challenges that require strategic optimization:
- Interferon Response: Unmodified mRNA can activate pattern recognition receptors (TLR7, TLR8), triggering type I interferon secretion that inhibits reprogramming. This is mitigated through nucleoside modifications (ψU, 5mC) [9].
- Cytotoxicity: Repeated transfections and robust transgene expression can induce cellular stress and apoptosis. Optimization of mRNA dosage and transfection intervals is critical [11].
- Labor Intensity: The requirement for daily transfections over 12+ days makes the process more hands-on than viral methods. Partial automation and suspension culture formats can alleviate this constraint [10].
- Cell Type Variability: Certain somatic cell types (especially blood cells) exhibit lower transfection efficiency. Pre-conditioning strategies and alternative delivery systems may be required [11].
Emerging Applications and Future Directions
mRNA-mediated iPSC generation serves as a foundational technology enabling numerous advanced applications:
Disease Modeling and Drug Discovery
iPSCs generated via mRNA reprogramming provide unique opportunities for human disease modeling:
- Neurological Disorders: Patient-specific iPSCs have been differentiated into motor neurons to study amyotrophic lateral sclerosis (ALS) mechanisms and screen potential therapeutics [13].
- Cardiac Conditions: iPSC-derived cardiomyocytes enable modeling of inherited arrhythmias and functional drug testing in human-relevant systems [8].
- Personalized Medicine: mRNA-reprogrammed iPSCs from individual patients facilitate development of patient-specific disease models and therapeutic screening platforms [14].
Clinical Translation and Therapeutic Applications
The footprint-free nature of mRNA-reprogrammed iPSCs positions them ideally for clinical applications:
- Regenerative Medicine: Multiple clinical trials are underway using iPSC-derived dopaminergic progenitors for Parkinson's disease treatment, demonstrating safety and functional integration [8].
- Retinal Therapies: iPSC-derived retinal pigment epithelial (RPE) products like Eyecyte-RPE have received regulatory approval for geographic atrophy associated with age-related macular degeneration [8].
- Personalized Cell Therapies: Autologous iPSC-derived dopamine neuron trials pioneer the use of patient-specific cells without immune suppression [8].
Technological Convergence
The future of mRNA reprogramming lies in its integration with other cutting-edge technologies:
- CRISPR-Cas9 Integration: Combined mRNA and CRISPR systems enable correction of genetic defects in patient-derived iPSCs before differentiation and transplantation [8] [15].
- AI-Guided Optimization: Machine learning approaches are being applied to predict differentiation outcomes, classify colony morphology, and enhance standardization in iPSC manufacturing [8] [15].
- Organoid Generation: mRNA-reprogrammed iPSCs facilitate development of complex 3D organoid models that better recapitulate tissue architecture and disease pathology [15].
mRNA-based reprogramming represents a paradigm shift in iPSC generation, effectively overcoming the fundamental safety limitations of viral vector systems. Through its footprint-free mechanism, precise temporal control, and high reprogramming efficiency, this technology has established itself as the gold standard for clinical-grade iPSC generation. The continued refinement of mRNA design, delivery systems, and culture protocols will further enhance the accessibility and applicability of this powerful technology. As mRNA reprogramming converges with advances in gene editing, bioengineering, and computational biology, it promises to accelerate the development of personalized regenerative therapies and human disease models, ultimately fulfilling the transformative potential of iPSC technology in both research and clinical domains.
The development of mRNA-based technologies for cell fate reprogramming represents a paradigm shift in regenerative medicine. The core of this technology lies in the sophisticated engineering of mRNA structural elements: the 5' cap, 5' and 3' untranslated regions (UTRs), open reading frame (ORF), and poly(A) tail. Together, these components determine the stability, translational efficiency, and immunogenicity of mRNA transcripts, enabling safe and efficient reprogramming of somatic cells without genomic integration. This technical guide examines the precise function and optimization strategies for each mRNA component within the context of induced pluripotent stem cell (iPSC) generation, providing researchers with a foundation for developing advanced mRNA therapeutics for cell fate conversion.
mRNA-based technology has emerged as a powerful platform for cell fate reprogramming since the landmark discovery that synthetic mRNA can encode transcription factors to convert somatic cells into induced pluripotent stem cells (iPSCs) [16]. Unlike viral vectors that pose risks of genomic integration and insertional mutagenesis, mRNA offers a transient, non-integrating approach for expressing reprogramming factors such as OCT4, SOX2, KLF4, and c-Myc (OSKM) [9] [16]. The successful application of mRNA for somatic reprogramming depends on the careful engineering of its structural components to maximize protein expression while minimizing innate immune responses [17] [18]. This guide provides an in-depth examination of these structural elements and their optimization for therapeutic applications, particularly in cell fate reprogramming research.
Structural Components of Therapeutic mRNA
The 5' Cap
Function and Importance: The 5' cap is a modified guanine nucleotide (m7G) added to the 5' end of mRNA via a 5'-5'-triphosphate linkage [9] [19]. This structure is critical for nuclear export, protection from RNase degradation, and recruitment of translation initiation factors [18] [19]. Perhaps most importantly, the cap structure allows differentiation between self and non-self mRNA molecules, with specific cap configurations evading innate immune recognition [19].
Optimization Strategies: Cap 0 (m7GpppN) undergoes methylation to form Cap 1, which evades recognition by cytosolic innate immune receptors like RIG-I [19]. Anti-reverse cap analogs (ARCAs) are designed to incorporate exclusively in the correct orientation, preventing degradation by Dcp2 and significantly enhancing translation efficiency and mRNA stability [9] [19]. Recent advances include trimeric cap analogs that yield Cap 1 structures with improved capping efficiency and gene expression profiles ideal for therapeutic applications [19].
Table 1: 5' Cap Structures and Their Characteristics
| Cap Type | Structure | Immune Recognition | Translation Efficiency |
|---|---|---|---|
| Cap 0 | m7GpppN | High (RIG-I recognized) | Moderate |
| Cap 1 | m7GpppNmN | Low (evades RIG-I) | High |
| ARCA Cap | Modified m7GpppN | Low | Very High |
| Trimetric Cap | Complex m7GpppNmN | Very Low | Very High |
5' and 3' Untranslated Regions (UTRs)
Function and Importance: UTRs flank the coding region and contain regulatory elements that influence mRNA stability, subcellular localization, and translational efficiency [9] [18]. The 5' UTR facilitates ribosome binding and scanning, while the 3' UTR contains binding sites for regulatory proteins and miRNAs that influence mRNA half-life [9].
Optimization Strategies: Effective UTR optimization involves several key principles: (1) eliminating upstream start codons that might disrupt ORF translation, (2) avoiding highly stable secondary structures that impede ribosome scanning, and (3) implementing shorter UTRs that generally enhance translation efficiency [18]. For therapeutic mRNA, UTRs from highly expressed genes like α-globin and β-globin are frequently employed because they contain sequence features that significantly enhance both translation and stability [20] [18]. Some advanced designs incorporate two consecutive 3' UTRs in a head-to-tail orientation to further enhance stability and expression levels [18].
Coding Region (Open Reading Frame)
Function and Importance: The open reading frame (ORF) contains the protein-coding sequence, beginning with a start codon (AUG) and ending with a stop codon [9]. For reprogramming applications, the ORF typically encodes transcription factors such as the Yamanaka factors (OCT4, SOX2, KLF4, c-Myc) or their optimized variants [21].
Optimization Strategies:
- Codon Optimization: Replacing rare codons with frequently used synonymous codons enhances translation efficiency by matching tRNA abundance in target cells [18] [19]. This optimization must also consider that some proteins require delayed translation facilitated by rare codons for proper folding [18].
- Nucleotide Modification: Incorporating modified nucleosides such as pseudouridine (ψ), 5-methylcytidine (m5C), and N1-methylpseudouridine (m1ψ) reduces immunogenicity by evading Toll-like receptor recognition while increasing translation efficiency [9] [18] [19]. These modifications were crucial in the development of successful mRNA reprogramming protocols [16].
- GC Content: Increasing GC-content can enhance translation efficiency, though excessive GC-richness may create problematic secondary structures [18].
Poly(A) Tail
Function and Importance: The poly(A) tail is a stretch of adenosine residues (typically 70-200 nucleotides in length) at the 3' end of mRNA that plays critical roles in stability, nuclear export, and translation efficiency [22] [23]. In the cytoplasm, poly(A) binding protein (PABPC) binds the tail and interacts with translation initiation factors at the 5' end, forming a closed-loop complex that enhances ribosomal recycling and protects mRNA from degradation [22] [23].
Optimization Strategies: Poly(A) tail length significantly influences translational efficiency, with optimal lengths typically ranging from 100-150 nucleotides for therapeutic applications [23]. While longer tails generally enhance stability and translation, excessively long tails may interfere with closed-loop formation [23]. Two primary methods exist for adding poly(A) tails to in vitro transcribed (IVT) mRNA: template-encoded polyadenylation (which produces consistent tail lengths) and enzymatic polyadenylation (which offers flexibility but yields variable lengths) [23].
Table 2: Impact of Poly(A) Tail Length on mRNA Function
| Tail Length (nt) | Stability | Translation Efficiency | Production Considerations |
|---|---|---|---|
| <50 | Low | Low | Easy to produce |
| 70-100 | Moderate | Moderate | Standard production |
| 100-150 | High | High (optimal) | Ideal balance |
| >150 | Very High | Potential decrease | Difficult to clone and maintain |
Experimental Protocols for mRNA Reprogramming
mRNA Synthesis and Modification
Template Design: Clone the gene of interest (e.g., OCT4, SOX2, KLF4, c-Myc) into an IVT vector containing optimized 5' and 3' UTRs (e.g., α-globin or β-globin UTRs) and a template-encoded poly(A) tail of defined length (100-120 nucleotides) [23]. For large-scale production, segmented poly(A) approaches with spacer sequences between poly(A) segments reduce plasmid recombination in E. coli [19].
In Vitro Transcription: Perform IVT reactions using T7 RNA polymerase with modified nucleotides (e.g., N1-methylpseudouridine instead of uridine) to reduce immunogenicity [9] [19]. Include ARCA cap analogs or use enzymatic capping methods post-transcription to ensure proper Cap 1 formation [19]. Purify the resulting mRNA using HPLC or cellulose-based methods to remove double-stranded RNA contaminants that trigger innate immune responses [20].
Cell Transfection and Reprogramming
Cell Preparation: Culture human fibroblasts or other target somatic cells in appropriate growth media. For reprogramming, cells should be at early passages and 50-80% confluent at the time of first transfection [16] [21].
mRNA Transfection: Transfect cells daily with 0.5-1 µg/mL of each modified mRNA encoding reprogramming factors using appropriate transfection reagents [16]. Daily transfections are necessary to maintain sufficient levels of reprogramming factors due to mRNA's relatively short half-life [16].
Immunogenicity Management: Include interferon inhibitors such as B18R in the culture medium to suppress antiviral responses triggered by exogenous mRNA [16]. The combination of nucleotide modifications and interferon suppression enables sustained protein expression without excessive cytotoxicity [16].
Reprogramming Timeline: Continue daily transfections for approximately 2-3 weeks, monitoring for emergence of embryonic stem cell-like colonies expressing pluripotency markers (SSEA-4, TRA-1-60, NANOG) [16] [21]. The entire process using optimized mRNA factors is significantly faster (about two times) and more efficient (35-fold higher) than viral methods [16].
Visualization of mRNA Structure and Function
mRNA Closed-Loop Model and Translation
Diagram 1: mRNA Closed-Loop Model for Efficient Translation. This illustrates how the 5' cap and 3' poly(A) tail interact via protein complexes to form a circular structure that enhances translational efficiency by promoting ribosomal recycling.
mRNA Reprogramming Workflow
Diagram 2: mRNA-Based Cellular Reprogramming Workflow. This chart outlines the key steps in converting somatic cells to induced pluripotent stem cells using modified mRNA, highlighting the cyclical nature of daily transfections required for successful reprogramming.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for mRNA Reprogramming Research
| Reagent / Material | Function | Application Notes |
|---|---|---|
| IVT Vector with UTRs | Template for mRNA synthesis | Contains optimized 5' and 3' UTRs (e.g., β-globin) |
| Modified Nucleotides | Reduce immunogenicity, enhance stability | N1-methylpseudouridine, 5-methylcytidine |
| ARCA Cap Analogs | Ensure proper 5' capping | Prevent reverse incorporation, enhance translation |
| T7 RNA Polymerase | In vitro transcription | High-yield mRNA synthesis |
| Poly(A) Polymerase | Enzymatic polyadenylation | Alternative to template-encoded tails |
| B18R Interferon Inhibitor | Suppress innate immune response | Critical for repeated transfections |
| Lipid Nanoparticles | mRNA delivery vehicles | Protect mRNA, enhance cellular uptake |
| Pluripotency Markers | Assess reprogramming success | Antibodies for SSEA-4, TRA-1-60, NANOG |
Applications in Cell Fate Reprogramming
The structural optimization of mRNA components has been crucial for advancing cell fate reprogramming technologies. Modified mRNA encoding Yamanaka factors has demonstrated remarkable efficiency in generating iPSCs from various somatic cell types, including fibroblasts, mesenchymal stem cells, and amniotic fluid stem cells [9]. Recent advances include AI-assisted engineering of enhanced reprogramming factors, such as RetroSOX and RetroKLF variants, which have shown a 50-fold increase in expression of stem cell reprogramming markers compared to wild-type factors [21]. These optimized factors also demonstrate enhanced DNA damage repair capabilities, suggesting improved rejuvenation potential [21].
The versatility of mRNA technology extends beyond iPSC generation to direct lineage conversion, where specialized cell types are produced without transitioning through a pluripotent state [16]. Furthermore, mRNA-synthesized chimeric antigen receptors (CARs) enable the production of safer CAR-T cells without genomic integration risks [17]. The transient nature of mRNA expression is particularly advantageous for precise control over reprogramming dynamics, allowing researchers to fine-tune the duration and levels of transcription factor expression [16].
The precise engineering of mRNA structural elements—5' cap, UTRs, coding region, and poly(A) tail—has transformed mRNA from a simple genetic messenger to a powerful therapeutic tool for cell fate reprogramming. Through strategic modifications that enhance stability, translation, and immune evasion, researchers have developed mRNA platforms that safely and efficiently reprogram somatic cells to pluripotency. As optimization strategies continue to evolve, including AI-assisted protein design and novel nucleotide modifications, mRNA technology holds unprecedented potential for regenerative medicine, disease modeling, and personalized cell therapies. The continued refinement of each mRNA component promises to further enhance the efficacy and applicability of this transformative technology.
The groundbreaking work of Katalin Karikó and Drew Weissman, awarded the 2023 Nobel Prize in Physiology or Medicine, addressed a fundamental barrier that had hindered mRNA therapeutic development for decades: the innate immune system's violent reaction to externally delivered mRNA [24] [25]. Their discovery that nucleoside base modifications could render synthetic mRNA non-immunogenic has not only enabled the development of effective COVID-19 vaccines but has also created a powerful new platform for cell fate reprogramming research. This whitepaper details the technical underpinnings of this breakthrough, its application in somatic cell reprogramming, and the essential toolkit for researchers leveraging this technology.
The conceptual appeal of using messenger RNA (mRNA) as a therapeutic or research tool is profound. It enables the cell's own machinery to produce almost any protein of interest, without the risk of genomic integration posed by viral vectors [26]. However, early attempts to harness this potential were thwarted by a critical obstacle. In vitro transcribed (IVT) mRNA was recognized as foreign by the host's immune system, triggering robust inflammatory responses and leading to the suppression of protein translation [24] [2].
This innate immune detection occurs through various pattern recognition receptors (PRRs), including Toll-like receptors (TLR7, TLR8) in endosomes and cytosolic sensors like RIG-I and PKR [27] [26]. Upon binding exogenous RNA, these receptors activate signaling cascades that result in the production of type I interferons and pro-inflammatory cytokines, effectively halting the practical application of mRNA technologies [27]. Karikó and Weissman's seminal insight was to investigate how mammalian cells distinguish their own native mRNA, which is not inflammatory, from synthetic IVT mRNA.
The Breakthrough Discovery: Mimicking Self
Core Hypothesis and Experimental Rationale
Karikó and Weissman hypothesized that the key distinction lay in the chemical composition of the nucleosides. They observed that bases in RNA from mammalian cells are frequently chemically modified, whereas standard IVT mRNA incorporates only natural, unmodified nucleosides [24]. This led them to investigate whether the absence of these altered bases in IVT mRNA explained its unwanted immunogenicity.
Key Experimental Methodology
To test their hypothesis, they produced multiple variants of mRNA, each with unique chemical alterations in their bases. They focused on modifications naturally found in mammalian RNA, particularly the replacement of uridine with pseudouridine (Ψ) [24]. The experimental workflow involved:
- In Vitro Transcription: Synthesis of mRNA using modified nucleoside triphosphates (e.g., pseudouridine-5'-triphosphate).
- Cell Transfection: Delivery of these base-modified mRNA variants to dendritic cells, which are key sentinels of the immune system.
- Immune Response Assessment: Measurement of inflammatory cytokine release and interferon signaling to quantify the immune activation.
- Protein Expression Analysis: Evaluation of the translational capacity of the modified mRNA by detecting the encoded protein.
Seminal Results and Impact
Their results, published in the landmark 2005 paper, were striking. The inflammatory response was almost abolished when base modifications like pseudouridine were incorporated into the mRNA [24]. This represented a paradigm shift in understanding how cells recognize and respond to different forms of RNA. Subsequent work in 2008 and 2010 demonstrated that base-modified mRNA not only evaded immune detection but also exhibited markedly increased protein production due to reduced activation of enzymes like PKR that suppress translation [24].
Table 1: Key Nucleoside Modifications and Their Effects
| Nucleoside Modification | Replaces | Key Immunogenic Effects | Impact on Translation |
|---|---|---|---|
| Pseudouridine (Ψ) | Uridine | Reduces activation of TLR7, TLR8, PKR, and RIG-I [27] [28] | Increases protein production [24] |
| N1-methylpseudouridine (m1Ψ) | Uridine | Superior reduction of immunogenicity; used in COVID-19 vaccines [27] [28] | Further enhances translational capacity [28] |
| 5-methylcytidine (m5C) | Cytidine | Attenuates innate immune sensing [29] | Improves mRNA stability and expression |
| 5-methyluridine (m5U) | Uridine | Reduces recognition by immune receptors [28] | Can improve stability and translation |
This breakthrough provided the foundational technology to finally realize the potential of mRNA therapeutics, paving the way for vaccines and new approaches to cell reprogramming.
Application in Cell Fate Reprogramming
The ability to safely and efficiently express specific proteins in cells is the cornerstone of directed cell fate change. The nucleoside modification technology enabled a new, superior method for generating induced pluripotent stem (iPS) cells.
Overcoming Limitations of Traditional Reprogramming
The original method for creating iPS cells, pioneered by Shinya Yamanaka, involved introducing four transcription factors (OCT4, SOX2, KLF4, c-MYC) using integrating retroviral vectors [2] [30]. This approach carries the risk of insertional mutagenesis and potential tumorigenesis, as the oncogenes c-Myc and Klf4 are integrated into the genome [2] [29]. While subsequent non-integrating methods were developed, they often suffered from very low reprogramming efficiencies [29].
mRNA-Based Reprogramming Protocol
The modified mRNA platform offers a safe and highly efficient alternative. A seminal study demonstrated that repeated transfections of modified mRNA encoding the reprogramming factors could reprogram human somatic cells to pluripotency [29]. The key procedural steps are outlined below.
The critical technical considerations for this protocol include:
- Modified Nucleosides: The mRNA is synthesized to incorporate pseudouridine (Ψ) and 5-methylcytidine (5mC), which dramatically reduce cytotoxicity and interferon responses, allowing for sustained daily transfections [29].
- Interferon Suppression: The culture medium is supplemented with the B18R protein, a decoy receptor for type I interferons, to further dampen any residual innate immune signaling [29].
- Transfection Regimen: A daily, repeated transfection schedule is required to maintain high levels of protein expression because the mRNA and its protein products are transiently expressed [29].
Outcomes and Advantages
This mRNA-based strategy results in reprogramming efficiencies and kinetics substantially superior to established viral protocols [29]. The resulting RNA-induced pluripotent stem (RiPS) cells are genomically pristine, free of vector integration, and can be further differentiated into desired cell lineages using the same modified mRNA technology to express differentiation factors [29]. This creates a completely non-integrating, highly controllable pipeline for regenerative medicine research.
Implementing nucleoside-modified mRNA technology requires a suite of key reagents, each with a specific function crucial for success.
Table 2: Key Research Reagent Solutions for mRNA-Based Reprogramming
| Reagent / Material | Function | Technical Notes |
|---|---|---|
| Modified Nucleoside Triphosphates (e.g., N1-methylpseudouridine-5'-TP) | Incorporation into IVT mRNA to evade innate immune recognition and enhance translation [24] [28]. | Total replacement of uridine is standard. Co-modification with 5-methylcytidine can offer further benefits [29]. |
| In Vitro Transcription Kit | Enzymatic synthesis of mRNA from a linear DNA template using T7, T3, or SP6 RNA polymerase [28]. | Must be compatible with modified NTPs. Critical to minimize double-stranded RNA (dsRNA) contaminants [26]. |
| 5' Capping Reagent | Adds a 5' cap structure (e.g., Cap 1) to mRNA, essential for translation initiation and reducing immune sensing by RIG-I [2] [26]. | Co-transcriptional capping (e.g., CleanCap) or enzymatic post-transcription capping (e.g., VCE) can be used. |
| Lipid Nanoparticles (LNPs) | Formulate mRNA for efficient cellular delivery; protect mRNA from degradation; facilitate endosomal escape [27] [2]. | Ionizable lipids are key for adjuvant activity and endosomal disruption. Tissue-specific LNP formulations are an area of active research. |
| B18R Recombinant Protein | Inhibits type I interferons by acting as a decoy receptor, mitigating residual interferon-driven cytotoxicity during repeated transfections [29]. | Particularly important in sensitive primary cell types. |
| Cationic Transfection Reagent | Complexes with negatively charged mRNA to facilitate cellular uptake via endocytosis for in vitro applications. | Suitable for repeated transfections with low baseline cytotoxicity. |
Karikó and Weissman's discovery of nucleoside base modifications was a transformative event in biomedical science. By solving the problem of mRNA immunogenicity, they unlocked a versatile and powerful platform that extends far beyond vaccines. In the field of cell fate reprogramming, this technology provides researchers with an unprecedentedly safe, efficient, and precise tool to manipulate cellular identity for basic research, disease modeling, and the development of future regenerative therapies. As innovations in mRNA design, delivery, and application continue to emerge, this platform is poised to remain at the forefront of biological research and therapeutic development.
The advent of mRNA-based technology has revolutionized the field of cell fate reprogramming, offering two primary pathways for engineering cellular identity: induced pluripotency, which reverts somatic cells to a pluripotent state, and direct lineage conversion, which transdifferentiates one somatic cell type directly into another. This whitepaper provides a technical guide to these endpoints, detailing the underlying mechanisms, experimental protocols, and key advantages of mRNA reprogramming. Framed within the broader context of mRNA technology for cell fate engineering, we highlight how synthetic mRNA, through its precision, safety, and transience, enables efficient reprogramming without genomic integration. For researchers and drug development professionals, this document synthesizes current methodologies, quantitative data, and essential reagent toolkits to advance therapeutic applications in regenerative medicine and disease modeling.
Messenger RNA (mRNA) technology has emerged as a transformative platform for programming and reprogramming human cell fate. Its application in reprogramming leverages the cell's own translational machinery to transiently express proteins that dictate cellular identity, a process that is both non-integrating and highly controllable [29] [31]. In the context of a broader thesis on mRNA-based cell fate research, this technology represents a pivotal tool for manipulating cellular function for basic research, disease modeling, and regenerative medicine [32].
Cellular reprogramming approaches can be broadly categorized. Reprogramming to induced pluripotency reverses mature, specialized cells to a pluripotent state, regaining the potential to differentiate into any cell type. In contrast, direct reprogramming or transdifferentiation converts one somatic cell type directly into another without passing through an intermediate pluripotent stage [33]. mRNA technology has proven applicable to both strategies, enabling efficient derivation of RNA-induced pluripotent stem (RiPS) cells and direct differentiation into terminally differentiated cells, such as myogenic cells [29].
The fundamental advantage of mRNA technology lies in its safety profile and efficiency. Unlike DNA-integrative methods (e.g., retroviruses, lentiviruses), mRNA does not pose a risk of insertional mutagenesis [31]. Furthermore, the use of modified ribonucleosides (e.g., pseudouridine, 5-methylcytidine) dramatically reduces the immunogenicity of synthetic mRNA by evading innate antiviral defenses, allowing for sustained protein expression through repeated transfections and resulting in reprogramming efficiencies that surpass established viral protocols [29] [34].
Comparative Analysis: Induced Pluripotency vs. Direct Lineage Conversion
The choice between reprogramming to pluripotency or directly to another lineage depends on the application's specific requirements for developmental potential, safety, and time. The table below summarizes the core characteristics of these two endpoints.
Table 1: Defining the Endpoints of mRNA Reprogramming
| Feature | Induced Pluripotency | Direct Lineage Conversion |
|---|---|---|
| Definition | Reprogramming of differentiated cells to a pluripotent state, capable of generating all embryonic lineages [33] [31]. | Direct conversion of one somatic cell type into another without an intermediate pluripotent state [33]. |
| Also Known As | Reprogramming | Transdifferentiation, Direct Reprogramming [33] [31] |
| Key mRNA Cargo | Pluripotency transcription factors (e.g., OCT4, SOX2, KLF4, c-MYC, NANOG, LIN28) [29] [31]. | Lineage-specific transcription factors and/or miRNAs (e.g., MyoD for myogenesis) [29] [31]. |
| Developmental Path | Multistep process erasing somatic memory and establishing pluripotency [33]. | Often a shortcut, though may involve a plastic intermediate progenitor state [31]. |
| Key Advantage | Unlimited self-renewal and differentiation potential; ideal for generating diverse cell types from a single source. | Avoids the theoretical risk of teratoma formation from residual pluripotent cells; can be faster. |
| Primary Challenge | Requires a subsequent, often complex, differentiation step to obtain target somatic cells; risk of incomplete differentiation. | May yield immature cells that lack full functional maturity of the target tissue; efficiency can be variable [33]. |
| Therapeutic Application | Disease modeling, drug screening, generation of autologous cells for regenerative therapies. | In situ regeneration, direct cell replacement therapies, rapid generation of specific cell types. |
Core mRNA Technology and Experimental Design
The efficacy of mRNA reprogramming hinges on sophisticated molecular design and delivery strategies to ensure high protein expression while minimizing cellular toxicity.
mRNA Construct Design and Synthesis
The synthetic mRNA used for reprogramming is engineered to mimic mature eukaryotic mRNA, incorporating specific structural elements to enhance stability, translation, and safety [34].
- 5' Cap: A modified guanine nucleotide (7-methylguanosine) added to the 5' end is essential for ribosome recognition and protects the mRNA from exonuclease degradation [29] [34] [35].
- 5' and 3' Untranslated Regions (UTRs): These flanking regions impact translation efficiency, localization, and stability. They can be adapted (e.g., using alpha-globin 3' UTR) to improve protein expression [29] [34].
- Coding Sequence (CDS): This open reading frame contains the gene of interest. Codon optimization and nucleotide modification are critical. The incorporation of modified nucleosides like pseudouridine (ψ) and 5-methylcytidine (5mC) is a key innovation that reduces immunogenicity by preventing activation of pattern recognition receptors such as TLRs, RIG-I, and PKR [29] [34].
- Poly(A) Tail: A 3' tail of adenosine residues is crucial for mRNA stability and translational efficiency. Its length can be optimized for performance [29] [34].
The manufacturing process involves in vitro transcription (IVT) from a linearized DNA template, followed by capping, tailing, and purification to remove contaminants like double-stranded RNA (dsRNA), a potent inducer of interferon responses [29] [34].
Workflow for mRNA Reprogramming
The following diagram illustrates the generalized workflow for conducting mRNA reprogramming experiments, from mRNA preparation to the final fate endpoints.
Overcoming Innate Immune Recognition
A central challenge in mRNA reprogramming is the innate immune system's robust response to exogenous RNA. The successful strategy involves:
- Nucleoside Modification: Using pseudouridine and 5-methylcytidine to make the mRNA "invisible" to immune sensors [29].
- Phosphatase Treatment: Removing 5' triphosphates from IVT RNA to avoid RIG-I activation [29].
- Interferon Inhibition: Supplementing culture media with a recombinant interferon inhibitor (e.g., B18R protein) to block the positive-feedback loop of interferon signaling [29].
These modifications are critical for enabling the repeated transfections needed over days or weeks to sustain factor expression and achieve successful reprogramming without significant cytotoxicity [29].
Quantitative Data and Protocol Efficiency
The performance of mRNA reprogramming is quantified by its efficiency, kinetics, and the functional quality of the resulting cells.
Table 2: Quantitative Metrics of mRNA Reprogramming Protocols
| Reprogramming Endpoint | Reprogramming Factors | Reported Efficiency | Time to Emergence | Key Functional Validation |
|---|---|---|---|---|
| Induced Pluripotency (RiPS) | KLF4, c-MYC, OCT4, SOX2 (KMOS) [29] | Efficiencies that "greatly surpass" established viral protocols (0.01-0.02%) [29] [31] | Can be completed in as little as one week [36] | Pluripotency marker expression (NANOG), teratoma formation, in vitro differentiation into all three germ layers [29] |
| Direct Myogenic Conversion | Not specified, but demonstrated as feasible using mRNA technology [29] | Described as "efficient" terminal differentiation [29] | Not explicitly quantified | Expression of terminal differentiation markers, formation of contractile myotubes, functional response to stimuli [29] [33] |
The Scientist's Toolkit: Essential Reagents for mRNA Reprogramming
Successful implementation of mRNA reprogramming protocols requires a suite of specialized reagents and tools.
Table 3: Key Research Reagent Solutions for mRNA Reprogramming
| Reagent / Tool | Function / Explanation | Example/Note |
|---|---|---|
| Modified Nucleosides | Reduces immunogenicity of synthetic mRNA by evading innate immune sensors. | Pseudouridine (ψ), 1-methylpseudouridine, 5-methylcytidine (5mC) [29] [34] |
| In Vitro Transcription (IVT) Kit | Synthesizes mRNA from a linearized DNA template. | Must support co-transcriptional capping and incorporation of modified NTPs. Scalability from research to GMP is key [34]. |
| Lipid Nanoparticles (LNPs) | Delivery system that protects mRNA and facilitates cellular uptake via endocytosis. | Biodegradable, lower toxicity than viral vectors; essential for efficient in vivo delivery [37] [34]. |
| Interferon Inhibitor | Suppresses the innate immune response to repeated mRNA transfections, improving cell viability. | Recombinant B18R protein (a vaccinia virus decoy receptor for Type I interferons) [29] |
| cGMP-Grade Reprogramming mRNA | Clinically relevant, quality-controlled mRNA for therapeutic development. | Commercially available services offer end-to-end cGMP reprogramming and iPSC banking with a validated quality management system [36]. |
Signaling Pathways in Cell Fate Conversion
Cell fate decisions, whether toward pluripotency or a specific lineage, are governed by complex intracellular signaling networks. The following diagram maps the core signaling pathways and regulatory logic involved in these processes.
mRNA technology provides a powerful, versatile, and clinically relevant platform for programming human cell fate. The choice between induced pluripotency and direct lineage conversion as an endpoint depends on the specific research or therapeutic goal, balancing factors such as developmental potential, time, and safety. The high efficiency and non-integrating nature of mRNA reprogramming, achieved through modified nucleosides and sophisticated delivery systems, position it as a cornerstone technology for the future of regenerative medicine, disease modeling, and drug development. As single-cell technologies and computational models advance, they will further refine our ability to guide these reprogramming processes with unprecedented precision, enabling the generation of functionally mature cell types for a new era of cellular therapies.
From Synthesis to Delivery: Methodologies and Clinical Applications of mRNA Reprogramming
The choice of mRNA capping strategy is a critical determinant of success in cell fate reprogramming research, directly influencing translational yield, transcript stability, and immunogenic profile. This technical guide provides an in-depth comparison between two predominant capping methodologies: co-transcriptional capping using CleanCap technology and post-transcriptional capping employing the Vaccinia Capping Enzyme (VCE). For researchers developing mRNA-based reprogramming factors, the selection between these methods balances trade-offs in capping efficiency, structural fidelity, process simplicity, and scalability. Quantitative data and protocol details herein are synthesized to inform experimental design, ensuring the production of high-quality mRNA that maintains precise control over protein expression—a foundational requirement for deterministic cell fate manipulation.
Table 1: Key Characteristics of Co-transcriptional and Post-Transcriptional Capping Methods
| Feature | Co-transcriptional Capping (CleanCap) | Post-Transcriptional Capping (VCE) |
|---|---|---|
| Primary Mechanism | Cap analog incorporated during IVT by RNA polymerase [39] [40] | Enzymatic addition of cap to purified, uncapped mRNA post-IVT [39] [41] |
| Typical Cap Structure | Cap 1 (m7GpppNm) [42] [40] | Cap 0 (m7GpppN), requires a separate methyltransferase (e.g., 2'-O-MTase) for Cap 1 [39] [43] |
| Reported Capping Efficiency | >95% [42] [40] | High, but dependent on enzyme purity and reaction optimization [41] [43] |
| Workflow Complexity | Simplified, "one-pot" reaction; fewer steps [39] [42] | Multi-step process requiring additional enzymatic reaction and purification [39] [41] |
| Key Advantage | High efficiency and simplicity; streamlined workflow [42] [40] | Control over cap structure; all caps are incorporated in the correct orientation [39] [43] |
| Key Disadvantage | Cost of cap analog; patent/licensing considerations [40] | Longer process time; requires additional reagents and purification steps [39] [41] |
| Ideal Application Context | High-throughput production of mRNAs where Cap 1 structure and high yield are priorities [42] | Applications requiring specific, non-standard cap structures or stringent control over capping [39] |
In the realm of mRNA-based cell fate reprogramming, the 5' cap is not merely a protective modification but a master regulator of transcript function and cellular perception. The cap structure, a 7-methylguanosine (m7G) linked to the first nucleotide via a 5'-5' triphosphate bridge (m7GpppN), is essential for mRNA stability, efficient translation initiation, and nuclear export [39] [41]. For cell reprogramming applications, where the precise and robust expression of transcription factors like Oct4, Sox2, Klf4, and c-Myc is paramount, an inadequate cap leads to poor protein yield and failed experiments.
The cap structure evolves from Cap 0 (m7GpppN) to Cap 1 (m7GpppNm) through 2'-O-methylation of the first transcribed nucleotide. This Cap 1 structure is particularly crucial for immune evasion. It enables the synthetic mRNA to be recognized as "self" by the cell, thereby avoiding potent innate immune sensors such as RIG-I and MDA5 [39] [41]. Unwanted immune activation can derail reprogramming by altering the cellular transcriptome, inducing apoptosis, or provoking an inflammatory state that is incompatible with precise lineage conversion. Consequently, achieving a near-homogeneous Cap 1 structure is a non-negotiable objective for reliable reprogramming protocols.
Technical Deep Dive: Capping Methodologies
Co-transcriptional Capping with CleanCap Technology
Mechanism and Workflow
Co-transcriptional capping introduces a cap analog directly into the in vitro transcription (IVT) reaction mixture. The RNA polymerase incorporates this analog at the initiation of transcription, capping the mRNA as it is synthesized [39] [40]. CleanCap technology represents a third-generation advance in cap analogs. It is a trinucleotide analog (e.g., m7GpppA2'pG) that is recognized by the polymerase and results in the direct synthesis of a natural Cap 1 structure in a single step [42] [40]. This process is highly efficient because the analog is designed to be the preferred initiator of transcription.
Performance and Protocol Considerations
- Efficiency: The primary advantage of CleanCap is its exceptional capping efficiency, consistently reported at >95% Cap 1 structure formation [42] [40]. This high efficiency minimizes the population of uncapped, immunostimulatory transcripts.
- Yield and Simplicity: The protocol is streamlined, eliminating post-transcription enzymatic steps and subsequent purifications. This "one-pot" reaction saves time and reduces the risk of mRNA degradation or loss during handling [39]. Yields can be very high, with protocols reporting up to 5 mg/mL of IVT mRNA [42].
- Template Design: A critical requirement for using CleanCap AG reagent is a DNA template with a transcription start site sequence of AGG [40]. This sequence complements the trinucleotide analog to ensure efficient incorporation.
Post-Transcriptional Capping with Vaccinia Capping Enzyme (VCE)
Mechanism and Workflow
Post-transcriptional capping is a two-step process. First, standard IVT is performed to produce uncapped mRNA. Second, the purified mRNA is used as a substrate for a multi-enzyme reaction. The Vaccinia Capping Enzyme (VCE), a viral enzyme widely adopted for in vitro use, possesses both RNA triphosphatase and guanylyltransferase activities. It catalyzes the cleavage of the 5' γ-phosphate and the addition of a GMP moiety to form the Cap 0 structure. To achieve the critical Cap 1 structure, a second enzyme, mRNA Cap 2'-O-Methyltransferase (2'-O-MTase), must be added to the reaction to methylate the 2'-O position of the first nucleotide [39] [43].
Performance and Protocol Considerations
- Control and Fidelity: A key strength of the enzymatic method is that it guarantees the cap is added in the biologically correct orientation, as the enzyme is specific for its RNA substrate [39] [43]. This provides robust control over the final cap structure.
- Workflow and Time: The requirement for a separate enzymatic reaction and at least one additional purification step post-IVT makes this a more time-consuming and labor-intensive process compared to co-transcriptional capping [39] [41].
- Enzyme Considerations: The reaction requires multiple enzymes and co-factors (GTP, S-adenosylmethionine (SAM)). While VCE is common, the Faustovirus Capping Enzyme (FCE) is a noted alternative with broader temperature tolerance and higher reported activity on some substrates [39] [43].
Quantitative Comparison and Method Selection
Table 2: Direct Comparison of Capping Methods for Reprogramming Applications
| Performance Metric | CleanCap | VCE + 2'-O-MTase |
|---|---|---|
| Capping Efficiency (Cap 1) | >95% [42] [40] | High, but variable; requires optimization [41] |
| Typical IVT mRNA Yield | Very High (e.g., 5 mg/ml) [42] | High, but can be reduced by purification losses [39] |
| Immune Evasion Profile | Excellent (Low immunogenicity due to high Cap 1 purity) [42] | Excellent, if 2'-O-methylation is complete [39] |
| Process Scalability | Highly scalable; simplified workflow is amenable to automation [42] | Scalable, but complex multi-step process can be a bottleneck [39] |
| Development Time | Shorter (Single-step reaction) | Longer (Multiple steps and purifications) |
| Relative Cost | Higher reagent cost per reaction | Lower reagent cost, but higher labor/process cost |
Selection Guide for Reprogramming Workflows
The decision between CleanCap and VCE capping should be guided by the specific goals and constraints of the research program.
For most reprogramming applications, CleanCap is the recommended choice. The primary rationale is its superior efficiency and simplicity. The guarantee of >95% Cap 1 formation directly translates to a more potent and consistent reprogramming mRNA, minimizing batch-to-batch variability and the confounding effects of the innate immune response. The streamlined workflow allows researchers to focus on the biological assessment of their mRNA constructs rather than complex production processes [42] [40].
Select VCE-based enzymatic capping in the following scenarios:
- Template Incompatibility: When the DNA template or experimental design necessitates a transcription start site that is incompatible with CleanCap trinucleotide analogs.
- Requirement for Exotic Cap Analogs: When research demands the use of specialized, non-standard cap structures (e.g., for pull-down assays or specific mechanistic studies) that cannot be incorporated co-transcriptionally [39].
- Established GMP Processes: For translational work where existing, validated manufacturing processes are built around enzymatic capping methods [43].
The Scientist's Toolkit: Essential Reagents and Protocols
Research Reagent Solutions
Table 3: Key Reagents for mRNA Capping Workflows
| Reagent / Kit | Function | Example Provider(s) |
|---|---|---|
| CleanCap Reagent AG | Trinucleotide cap analog for co-transcriptional synthesis of Cap 1 mRNA. | TriLink BioTechnologies [42], Takara Bio [40], NEB [43] |
| Vaccinia Capping Enzyme (VCE) | Adds Cap 0 structure to the 5' end of uncapped RNA via its triphosphatase and guanylyltransferase activities. | Takara Bio [40], New England Biolabs (NEB) [43] |
| mRNA Cap 2'-O-Methyltransferase | Converts Cap 0 to Cap 1 by methylating the 2'-O position of the first transcribed nucleotide. | Takara Bio [40], NEB [43] |
| HiScribe T7 mRNA Kit with CleanCap | All-in-one kit for IVT including T7 RNA polymerase, NTPs, buffer, and CleanCap reagent for simplified Cap 1 mRNA production. | NEB [43] |
| Takara IVTpro mRNA Synthesis System | A system for high-yield mRNA synthesis, compatible with both CleanCap and ARCA capping methods. | Takara Bio [40] |
| Faustovirus Capping Enzyme (FCE) | An alternative capping enzyme with high activity and a broader temperature range than VCE. | NEB [43] |
Detailed Experimental Protocols
Protocol for Co-transcriptional Capping with CleanCap
This protocol is adapted from commercial kit instructions and published methodologies [42] [40] [43].
- Template Design: Ensure your DNA template (linearized plasmid or PCR product) has a T7 promoter and a transcription start site of AGG.
- IVT Reaction Assembly: Combine the following components in a nuclease-free tube:
- 1 µg of DNA template
- 10 µL 2X IVT buffer (supplied with kit, typically contains rNTPs, DTT)
- 2 µL T7 RNA Polymerase mix
- 2 µL CleanCap Reagent AG (e.g., TriLink)
- Nuclease-free water to 20 µL total volume
- Incubation: Mix gently and incubate at 37°C for 2 hours.
- DNase I Treatment: Add 2 µL of DNase I (RNase-free) and incubate for 15 minutes at 37°C to digest the DNA template.
- mRNA Purification: Purify the mRNA using a LiCl precipitation method or a silica membrane-based purification kit. Validate yield and purity by spectrophotometry (NanoDrop) and integrity by agarose gel electrophoresis.
Protocol for Post-Transcriptional Capping with VCE
This protocol utilizes enzymes from suppliers like NEB and Takara Bio [40] [43].
- Produce Uncapped mRNA: Perform a standard IVT reaction without any cap analog. Purify the resulting uncapped mRNA.
- Capping Reaction Assembly: Combine the following:
- 10 µg of purified, uncapped mRNA
- 5 µL 10X Capping Buffer (supplied with enzyme)
- 5 µL 2 mM GTP (for VCE)
- 5 µL 2 mM S-adenosylmethionine (SAM) - (for methyltransferase activity)
- 5 µL Vaccinia Capping Enzyme (VCE)
- 2.5 µL mRNA Cap 2'-O-Methyltransferase (for Cap 1 formation)
- Nuclease-free water to 50 µL total volume
- Incubation: Mix gently and incubate at 37°C for 1 hour.
- Purification: Purify the capped mRNA using a standard method (e.g., LiCl precipitation or column purification) to remove enzymes and excess reagents.
Quality Control and Capping Efficiency Analysis
Rigorous QC is essential. Beyond standard yield and purity checks, analyzing capping efficiency is critical.
- LC-MS/MS: The gold-standard method for direct identification and quantification of Cap 0, Cap 1, and Cap 2 structures [39].
- Cap-Specific Assays: Techniques like reverse transcription-qPCR (RT-qPCR) can be used, as capped 5' ends are necessary for efficient reverse transcription priming [39]. Cap-specific antibodies in an ELISA format can also quantify cap structures [39].
In the rapidly advancing field of mRNA-based cell fate reprogramming, the reliability of the underlying technology is a prerequisite for biological discovery. The capping method forms a cornerstone of this reliability. While both CleanCap and VCE-based methods are capable of producing high-quality, functional mRNA for reprogramming factors, the co-transcriptional CleanCap method offers a compelling combination of superior efficiency, a simplified workflow, and exceptional performance that aligns with the needs of most research programs.
Future developments will likely focus on further enhancing capping efficiency and creating novel cap analogs with additional functionalities, such as improved translational kinetics or even greater immune silencing properties. As the field moves toward clinical translation of reprogramming therapies, the scalability and consistency afforded by technologies like CleanCap will become increasingly paramount. By selecting the appropriate capping strategy detailed in this guide, researchers can ensure their foundational mRNA tools are robust, enabling them to focus on the complex and rewarding challenge of orchestrating cell identity.
The field of mRNA-based technology for cell fate reprogramming research is fundamentally reliant on the quality and purity of the administered messenger RNA. The 5′ cap structure is not merely a decorative molecular addition but a critical functional component that spatially protects mRNA from exonuclease degradation and significantly enhances the initiation of translation reactions [44]. In the specific context of cell fate reprogramming, where the transient expression of transcription factors must be precisely controlled to direct cellular identity, the presence of a fully functional cap structure becomes paramount. Uncapped mRNA byproducts, which typically feature a 5′ triphosphate (ppp), are not just translationally inactive but can trigger innate immune responses via receptors such as RIG-I and MDA-5, potentially derailing delicate reprogramming processes and altering the resulting cell phenotype [45]. The standard capping methods used historically, including co-transcriptional capping with analogs like ARCA and enzymatic capping, have provided a solid foundation but face a significant limitation: maximum capping efficiencies of 80-90%, leaving a substantial fraction of immunogenic uncapped mRNA in the final product [46] [45]. This technical note explores the latest technological advancements, specifically the PureCap platform and other next-generation solutions, which are poised to overcome these limitations and provide researchers with the ultra-pure, fully capped mRNA required for precise and reliable cell fate reprogramming.
The Evolution and Limitations of Conventional mRNA Capping Methods
The journey to produce high-quality capped mRNA has traditionally navigated two primary pathways: co-transcriptional capping and enzymatic capping. Each method has distinct mechanisms, advantages, and inherent limitations that impact their utility in sensitive applications like reprogramming.
Co-transcriptional Capping with Cap Analogs
This method involves incorporating synthetic cap analogs directly into the in vitro transcription (IVT) reaction. The evolution of these analogs reflects a continuous effort to improve efficiency and fidelity.
- First-Generation Analog (mCap/m7GpppG): This Cap-0 analog suffers from bidirectional incorporation, resulting in approximately 50% of mRNAs being capped in the reverse orientation and thus untranslatable. It typically requires a high cap-to-GTP ratio (around 4:1), which reduces full-length mRNA yield and achieves capping efficiencies of only about 70% [46].
- Second-Generation Analog (ARCA): The Anti-Reverse Cap Analog was designed with a 3′-O-methyl group on the m7Guo moiety, ensuring incorporation exclusively in the correct orientation. This guarantees that all capped mRNAs are translatable, but the method still requires a high cap-to-GTP ratio, suppressing overall mRNA yield [46].
- Third-Generation Analog (CleanCap): This trinucleotide Cap-1 analog represents a significant step forward. It operates efficiently at a standard 1:1 ratio with GTP, enabling high yields of mRNA (>5 mg/mL) with dramatically improved capping efficiencies of over 95% [46]. A key consideration is its requirement for an AG initiation sequence in the DNA template instead of the more common GG.
Enzymatic Capping
As an alternative or complementary approach, enzymatic capping involves a post-transcription reaction where mRNA is treated with a capping enzyme (e.g., guanylyltransferase) and methyltransferase. This process can achieve near-100% capping efficiency with the cap added in the correct direction [44]. A recent comparative study screened seven viral-derived capping enzymes and identified the bluetongue virus capping enzyme (VP4) as a superior candidate, yielding a 38% higher transfection efficiency in human cells compared to the traditionally used vaccinia virus capping enzyme (VCE) [44]. Despite its high efficiency, a drawback of this method is the need for additional purification steps post-capping, which can increase production time and cost and risk mRNA degradation.
Table 1: Comparison of Conventional and Next-Generation mRNA Capping Methods
| Capping Method | Mechanism | Reported Capping Efficiency | Key Advantages | Key Limitations |
|---|---|---|---|---|
| mCap / m7GpppG | Co-transcriptional | ~70% [46] | Simple, one-step process | ~50% reverse incorporation; low yield |
| ARCA | Co-transcriptional | ~80-90% [45] | Unidirectional capping; all capped mRNA is translatable | High cap:GTP ratio lowers yield |
| CleanCap | Co-transcriptional | >95% [46] | High yield and efficiency; Cap 1 structure | Requires AG initiation sequence |
| Enzymatic Capping | Post-transcriptional | ~100% [44] [45] | Correct orientation; high efficiency | Multiple steps and purifications; higher cost |
| PureCap Technology | Co-transcriptional + Purification | 100% [45] | 100% efficiency; simultaneous removal of dsRNA impurities | Requires specialized cap analog and RP-HPLC |
A fundamental challenge common to all co-transcriptional methods is the competition between the cap analog and GTP for the initiation site of the RNA polymerase. This competition makes it theoretically impossible to achieve a 100% capping efficiency, consistently leaving a population of uncapped, immunogenic ppp-RNA in the final product [45]. These impurities are problematic for any therapeutic application but are particularly detrimental to cell reprogramming, where unintended immune activation can alter cell states and compromise experimental reproducibility.
PureCap Technology: A Paradigm Shift in mRNA Purification
The PureCap technology, developed and detailed in a 2023 Nature Communications publication, introduces a revolutionary approach that moves beyond merely improving capping efficiency to enabling the physical separation and purification of fully capped mRNA from its uncapped counterparts [45].
Core Principle and Workflow
The technology is built on a novel class of cap analogs, the PureCap analogs, which are modified with a hydrophobic, photocleavable tag. This tag, which includes a tert-butyl group within a 2-nitrobenzyl (Nb) photocaging molecule, dramatically alters the physicochemical properties of the capped mRNA during the IVT reaction [45].
The following diagram illustrates the streamlined workflow from synthesis to pure, functional mRNA.
The process involves several key stages, each designed to leverage the unique properties of the PureCap analog. First, the hydrophobic PureCap analog is incorporated into the growing mRNA strand during a standard IVT reaction. This results in a mixture of mRNA molecules, some containing the hydrophobic tag and others (the uncapped byproducts) that do not. The revolutionary step follows: the reaction mixture is subjected to Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC). Due to its added hydrophobicity, the capped mRNA exhibits a longer retention time than the uncapped mRNA, allowing for their clean physical separation. Finally, the collected, pure capped mRNA is exposed to UV light, which cleaves the photocleavable tag. This final step leaves behind a "footprint-free" native cap structure (Cap-0, Cap-1, or even Cap-2), ready for use in downstream applications [45].
Key Experimental Findings and Validation
The development and validation of the PureCap technology were backed by rigorous experimentation. A key achievement was the demonstration of 100% capping efficiency across different cap types (Cap-0, Cap-1, Cap-2) and mRNA lengths, ranging from a short 650 nt transcript to a long 4,247 nt mRNA [45]. This level of purity was previously unattainable with any standard method.
Furthermore, the technology enables the production of advanced cap structures with high purity. Notably, Cap-2-type mRNA produced via the PureCap method showed a 3- to 4-fold increase in translation activity in both cultured cells and animal models compared to Cap-1-type mRNA prepared by standard capping methods [45]. This is attributed to the Cap-2 structure's superior ability to reduce recognition by innate immune receptors like RIG-I, thereby minimizing immune activation and enhancing protein production. The RP-HPLC purification step also concurrently removes double-stranded RNA (dsRNA) impurities, a common byproduct of IVT that also triggers innate immunity and inhibits translation, thereby further enhancing the quality and performance of the final mRNA product [45].
The Scientist's Toolkit: Essential Reagents for Next-Generation mRNA Production
Implementing advanced mRNA capping and purification strategies requires a specific set of reagents and tools. The following table outlines key solutions for researchers building a next-generation mRNA workflow.
Table 2: Research Reagent Solutions for High-Purity mRNA Production
| Reagent / Tool | Function / Description | Key Utility in Reprogramming Research |
|---|---|---|
| PureCap Analogs [45] | Hydrophobic, photocleavable cap analogs for RP-HPLC purification of capped mRNA. | Ensures a defined, homogenous population of reprogramming factor mRNA with minimal immunogenic byproducts. |
| CleanCap Reagent AG [46] | A trinucleotide co-transcriptional capping analog for high-yield production of Cap 1 mRNA. | Rapid, high-yield production of high-quality mRNA for screening multiple transcription factor combinations. |
| Bluetongue Virus Capping Enzyme (VP4) [44] | A highly efficient viral capping enzyme identified for superior transfection efficiency. | Ideal for enzymatic capping of long or complex mRNA sequences used in reprogramming (e.g., transcription factors). |
| RP-HPLC System [45] | Chromatography system for separating biomolecules based on hydrophobicity. | Critical for the PureCap workflow to resolve capped from uncapped mRNA and remove dsRNA contaminants. |
| mMESSAGE mMACHINE T7 Kit with CleanCap [46] | A commercial kit optimized for high-yield mRNA synthesis with CleanCap technology. | A user-friendly, integrated system for reliable production of high-quality mRNA without specialized equipment. |
Integrated Workflow for Reprogramming-Grade mRNA and Future Outlook
Producing clinical-grade mRNA for sensitive applications like cell fate reprogramming requires an integrated approach that prioritizes purity and functionality. The following workflow diagram outlines a robust protocol from template design to final validation.
The future of mRNA-based cell fate reprogramming is inextricably linked to advancements in mRNA production technology. The PureCap platform represents a significant leap forward, but the innovation ecosystem is vibrant. Concurrent developments in delivery technologies, such as next-generation lipid nanoparticles (LNPs) designed for extrahepatic targeting and improved potency, are crucial for efficiently delivering reprogramming factors to specific cell types [47]. Furthermore, the integration of AI-driven sequence optimization and the exploration of novel modalities like self-amplifying mRNA and circular mRNA are poised to further enhance the efficacy, durability, and application scope of mRNA therapeutics [48]. As these technologies mature and converge, they will empower researchers to achieve unprecedented control and precision in cell fate reprogramming, accelerating the development of new regenerative medicines and personalized cell therapies.
The emergence of messenger RNA (mRNA) as a therapeutic modality has revolutionized the potential for treating a wide spectrum of diseases, from infectious diseases to cancer and genetic disorders. Within the specific context of cell fate reprogramming research—a cornerstone of regenerative medicine—mRNA technology offers a non-integrative, controllable strategy for expressing transcription factors that direct cellular behavior toward desired lineages. However, the clinical success of mRNA-based therapeutics is intrinsically dependent on a delivery vehicle that can overcome substantial biological barriers. Naked mRNA is inherently unstable, susceptible to enzymatic degradation, and shows poor cellular uptake, resulting in low protein production levels. Lipid nanoparticles (LNPs) have emerged as the preeminent solution to these challenges, providing a protective, efficient vehicle for mRNA transfection that was conclusively validated by the rapid development and deployment of mRNA-based COVID-19 vaccines.
The "delivery challenge" encompasses the entire journey of the therapeutic mRNA, from systemic administration to cytosolic release. LNPs must protect their mRNA cargo from nucleases, facilitate cellular uptake, and critically, enable escape from the endosomal compartment to release the mRNA into the cytoplasm for translation. The composition and design of modern LNPs are finely tuned to meet these challenges. This technical guide delves into the core aspects of LNP technology, examining its formulation, mechanism of action, key experimental methodologies for evaluation, and its specific applicability to advanced cell fate reprogramming research. By providing a detailed overview of the current landscape, this document aims to equip researchers and drug development professionals with the knowledge to harness LNPs effectively for next-generation mRNA therapeutics.
LNP Formulation: Composition and Design Rationale
The efficacy and safety of LNPs are dictated by their individual components, each playing a critical and synergistic role. A deep understanding of this formulation rationale is essential for designing LNPs tailored for specific applications, such as reprogramming.
Core Components and Their Functions
Modern mRNA-LNPs are typically composed of four key lipidic components, whose molar ratios are carefully optimized to achieve the desired characteristics for nucleic acid delivery [49] [50].
- Ionizable Lipids: These are the cornerstone of modern LNP technology. They are neutral at physiological pH (7.4) but become positively charged in acidic environments like the endosome (pH 5.5-6.5). This pH-dependent behavior serves a dual purpose: it minimizes toxicity and non-specific interactions in the bloodstream while facilitating endosomal escape after cellular uptake. The positive charge allows the lipid to interact with and disrupt the anionic endosomal membrane, releasing the mRNA cargo into the cytosol. Ionizable lipids typically constitute 30-50% of the total lipid mass. Examples include SM-102 (used in Moderna's Spikevax), ALC-0315 (used in Pfizer-BioNTech's Comirnaty), DLin-MC3-DMA (MC3, used in the siRNA drug Onpattro), and C12-200 [51] [52] [49].
- Phospholipids (Helper Lipids): These lipids, such as DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) or DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), provide structural integrity to the LNP bilayer, contributing to its stability and morphology. DOPE, with its cone-shaped structure, is particularly known to enhance membrane fusion and facilitate endosomal escape [52] [50]. Helper lipids generally make up about 10-15% of the formulation.
- Cholesterol: A key stabilizing agent, cholesterol is integrated into the lipid bilayer to enhance its rigidity, fluidity, and stability. It improves the encapsulation efficiency of mRNA, reduces drug leakage, and aids in the fusion of the LNP with cellular membranes. Recent studies have shown that modifying cholesterol, for instance by incorporating hydroxycholesterols or cationic cholesterol derivatives, can significantly improve mRNA delivery efficiency and even alter organ tropism [52].
- PEGylated Lipids: These lipids, positioned on the surface of the LNP, are crucial for controlling particle size, preventing nanoparticle aggregation during storage and in circulation, and reducing rapid clearance by the mononuclear phagocyte system (MPS). By forming a hydrophilic shield, PEG prolongs the circulation time of LNPs. However, the immunogenicity of PEG and the potential development of anti-PEG antibodies upon repeated dosing remain challenges. PEGylated lipids are typically used at low molar ratios (1.5-2%) [52] [53].
Table 1: Key Components of mRNA-LNPs and Their Functions
| Component | Key Function | Common Examples | Typical Molar % |
|---|---|---|---|
| Ionizable Lipid | mRNA encapsulation; endosomal escape | SM-102, ALC-0315, MC3, C12-200 | 30-50% |
| Phospholipid | Structural support; membrane integrity | DSPC, DOPE | 10-15% |
| Cholesterol | Stabilizes bilayer; enhances fusion | Cholesterol, Hchol derivatives | 30-40% |
| PEGylated Lipid | Controls size; reduces aggregation & clearance | DMG-PEG2000, ALC-0159 | 1.5-2% |
Manufacturing Methods: Achieving Reproducibility and Scale
The method of LNP formulation is a Critical Process Parameter (CPP) that directly impacts Critical Quality Attributes (CQAs) such as particle size, polydispersity index (PDI), and encapsulation efficiency [49].
- Microfluidics: This is considered the gold standard for LNP manufacturing, especially during research and development. It offers superior control over mixing conditions (e.g., total flow rate, flow rate ratio) by precisely combining an aqueous buffer containing the mRNA with an ethanolic solution of lipids in a microfluidic chip. This results in highly reproducible, homogeneous nanoparticles (PDI < 0.2) with high encapsulation efficiency (often >90%) and tunable sizes (typically 50-200 nm). Its ability to handle small volumes makes it ideal for screening numerous formulations [51] [49] [50].
- Macrofluidics/T-Junction Mixing: This scalable nanoprecipitation method, used in industrial-scale production like COVID-19 vaccine manufacturing, employs impingement jet mixers for rapid mixing. While excellent for large volumes, it requires substantial liquid volumes, limiting its utility in early-stage discovery [49].
- Other Methods: Historical methods like thin-film hydration and manual pipette mixing offer poor control over nanoparticle characteristics, low encapsulation efficiency, and poor reproducibility, making them unsuitable for robust therapeutic development [49].
The Mechanism of mRNA Transfection: From Uptake to Protein Expression
The journey of an mRNA-LNP from administration to protein production is a multi-step process, each stage presenting its own set of challenges that the LNP must overcome.
- Systemic Administration and Biodistribution: Upon intravenous injection, LNPs must evade the mononuclear phagocyte system and renal clearance. A significant challenge is the natural tropism of many standard LNPs for the liver, where up to 80-90% can accumulate, leaving little for extrahepatic targets. This is a major hurdle for reprogramming therapies targeting other tissues like the heart, lungs, or brain. Strategies to overcome this include incorporating selective organ targeting (SORT) molecules or modifying lipid chemistry to alter tropism [52] [54].
- Cellular Uptake: LNPs are typically internalized by cells via energy-dependent endocytic pathways, such as clathrin-mediated endocytosis or macropinocytosis. The specific pathway can depend on the LNP's properties (size, surface charge) and the cell type [51].
- Endosomal Escape: This is the most critical bottleneck for efficient transfection. After uptake, the LNP is trapped in an endosome, which matures and acidifies. In this acidic environment (pH 5.5-6.5), the ionizable lipids become protonated, gaining a positive charge. This leads to a structural destabilization of the LNP and interaction with the anionic endosomal membrane, promoting membrane disruption and the release of mRNA into the cytoplasm. Failure results in the LNP and mRNA being trafficked to lysosomes for degradation [51] [55] [53].
- Protein Expression and Application: Once in the cytosol, the released mRNA is translated by ribosomes into the encoded protein. In the context of cell fate reprogramming, this protein could be a transcription factor (e.g., for generating induced pluripotent stem cells), a cytokine, or a membrane receptor that directs the cell toward a new identity or function for regenerative applications [32].
Experimental Evaluation: Key Methodologies and Protocols
Rigorous preclinical evaluation is essential to establish the quality, efficacy, and safety of LNP formulations. This involves a series of standardized experiments.
Physicochemical Characterization
These tests define the Critical Quality Attributes (CQAs) of LNPs [51] [49].
- Particle Size and Polydispersity Index (PDI): Measured by Dynamic Light Scattering (DLS). Size influences biodistribution and cellular uptake, with 50-200 nm being typical. PDI indicates the homogeneity of the sample; a value below 0.2 is generally considered monodisperse.
- Zeta Potential: Measulated by Laser Doppler Micro-electrophoresis. It reflects the surface charge of particles in a solution. A near-neutral zeta potential at physiological pH is desirable for reduced non-specific interactions.
- Encapsulation Efficiency (EE%): Quantifies the percentage of mRNA successfully encapsulated within the LNPs. This is typically measured using a dye exclusion assay (e.g., RiboGreen). A sample is diluted, and the dye is added. The fluorescence of the free mRNA in solution is measured. Then, a detergent is added to disrupt the LNPs and release all mRNA, and the total fluorescence is measured. EE% = (1 - (Free mRNA / Total mRNA)) * 100. High EE% (>90% with microfluidics) is crucial for efficacy and minimizing immune activation.
In Vitro and In Vivo Performance Assessment
- In Vitro Transfection Efficiency: LNPs are incubated with relevant cell lines (e.g., HEK293, HeLa, THP-1, or primary cells). Protein expression is quantified 24-48 hours post-transfection using methods like luciferase assays (for luminescent reporters) or flow cytometry (for fluorescent proteins like GFP) [51].
- In Vivo Performance: This is critical as in vitro data often does not adequately predict in vivo behavior [51] [56]. Studies involve administering mRNA-LNPs to animal models (e.g., mice) and assessing:
- Protein Expression Kinetics: Measuring luciferase activity via IVIS imaging or quantifying specific proteins in tissues over time via ELISA.
- Biodistribution: Using radiolabeled or fluorescently labeled LNPs/mRNA to track their distribution in various organs.
- Functional Efficacy: For vaccines, this means measuring antigen-specific antibody titers or T-cell responses. For reprogramming, this would involve assessing markers of the target cell type or functional recovery in a disease model.
Table 2: Comparison of Common Ionizable Lipids in Model Experiments
| Ionizable Lipid | In Vitro Performance (e.g., HEK293) | In Vivo Performance (mRNA Expression) | Key Characteristics & Notes |
|---|---|---|---|
| SM-102 | Significantly higher protein expression than MC3, C12-200 [51] | High protein expression; no significant difference from ALC-0315 [51] | Used in Moderna's Spikevax; good balance of efficacy and safety. |
| ALC-0315 | Variable, lower than SM-102 in some cell lines [51] | High protein expression; strong vaccine immune responses [51] | Used in Pfizer-BioNTech's Comirnaty. |
| MC3 | Lower protein expression than SM-102 [51] | Lower expression levels than SM-102/ALC-0315 [51] | First approved ionizable lipid (Onpattro); known for established safety profile. |
| C12-200 | Lower protein expression in immortalized cells [51] | Lower expression levels [51] | Used in research; stereochemistry (C12-200-S) can improve delivery [52]. |
For researchers embarking on LNP development for mRNA delivery, a core set of tools and reagents is required.
Table 3: Research Reagent Solutions for LNP Development
| Category / Item | Specific Examples | Function / Application |
|---|---|---|
| Ionizable Lipids | SM-102, ALC-0315, DLin-MC3-DMA (MC3), C12-200 | The primary functional component for mRNA binding and endosomal escape. Available from specialty chemical suppliers (e.g., BroadPharm, Avanti Polar Lipids) [51]. |
| Structural Lipids | DSPC, DOPE, Cholesterol | Provide LNP structure and stability. Sourced from vendors like Avanti Polar Lipids and Lipoid [51] [50]. |
| PEGylated Lipids | DMG-PEG2000, ALC-0159, DMPE-PEG2000 | Control particle size, stability, and circulation time. Available from Avanti Polar Lipids [51]. |
| mRNA Cargo | EZ Cap Luciferase mRNA, Green Lantern mRNA, OVA mRNA | Model mRNAs for optimizing transfection efficiency and tracking protein expression in R&D [51]. |
| Microfluidic Instrument | NanoAssemblr Ignite, PreciGenome NanoGenerator | Enables reproducible, high-efficiency LNP formulation with precise control over particle characteristics [51] [50]. |
| Analytical Kits | RiboGreen Assay Kit, One-Glo Luciferase Assay System | For quantifying mRNA encapsulation efficiency and measuring transfection efficacy in cell-based assays [51]. |
Lipid nanoparticles have undeniably solved the primary delivery challenge for mRNA transfection, enabling a new class of therapeutics and research tools. Their success is built on a foundation of rational design, with each lipid component playing a deliberate role in navigating biological barriers to ensure the mRNA payload reaches its cytosolic destination.
For the field of cell fate reprogramming, LNPs offer a particularly powerful and transient means to express reprogramming factors without the risks of genomic integration associated with viral vectors. The future of LNPs in this context will be shaped by overcoming persistent challenges. A major focus is on extrahepatic targeting to direct reprogramming factors to specific tissues like the heart, brain, or pancreas, moving beyond the liver's dominance. This involves developing novel ionizable lipids and incorporating SORT molecules. Furthermore, managing the reactogenicity and immunogenicity of LNPs, potentially driven by both the ionizable lipids and PEG components, is crucial for therapeutic safety, especially in chronic regenerative applications requiring repeated dosing [52] [54].
As research advances, the synergy between increasingly sophisticated LNP delivery systems and the expanding toolkit of mRNA technologies—including self-replicating RNA and CRISPR-based gene editing—will unlock unprecedented precision in controlling cell fate. This progression promises to accelerate the development of personalized regenerative therapies to address complex degenerative diseases and injury-related conditions.
Tissue Nanotransfection (TNT) is a novel, non-viral nanotechnology platform that enables in vivo gene delivery and direct cellular reprogramming through localized nanoelectroporation [57]. As regenerative medicine increasingly turns toward gene-based approaches to repair or replace damaged tissues, TNT addresses substantial barriers faced by conventional gene delivery systems, particularly viral vectors, including immunogenicity, off-target effects, and limited in vivo applicability [57] [58]. This technology represents a significant advancement in the field of mRNA-based cell fate reprogramming by providing a physical delivery method that is non-integrative, highly specific, and demonstrates minimal cytotoxicity compared to biological and chemical alternatives [57] [59].
TNT exemplifies an interdisciplinary approach by integrating bioengineering, molecular biology, biotechnology, regenerative medicine, and immunology to enable in vivo cellular reprogramming and gene delivery [57] [58]. The technology leverages the advantages of mRNA transfection, which allows for direct protein translation in the cytoplasm without requiring nuclear entry, making it simpler, faster, and more efficient than DNA plasmid transfection [57]. This characteristic is particularly valuable for cell fate reprogramming applications where transient expression of reprogramming factors is often desirable to avoid permanent genetic alterations [57] [60].
TNT Device Architecture and Mechanism of Action
Structural Components and Design Evolution
The TNT device architecture has evolved through several generations to optimize delivery efficiency. The core system consists of a hollow-needle silicon chip mounted beneath a cargo reservoir containing genetic material [57] [59]. This device is placed directly on the skin or target tissue, with the cargo reservoir connected to the negative terminal of an external pulse generator, while a dermal electrode connected to the tissue serves as the positive terminal [57] [58].
- TNT 1.0: The first generation utilizes the mechanism of nanoelectroporation via nanochannels [59] [61].
- TNT 2.0: The second generation features a hollow microneedle array designed to enhance physical contact between the TNT chip and skin to accommodate nonuniform topography across the skin, thereby improving gene delivery efficiency [59] [61].
The sterilization of TNT devices is essential for biological and medical applications, with ethylene oxide gas sterilization and gamma irradiation being the most frequently applied processes as they preserve the interior architecture of the nanodevices [57] [58].
Figure 1: TNT Device Architecture and Core Components
Electroporation-Based Delivery Mechanism
TNT employs a highly localized and transient electroporation stimulus through nanochannel interfaces designed to create reversible nanopores in the plasma membrane [57] [58]. The mechanism involves several precisely controlled stages:
Electric Field Application: When electrical pulses are applied, the hollow needles concentrate the electric field at their tips [57]. The optimization of electrical pulse parameters—including voltage amplitude, pulse duration, and inter-pulse intervals—is critical for maximizing delivery efficiency while preserving cellular viability [57] [58].
Membrane Poratio: The electric field produces thermal fluctuations that rearrange the molecules in the phospholipid bilayer and form hydrophilic pores to allow molecules and ions to cross in both directions [57] [58]. These nanopores typically reseal within milliseconds or a few seconds, depending on cell type and membrane characteristics, which limits opportunities for cell damage and cytotoxicity [57].
Cargo Delivery: The temporary pores enable the targeted delivery of charged genetic material into the tissue, allowing for precise, localized, non-viral, and efficient in vivo gene delivery [57] [59].
This physical delivery mechanism offers significant advantages over biological delivery systems (such as viral vectors) that face challenges with immunotoxicity and unintended gene expression, and chemical delivery systems that struggle with low transfection efficiency in vivo due to poor cellular uptake and inefficient endosomal escape [57] [58].
Genetic Cargo for Reprogramming: Options and Considerations
TNT technology supports the delivery of various genetic cargo types, each with distinct characteristics, advantages, and considerations for reprogramming applications. The selection of cargo depends on factors such as desired expression duration, safety profile, and specific application requirements.
Table 1: Genetic Cargo Options for TNT-Mediated Reprogramming
| Cargo Type | Mechanism of Action | Key Advantages | Limitations/Considerations | Reprogramming Applications |
|---|---|---|---|---|
| Plasmid DNA | Requires nuclear entry for gene expression [57] | - Circular plasmids resistant to exonucleases [57]- Established production protocols | - Lower efficiency due to nuclear barrier [57]- Potential for genomic integration (low risk) | - Direct lineage conversion [57]- Induced pluripotency [57] |
| mRNA | Direct protein translation in cytoplasm [57] | - No nuclear entry required [57]- Faster, more efficient protein expression- Non-integrative, minimal risk of genomic alteration [57] [60] | - Transient expression requires repeated administration- Can trigger innate immune response (can be mitigated by modifications) [60] | - Protein supplementation [32]- Partial cellular rejuvenation [57]- Direct reprogramming [60] |
| CRISPR/Cas9 Components | Targeted genome or epigenome editing [57] | - Programmable, precise targeting- Catalytically inactive dCas9 enables transcriptional/ epigenetic control without DNA cutting [57] | - Delivery challenges for RNP complexes- Potential off-target effects (reduced with RNP delivery) [62] | - Gene knockout/knockin- Epigenetic remodeling [57]- Transcriptional activation/repression [57] |
For mRNA-based reprogramming applications, which are particularly relevant to cell fate control, significant advances have been made in mRNA design and delivery. Modified mRNA can overcome the natural cellular immunity to foreign RNA, which typically triggers an interferon response that shuts down cellular function [60]. The use of synthetic mRNA for reprogramming human adult skin cells into induced pluripotent stem (iPS) cells has demonstrated efficiencies of 1-4%, orders of magnitude higher than conventional iPS methods [60].
Cellular Reprogramming Strategies via TNT
TNT enables multiple reprogramming strategies by delivering specific combinations of transcription factors or gene editing components to target cells in vivo. Each approach offers distinct advantages for regenerative medicine applications.
Direct Lineage Conversion (Transdifferentiation)
Direct reprogramming, also referred to as transdifferentiation, involves the conversion of one somatic cell type into another without passage through a pluripotent state [57] [58]. This approach offers a more direct, rapid, and potentially safer strategy for cell replacement therapies and regenerative medicine without inducing uncontrolled proliferation or dedifferentiation [57]. In vivo, the overexpression of genetic factors can stimulate cell lineages to repair damaged tissue without tumorigenesis, risk of contamination, or cell transplantation [57] [58]. TNT has successfully demonstrated direct reprogramming of fibroblast cells in skin in vivo into neuronal and endothelial cells in preclinical studies to assist in the recovery of injured limbs and damaged brain tissue [59] [61].
Partial Cellular Reprogramming (Rejuvenation)
Partial reprogramming through transient activation of reprogramming factors (such as OSKM factors - Oct4, Sox2, Klf-4, c-Myc) has demonstrated the ability to reverse aging-related changes in senescent cells without altering cell identity [57] [58]. This approach resets epigenetic markers like DNA methylation clocks, reduces aging-associated transcriptional dysregulation, and restores serum metabolites to youthful levels [57]. Additionally, mitochondrial rejuvenation enhances oxidative phosphorylation and mitochondrial function in aged fibroblasts [57]. Telomerase activation and telomere lengthening are key outcomes of partial cellular reprogramming, resulting in improved genomic stability and reduced markers of cellular aging [57] [58].
Induced Pluripotency
While induced pluripotent stem cell (iPSC) reprogramming involves transforming somatic cells into a pluripotent state using transcription factors, this approach may be associated with risks of immunogenicity, tumorigenicity, and epigenetic abnormalities [57]. TNT can deliver the necessary factors for iPSC generation but with the safety advantage of a non-integrative approach [57] [60].
Figure 2: Cellular Reprogramming Strategies Enabled by TNT
Therapeutic Applications and Experimental Evidence
TNT demonstrates transformative therapeutic potential across diverse biomedical applications, with growing evidence from preclinical studies supporting its efficacy in regeneration, antimicrobial therapy, and disease treatment.
Tissue Regeneration and Repair
TNT has shown remarkable success in various tissue regeneration applications:
- Ischemic Stroke Recovery: TNT-mediated reprogramming has been applied to promote recovery in ischemic stroke models, demonstrating the technology's potential for neurological applications [59] [61].
- Wound Healing: In extreme chronic wound healing models, TNT has successfully promoted tissue repair and regeneration [59] [61]. The application of TNT chips has been extended to the area of exosomes, which are vital for intracellular communication to track their functionality during the wound healing process [59].
- Limb Salvage: Preclinical studies have demonstrated TNT's ability to reprogram fibroblast cells in skin in vivo into vascular cells to assist in the recovery of injured limbs [59] [61].
Antimicrobial Applications
Recent research has explored TNT for antimicrobial therapy, particularly against antibiotic-resistant pathogens:
- Biofilm Eradication: TNT of antimicrobial genes drives bacterial biofilm killing in wounds [63]. This approach has been shown to effectively reduce bacterial burden in Staphylococcus aureus-infected wounds [63].
- Mechanism of Action: TNT-mediated delivery of plasmids encoding for antimicrobial peptides (e.g., mouse cathelicidin CAMP) enhances host defense mechanisms [63]. The technology likely promotes antimicrobial effects through multiple mechanisms, including increased production of antimicrobial peptides (LL-37), enhanced tissue vascularization, and recruitment of macrophages to the infection site [63].
- Extracellular Vesicle Mediated Effects: The therapeutic effect appears to be potentially mediated by extracellular vesicles derived from transfected cells, which propagate the treatment to surrounding parts of the tissue and provide a means for signal amplification [63].
Cancer Therapy
TNT applications have been extended to cancer treatment, with studies demonstrating:
- Tumor Regression: TNT has been successfully employed for tumor regression in preclinical models [59] [61].
- RNA-Based Cancer Vaccines: While not exclusively using TNT delivery, RNA-based cancer vaccines represent a related approach that has shown impressive results, with one melanoma treatment reducing cancer recurrence by 44% when combined with existing immunotherapy [64]. These advances support the potential of nucleic acid delivery technologies in oncology applications.
Table 2: Experimental Evidence for TNT Therapeutic Applications
| Application Area | Model System | Genetic Cargo | Key Outcomes | Reference |
|---|---|---|---|---|
| Wound Healing & Antimicrobial | Mouse infected wound model | Plasmid DNA encoding antimicrobial peptide (CAMP) | Significant decrease in bacterial biofilm burden; increased vascularization and macrophage infiltration | [63] |
| Tissue Regeneration | Preclinical limb injury models | Reprogramming factors for vascular cells | Improved blood flow and tissue recovery in injured limbs | [59] [61] |
| Neurological Repair | Ischemic stroke models | Factors for neuronal reprogramming | Functional recovery in damaged brain tissue | [59] [32] |
| Cancer Therapy | Tumor models | Therapeutic genes for tumor regression | Successful tumor regression demonstrated | [59] [63] |
Detailed Experimental Protocols
TNT-Mediated In Vivo Reprogramming for Wound Healing
The following protocol outlines the methodology for TNT-mediated delivery of antimicrobial genes to infected wounds, based on established experimental procedures [63]:
Materials Required:
- TNT device (silicon chip with nanochannels or hollow microneedles)
- Pulse generator system
- Plasmid DNA or mRNA encoding therapeutic genes (e.g., CAMP gene for antimicrobial peptide)
- Sterile surgical supplies
- Anesthesia equipment appropriate for animal model
- Bacterial strain for infection model (e.g., Staphylococcus aureus)
Procedure:
Wound Creation and Infection:
- Create full-thickness wounds on the dorsum of experimental animals using a sterile biopsy punch.
- Infect wounds with bacterial suspension (e.g., 1×10^7 CFU of Staphylococcus aureus) to establish biofilm-associated infection.
TNT Treatment:
- Apply genetic cargo (e.g., 20 μL of plasmid DNA solution at 100-200 μg/mL) to the cargo reservoir of the TNT device.
- Position the TNT device directly on the wound edge tissue.
- Apply optimized electrical pulse parameters (typical range: 50-200 V/cm, pulse duration of 10-100 ms, multiple pulses with specific intervals).
- Repeat treatment daily for the required duration (e.g., 3-7 days based on experimental design).
Assessment and Analysis:
- Monitor bacterial burden through tissue homogenization and plating.
- Evaluate biofilm formation using scanning electron microscopy.
- Assess host response through histology, immunohistochemistry, and gene expression analysis.
Direct Lineage Conversion via TNT
This protocol describes the methodology for direct reprogramming of fibroblasts to other cell lineages using TNT technology [57] [59]:
Materials Required:
- TNT device with appropriate chip configuration
- Reprogramming factors (mRNA or plasmid DNA encoding lineage-specific transcription factors)
- Target tissue or in vivo model
- Electroporation buffer system
Procedure:
Cargo Preparation:
- Prepare specific combinations of reprogramming factors based on target cell type. For neuronal conversion: Ascl1, Brn2, Myt1l; for endothelial conversion: Etv2, Foxo1, Fli1.
- Formulate genetic cargo at optimized concentrations in appropriate buffer.
TNT Delivery:
- Apply cargo solution to TNT device reservoir.
- Position device on target tissue (e.g., skin for fibroblast reprogramming).
- Administer optimized electrical pulses for nanoelectroporation.
Validation and Characterization:
- Analyze conversion efficiency through immunohistochemistry for cell-type specific markers.
- Assess functional properties of reprogrammed cells (electrophysiology for neurons, tube formation for endothelial cells).
- Evaluate tissue integration and persistence of reprogrammed cells.
Research Reagent Solutions
The following table outlines essential research reagents and materials for implementing TNT-based reprogramming studies:
Table 3: Essential Research Reagents for TNT-Based Reprogramming Studies
| Reagent/Material | Function/Purpose | Examples/Specifications | Key Considerations |
|---|---|---|---|
| TNT Silicon Chips | Physical interface for nanoelectroporation | - TNT 1.0 (nanochannel design)- TNT 2.0 (hollow microneedle array) | Selection depends on target tissue topography and delivery depth requirements [59] [61] |
| Genetic Cargo | Reprogramming factors | - Plasmid DNA- Modified mRNA- CRISPR/Cas9 components | mRNA offers transient expression; plasmids for longer duration; CRISPR for precision editing [57] [60] |
| Electroporation System | Pulse generation | - Customizable pulse generators- Commercial electroporation systems | Must deliver precise microsecond pulses with controlled voltage/current parameters [57] [59] |
| Reprogramming Factors | Cell fate determination | - OSKM factors (Oct4, Sox2, Klf4, c-Myc)- Lineage-specific factors (Ascl1, Brn2, Ngn2 for neurons) | Specific combinations determine reprogramming outcome [57] [60] |
| Sterilization Supplies | Device preparation | - Ethylene oxide gas- Gamma irradiation | Preserves nanodevice integrity while ensuring sterility [57] [58] |
| Cell Culture Materials | In vitro validation | - Primary cells (fibroblasts, epithelial cells)- Appropriate culture media | Essential for preliminary testing before in vivo applications [63] |
Tissue Nanotransfection represents a conceptual and technological advance in regenerative medicine and targeted gene therapy, offering a roadmap for future research and clinical implementation [57] [58]. As a physical delivery system, TNT addresses critical limitations of viral and chemical delivery methods while enabling precise in vivo reprogramming with minimal genomic integrity concerns [57] [60].
The integration of TNT with mRNA-based technologies presents particularly promising opportunities for cell fate reprogramming research. The transient nature of mRNA expression complements TNT's non-integrative approach, reducing safety concerns while allowing controlled reprogramming outcomes [57] [32] [60]. Future directions will likely focus on enhancing phenotypic stability, improving scalability for clinical applications, and developing closed-system manufacturing platforms to streamline production [57] [64].
With ongoing advances in genetic cargo design, delivery optimization, and manufacturing processes, TNT is positioned to become a cornerstone technology in personalized regenerative medicine, offering transformative potential for treating complex degenerative diseases, injuries, and genetic disorders [57] [64]. The continued refinement of this platform will further establish its role in the broader landscape of mRNA-based cell fate reprogramming research and therapeutic development.
The discovery of induced pluripotent stem cells (iPSCs) by Shinya Yamanaka in 2006 represented a paradigm shift in regenerative medicine and disease modeling [65]. By introducing four transcription factors—OCT4, SOX2, KLF4, and c-Myc (collectively known as the OSKM factors)—his team demonstrated that somatic cells could be reprogrammed to an embryonic stem cell-like state, possessing the capacity for self-renewal and differentiation into any cell type [13]. This groundbreaking work, which earned the Nobel Prize in Physiology or Medicine in 2012, provided researchers with an ethically acceptable, patient-specific alternative to embryonic stem cells while simultaneously opening unprecedented opportunities for disease modeling, drug screening, and regenerative therapies [65].
The subsequent integration of mRNA-based technologies has further accelerated the clinical translation of iPSC applications by addressing critical safety concerns associated with early viral reprogramming methods [60]. Messenger RNA (mRNA) has emerged as a transformative tool in regenerative medicine, providing precision, safety, and transience in directing cellular behavior [32]. Unlike traditional gene therapy approaches, mRNA therapeutics offer a non-integrative and controllable strategy for expressing therapeutic proteins, effectively bypassing the risk of insertional mutagenesis that plagued viral vector systems [60]. Through advancements in mRNA chemistry, transcript engineering, and delivery platforms, mRNA therapeutics now enable efficient protein supplementation, cell reprogramming, and cell transdifferentiation, allowing for precise modulation of cell fate and function [32].
This technical guide examines the current state of iPSC technology within the broader context of mRNA-based cell fate reprogramming research, detailing methodologies from somatic cell reprogramming to advanced disease modeling and in vivo tissue regeneration strategies. We provide comprehensive experimental protocols, quantitative comparisons of emerging technologies, and visualization of key workflows to serve the research community in advancing this rapidly evolving field.
Core Principles of iPSC Generation and Reprogramming Technologies
Fundamental Reprogramming Factor Biology
The molecular machinery underlying cellular reprogramming centers on core transcription factors that orchestrate the epigenetic remodeling necessary for achieving pluripotency. The canonical OSKM factors function as master regulators of pluripotency networks, with each component playing distinct but complementary roles [13]. OCT4 (POU5F1) serves as a pivotal regulator of the pluripotency network, with studies demonstrating that its expression alone can generate iPSCs from human neural stem cells, highlighting its central position in the reprogramming hierarchy [13]. SOX2 collaborates with OCT4 to regulate numerous pluripotency-associated genes, while KLF4 contributes to the metabolic reprogramming necessary for pluripotency acquisition. The proto-oncogene c-Myc enhances reprogramming efficiency primarily through global chromatin modifications that promote a more open configuration conducive to reprogramming [13].
Research has identified several alternative factor combinations that can substitute for or augment the standard OSKM cocktail. The OSNL combination (OCT4, SOX2, NANOG, and LIN28) achieves reprogramming without c-Myc, thereby reducing tumorigenic potential [13]. Additional studies have revealed that KLF2 and KLF5 can substitute for KLF4, while SOX1 and SOX3 can replace SOX2, though often with reduced efficiency [13]. Non-transcription factor alternatives have also been identified, including small molecules such as RepSox (which replaces SOX2) and epigenetic modulators that enhance reprogramming efficiency through chromatin remodeling [13].
Comparative Analysis of Reprogramming Delivery Systems
The method of delivering reprogramming factors significantly influences the safety profile, efficiency, and potential clinical applicability of resulting iPSC lines. Early approaches relied heavily on integrating viral vectors, particularly retroviruses and lentiviruses, which offered high efficiency but carried significant risks of insertional mutagenesis and tumorigenesis [65]. Subsequent technological advances have focused on developing non-integrating methods that maintain genomic integrity while achieving satisfactory reprogramming efficiencies.
Table 1: Comparison of iPSC Reprogramming Delivery Systems
| Delivery System | Genetic Material | Genomic Integration | Reprogramming Efficiency | Key Advantages | Primary Limitations |
|---|---|---|---|---|---|
| Retrovirus/Lentivirus | DNA | Yes | 0.001-0.1% | High efficiency; stable expression | Insertional mutagenesis; transgene silencing |
| Sendai Virus (SeV) | RNA | No | ~0.1% | High efficiency; non-integrating | Requires dilution; persistent viral vectors |
| Episomal Plasmid | DNA | No | ~0.001% | Non-integrating; simple preparation | Low efficiency; requires multiple transfections |
| mRNA | RNA | No | 1-4% | Non-integrating; high efficiency; precise control | Requires multiple transfections; potential immune response |
| Recombinant Protein | Protein | No | <0.001% | Completely non-integrating; minimal safety concerns | Extremely low efficiency; technically challenging |
| Tissue Nanotransfection (TNT) | DNA/RNA | No | Varies | In vivo application; targeted delivery | Emerging technology; optimization ongoing |
Among these approaches, mRNA-based reprogramming has gained significant traction due to its favorable safety profile and enhanced efficiency. The Rossi group at Harvard demonstrated that synthetic mRNA encoding the OSKM factors could reprogram human fibroblasts with efficiencies of 1-4%—orders of magnitude higher than conventional viral methods [60]. A critical innovation was the modification of mRNA to evade innate immune recognition, thereby preventing the interferon response that previously limited RNA-based approaches [60]. This breakthrough enabled prolonged protein expression without triggering cellular antiviral defenses, making mRNA an ideal vehicle for reprogramming factors.
Somatic Cell Source Selection Considerations
The choice of somatic starting material significantly influences reprogramming efficiency, iPSC quality, and suitability for downstream applications. Multiple somatic cell types have been successfully reprogrammed, each with distinct advantages and limitations:
- Dermal Fibroblasts: Historically the first cell type used for iPSC generation, fibroblasts remain widely utilized due to their high genomic stability, reliable expansion protocols, and established banking procedures [65]. The primary limitation is the invasiveness of skin biopsy collection.
- Peripheral Blood Mononuclear Cells (PBMCs): These cells offer a less invasive collection procedure while maintaining reprogramming efficiency comparable to fibroblasts [65]. Blood-derived iPSCs are particularly valuable for translational studies where repeated sampling may be necessary.
- Urinary Epithelial Cells: This cell source provides a completely non-invasive, reproducible sampling method that enables generation of multiple iPSC lines from the same donor within short timeframes [65].
- Keratinocytes: Derived from plucked hair follicles, keratinocytes demonstrate higher reprogramming efficiency compared to fibroblasts, though the initial cell yield is typically lower [65].
Additional somatic cell sources including mesenchymal stromal cells from dental pulp, synovial tissue, and hepatocytes have been reprogrammed but remain primarily restricted to basic research contexts [65].
Advanced mRNA and Non-Viral Reprogramming Methodologies
mRNA Reprogramming Workflow and Protocol
The mRNA reprogramming protocol represents a significant advancement in generating clinical-grade iPSCs without genomic integration. The following detailed methodology is adapted from the Rossi protocol with subsequent refinements [60]:
Day 0: Plating of Somatic Cells
- Obtain human fibroblasts or other somatic cells and plate at 5×10^4 cells per well in a 6-well plate using standard culture medium.
- Ensure cells are at appropriate density (30-50% confluence) at the time of transfection.
Days 1-6: Daily mRNA Transfection
- Prepare mRNA cocktail containing modified (immune-evading) mRNAs encoding OCT4, SOX2, KLF4, and c-Myc (or alternative combinations such as OCT4, SOX2, KLF4, and L-MYC for reduced tumorigenicity).
- Complex mRNA with transfection reagent according to manufacturer's specifications.
- Replace medium with fresh medium containing mRNA-lipid complexes.
- Incubate cells for 4-6 hours before replacing with fresh standard medium.
- Repeat this process daily for 5-6 consecutive days.
Days 7-21: Emergence and Expansion of iPSC Colonies
- Transition to feeder-free iPSC culture system using defined medium such as mTeSR1 or E8.
- Monitor for emergence of compact, ESC-like colonies with defined borders.
- Manually pick and expand individual colonies for further characterization.
Critical Considerations:
- Modified nucleotides (pseudouridine, 5-methylcytidine) are essential to prevent innate immune activation [60].
- Efficiency can be enhanced 6.5-fold by combining 8-Br-cAMP with valproic acid [13].
- Multiple cycles may be required for complete reprogramming; typical efficiency ranges from 1-4% of starting cells [60].
Tissue Nanotransfection (TNT) for In Vivo Reprogramming
Tissue nanotransfection (TNT) represents a cutting-edge approach that enables in vivo cellular reprogramming through localized nanoelectroporation [57]. This non-viral nanotechnology platform utilizes a hollow-needle silicon chip mounted beneath a cargo reservoir containing genetic material (plasmid DNA, mRNA, or CRISPR/Cas9 components). When electrical pulses are applied, the hollow needles concentrate the electric field at their tips, temporarily porating nearby cell membranes and enabling targeted delivery of charged genetic material directly into tissues [57].
The TNT workflow for in vivo reprogramming includes:
- Device preparation and sterilization using ethylene oxide gas or gamma irradiation
- Application of TNT device to target tissue (skin, organ surface)
- Delivery of reprogramming factors via optimized electrical parameters (voltage, pulse duration, inter-pulse intervals)
- In situ cellular reprogramming without intermediate pluripotent state
TNT has demonstrated therapeutic potential in diverse applications including tissue regeneration, ischemic repair, wound healing, and antimicrobial therapy [57]. Its key advantages include high specificity, non-integrative approach, minimal cytotoxicity, and the ability to perform reprogramming in living organisms without cell transplantation.
Figure 1: Cellular Reprogramming Pathways via TNT. Tissue Nanotransfection enables multiple reprogramming trajectories from somatic cells to therapeutic cell types through partial reprogramming, direct lineage conversion, or complete pluripotency induction.
Quality Control and Characterization of iPSCs
Rigorous quality control is essential to verify the pluripotent state and genomic integrity of generated iPSC lines before their application in disease modeling or therapeutic development. The following multiparameter assessment strategy should be implemented:
Pluripotency Marker Verification:
- PCR Analysis: Confirm expression of endogenous pluripotency genes (OCT4, SOX2, NANOG) while ensuring silencing of exogenous reprogramming factors.
- Immunocytochemistry: Demonstrate protein expression of pluripotency markers (OCT4, SOX2, SSEA-4, TRA-1-60, TRA-1-81) with appropriate subcellular localization.
- Flow Cytometry: Quantify the percentage of cells expressing pluripotency-associated surface antigens.
Functional Pluripotency Assessment:
- In Vitro Differentiation: Directed differentiation into representatives of all three germ layers (ectoderm, mesoderm, endoderm) using established protocols.
- Teratoma Formation: Subcutaneous injection of iPSCs into immunodeficient mice followed by histological confirmation of differentiated tissues from all three germ layers (though this is being phased out in favor of in vitro methods).
Genomic Integrity Evaluation:
- Karyotype Analysis: G-banding cytogenetics to detect chromosomal abnormalities.
- Whole Genome Sequencing: Identification of point mutations, small insertions/deletions.
- Copy Number Variation Analysis: Detection of structural variations that may compromise safety.
Epigenetic Status Assessment:
- DNA Methylation Analysis: Bisulfite sequencing of pluripotency gene promoters to confirm appropriate epigenetic resetting.
- Transcriptome Profiling: RNA sequencing to verify similarity to reference embryonic stem cell lines.
Regular genomic monitoring is particularly crucial as reprogramming can introduce chromosomal abnormalities or epigenetic alterations that may compromise differentiation efficiency or predispose cells to malignant transformation [65].
Disease Modeling Applications Using iPSC Derivatives
Neurodegenerative Disease Modeling
iPSC-derived neuronal models have provided unprecedented insights into the pathophysiology of neurodegenerative disorders including Alzheimer's disease (AD), Parkinson's disease (PD), and amyotrophic lateral sclerosis (ALS) [65]. Patient-specific iPSCs enable the recapitulation of disease processes in vitro, allowing investigation of molecular mechanisms and evaluation of therapeutic interventions.
For ALS modeling, iPSC-derived motor neurons (iPSC-MNs) reproduce disease-specific pathology including TDP-43 proteinopathy, mitochondrial dysfunction, and axonal degeneration [13]. These models have enabled the identification of disease biomarkers and screening of therapeutic compounds. Similarly, AD models using iPSC-derived neurons and glia reproduce hallmark pathologies including tau hyperphosphorylation and β-amyloid deposition, providing platforms for targeted therapeutic development [65]. PD models have successfully recapitulated dopaminergic neuron degeneration and revealed the pathogenic role of α-synuclein aggregation, advancing understanding of both sporadic and familial forms of the disease [65].
Table 2: iPSC Applications in Disease Modeling and Drug Discovery
| Disease Category | iPSC-Derived Cell Types | Modeled Pathologies | Drug Screening Applications |
|---|---|---|---|
| Neurodegenerative | Motor neurons, cortical neurons, dopaminergic neurons | TDP-43 aggregation, α-synuclein accumulation, Aβ deposition | Neuroprotective compounds, protein aggregation inhibitors |
| Cardiovascular | Cardiomyocytes, endothelial cells, vascular smooth muscle cells | Arrhythmias, contractile dysfunction, hypertrophy | Ion channel modulators, inotropic agents |
| Metabolic | Hepatocytes, pancreatic β-cells, myocytes | CFTR dysfunction, copper accumulation, insulin deficiency | CFTR correctors, copper chelators, insulin secretagogues |
| Rare Genetic | Kidney organoids, retinal organoids, sensory neurons | Renal cyst formation, photoreceptor degeneration, neuronal-glial imbalance | Disease-modifying therapies, gene correction approaches |
Rare Disease Modeling using iPSC Platforms
iPSC technology has proven particularly transformative for rare disease research, where limited patient populations and scarcity of tissue samples have historically impeded therapeutic development [67]. Approximately 80% of rare diseases have genetic origins, making patient-derived iPSCs with isogenic controls uniquely powerful model systems [67].
Notable applications include:
- Juvenile Nephronophthisis (NPH): NPHP1-deficient iPSCs exhibit abnormal cell proliferation, primary cilia abnormalities, and renal cyst formation in kidney organoids—pathologies reversible upon NPHP1 reintroduction [67].
- RDH12-associated Retinitis Pigmentosa: Retinal organoids from patients with RDH12 mutations show reduced photoreceptor numbers, shortened photoreceptor length, and disruptions in retinol biosynthesis, modeling the late-onset, milder disease course seen in patients [67].
- Hereditary Sensory and Autonomic Neuropathy Type IV (HSAN IV): iPSC-derived dorsal root ganglion organoids from patients with NTRK1 mutations demonstrate disrupted balance between neuronal and glial differentiation, providing insights into developmental origins of sensory neuropathy [66].
These models exemplify how iPSC technology enables study of disease mechanisms in human cells and tissues previously inaccessible to researchers, particularly for disorders affecting internal organs or human-specific developmental processes.
In Vivo Tissue Regeneration and Therapeutic Applications
Cardiovascular Regeneration
iPSC-derived cardiomyocytes enable sophisticated modeling of arrhythmogenic disorders, heart failure, and myocardial injury while simultaneously providing cellular material for regenerative approaches [65]. For congenital arrhythmias linked to KCNQ1 mutations, iPSC-cardiomyocytes provide a platform for precision cardiology and drug testing [65]. In myocardial regeneration, iPSC-derived cardiomyocytes, fibroblasts, vascular smooth muscle cells, and endothelial cells have been explored for regenerative transplantation strategies, with promising improvements in cardiac function demonstrated in preclinical models [65].
Advanced tissue engineering approaches now combine iPSC-derived cardiovascular cells with biocompatible scaffolds to create functional cardiac patches with enhanced engraftment potential. These constructs demonstrate electrical integration, mechanical contraction, and vascularization capacity that surpass earlier cell suspension transplantation methods.
Wound Healing and Skin Regeneration
Cell-engineered technologies have emerged as a transformative frontier for wound healing and tissue regeneration [68]. iPSC-derived keratinocytes, fibroblasts, and endothelial cells can be combined into composite skin substitutes that actively promote tissue regeneration through multiple mechanisms:
- Re-epithelialization: iPSC-derived keratinocytes form stratified epidermis and restore barrier function.
- Dermal Reconstruction: iPSC-derived fibroblasts produce organized extracellular matrix and appropriate growth factors.
- Angiogenesis: iPSC-derived endothelial cells form functional vascular networks that integrate with host circulation.
Advanced delivery systems including hydrogels, 3D bioprinting, and bioactive dressings enhance cell survival, integration, and function at wound sites [68]. These technologies are particularly beneficial for chronic diabetic ulcers, burn wounds, and other conditions characterized by impaired healing capacity.
Figure 2: iPSC-Based Skin Regeneration Strategy. Patient-specific iPSCs are differentiated into multiple skin cell types then combined using tissue engineering approaches to create functional skin constructs for wound healing applications.
Clinical Translation and Ongoing Trials
The clinical translation of iPSC-based therapies has progressed rapidly, with multiple trials underway worldwide. The first iPSC-derived cell product was transplanted into a human patient in 2013 at the RIKEN Center in Kobe, Japan, where Dr. Masayo Takahashi investigated the safety of iPSC-derived retinal pigment epithelial cell sheets in patients with macular degeneration [69]. In 2016, Cynata Therapeutics received approval to launch the first formal clinical trial of an allogeneic iPSC-derived cell product (CYP-001) for steroid-resistant acute GvHD [69]. This historic trial met its clinical endpoints and produced positive safety and efficacy data, leading to advanced Phase 2 trials for COVID-19 complications, GvHD, and critical limb ischemia, as well as a Phase 3 trial in 440 patients with osteoarthritis—representing the world's first Phase 3 clinical trial involving an iPSC-derived cell therapeutic product [69].
These pioneering trials demonstrate the accelerating clinical translation of iPSC technologies and provide valuable insights into manufacturing, safety monitoring, and efficacy assessment requirements for future cell therapies.
The Scientist's Toolkit: Essential Research Reagents and Technologies
Table 3: Key Research Reagent Solutions for iPSC Technology
| Reagent Category | Specific Products | Function | Application Notes |
|---|---|---|---|
| Reprogramming Kits | mRNA Reprogramming Kit (Stemgent), CytoTune-iPS 2.0 Sendai | Non-integrating reprogramming | mRNA kits offer high efficiency; Sendai provides convenience |
| Culture Systems | mTeSR1, StemFlex, E8 medium | iPSC maintenance | Defined, xeno-free formulations enhance reproducibility |
| Differentiation Kits | Cardiomyocyte Differentiation Kit (iPSC-CM), Neural Induction Medium | Directed differentiation | Optimized protocols improve efficiency and purity |
| Extracellular Matrices | Matrigel, Recombinant Laminin-521, Vitronectin | Feeder-free culture | Recombinant matrices enhance consistency and scalability |
| Gene Editing Tools | CRISPR-Cas9 systems, TALEN, Zinc Finger Nuclease | Genetic modification | CRISPR offers highest efficiency and multiplexing capability |
| Characterization Kits | Pluripotency Flow Cytometry Kit, Trilineage Differentiation Kit | Quality control | Essential for validating iPSC lines pre-application |
| Delivery Technologies | Tissue Nanotransfection devices, Electroporation systems | In vivo/vitro delivery | TNT enables localized in vivo reprogramming |
The integration of iPSC technology with advanced reprogramming methodologies, particularly mRNA-based approaches and tissue nanotransfection, has created a powerful platform for disease modeling, drug discovery, and regenerative medicine. The field continues to evolve rapidly, with ongoing efforts focused on enhancing safety, efficiency, and clinical translatability.
Key future directions include:
- Manufacturing Standardization: Developing closed, automated systems for large-scale production of clinical-grade iPSCs and their derivatives.
- Maturation Technologies: Advancing methods to achieve full functional maturation of iPSC-derived cells, particularly for tissues with complex functional requirements such as brain and liver.
- Gene Editing Integration: Combining iPSC technology with precise gene correction approaches for autosomal dominant disorders.
- Personalized Medicine Applications: Utilizing patient-specific iPSC models for drug screening and therapy selection based on individual genetic backgrounds.
- In Vivo Reprogramming Optimization: Refining TNT and related technologies for direct tissue regeneration without intermediate cell transplantation.
As these technologies mature, iPSC-based approaches are poised to transform therapeutic development across a broad spectrum of human diseases, ultimately fulfilling the promise of personalized regenerative medicine that motivated Yamanaka's original discovery nearly two decades ago.
Navigating the Technical Landscape: Overcoming Immunogenicity, Inefficiency, and Delivery Hurdles
The clinical success of mRNA-based technologies hinges on the precise control of the molecule's immunostimulatory properties. This technical guide details two cornerstone strategies for tailoring mRNA-elicited immune responses: the manipulation of the 5' cap structure, specifically the Cap2 methylome, and the incorporation of modified nucleosides. Within the broader thesis of mRNA-based cell fate reprogramming research, these strategies are not merely for vaccine development but are fundamental to engineering precise transcriptional environments, modulating cell signaling, and ultimately directing cell fate. This whitepaper provides an in-depth analysis of the mechanisms, quantitative data, and detailed methodologies underpinning these approaches, serving as a resource for researchers and drug development professionals aiming to harness the full potential of mRNA therapeutics.
The application of mRNA technology for cell fate reprogramming extends beyond simple protein replacement. It involves the deliberate redirection of cellular identity and function, a process highly dependent on the transcriptional landscape and signaling environment. A critical, and often limiting, factor is the innate immune system's recognition of exogenous mRNA, which can trigger a potent type I interferon response. While potentially beneficial for vaccine adjuvanticity, this immunogenicity is typically undesirable for cell reprogramming as it can inhibit translation, activate pro-apoptotic pathways, and alter the intended cellular differentiation trajectory.
Consequently, "taming" this immune response is a prerequisite for efficient reprogramming. Two primary, complementary strategies have been developed:
- Cap2 Structures: Engineering the 5' terminus of mRNA to mimic mature, self-RNA, thereby evading cytosolic immune sensors.
- Modified Nucleosides: Incorporating naturally occurring modified nucleosides (e.g., N1-methylpseudouridine) to render the mRNA "invisible" to pattern recognition receptors.
The following sections dissect the biology, application, and experimental validation of these strategies, providing a framework for their implementation in next-generation cell reprogramming protocols.
The Cap2 Methylome: A Marker of mRNA Age and Self
Cap1 and Cap2 Biology
The 5' cap structure of eukaryotic mRNA is critical for stability, translation initiation, and immune recognition. The core cap structure consists of an N7-methylguanosine (m7G) linked to the first transcribed nucleotide. During mRNA biogenesis in the nucleus, the ribose of this first nucleotide is methylated by cap methyltransferase 1 (CMTR1) to form the Cap1 structure (m7G-ppp-Nm). After mRNA export to the cytosol, a subset of Cap1 mRNAs undergoes further methylation on the ribose of the second nucleotide by cap methyltransferase 2 (CMTR2) to form the Cap2 structure (m7G-ppp-Nm-Nm) [70].
The identity and function of Cap2 remained elusive for decades. Recent research has revealed that unlike other epitranscriptomic modifications, Cap2 is not sequence-specific but is a marker of mRNA age. Cap2 is formed through a slow, continuous conversion from Cap1 as mRNAs reside in the cytosol, making it enriched on long-lived mRNAs. This age-dependent methylation provides a powerful mechanism for the innate immune system to distinguish "self" from "non-self": newly transcribed viral RNAs are predominantly Cap1, while host mRNAs accumulate Cap2 over time [70].
Table 1: Cap2 Abundance Across Different Biological Contexts
| Organism/Cell Type | Relative Cap2 Abundance | Notes |
|---|---|---|
| HEK293T Cells | ~40% | Commonly used human cell line [70] |
| MCF-7 Cells | ~56% | Human breast cancer cell line [70] |
| Mouse Spleen | ~30% | Highest among profiled mouse tissues [70] |
| Mouse Brain | ~8% | Lowest among profiled mouse tissues [70] |
| C. elegans | 0.9% | Nearly absent in this model organism [70] |
Cap2's Role in Innate Immune Evasion
The key immunomodulatory function of the cap structure lies in its recognition by the innate immune sensor RIG-I. RIG-I is activated by RNA ligands containing a 5' triphosphate, which is characteristic of viral RNA. While the m7G cap of Cap0 and Cap1 structures somewhat shields the RNA, the conversion to Cap2 provides a superior level of immune evasion.
Research has demonstrated that Cap1 is intrinsically immunostimulatory and can bind to and activate RIG-I, especially when RIG-I expression is high. The methylation of Cap1 to Cap2 markedly reduces the ability of the RNA to activate RIG-I. Therefore, the slow rate of Cap2 methylation allows it to accumulate on long-lived host mRNAs (self), while newly expressed viral RNAs (non-self) remain primarily in the more immunogenic Cap1 state. This mechanism allows cells to dampen immune activation against self-mRNA while remaining vigilant to viral invasion [70].
Experimental Methods for Cap2 Analysis
CapTag-seq for Quantification
Purpose: To transcriptome-wide quantify the levels and dynamics of Cap1 and Cap2. Workflow:
- m7GDP Removal: mRNAs are enzymatically decapped, leaving 5'-monophosphorylated ends.
- Adapter Ligation: A 5' adapter composed of a 2'-O-methylated nucleotide (Nm) is ligated to the monophosphorylated mRNA end, rendering it resistant to RNase T2.
- RNase T2 Digestion: The sample is fully digested with RNase T2. This enzyme cleaves all phosphodiester bonds except those after Nm. Therefore:
- Cap1 mRNA is liberated as a two-nucleotide cap tag (m7G-ppp-Nm-N).
- Cap2 mRNA is liberated as a three-nucleotide cap tag (m7G-ppp-Nm-Nm-N).
- Library Construction & Sequencing: The adapter-linked cap tags are converted into a cDNA library and sequenced. The ratio of two-nucleotide to three-nucleotide tags provides a quantitative measure of Cap1 vs. Cap2 stoichiometry [70].
CLAM-Cap-seq for Mapping
Purpose: To identify which specific mRNA transcripts are modified with Cap2. Workflow:
- Decapping and Reverse Transcription: Decapped mRNA is reverse transcribed to create a cDNA-mRNA hybrid.
- Circligase-assisted Ligation: The 3' end of the cDNA is ligated to the first 5' nucleotide of the template mRNA, creating a cDNA-mRNA chimera.
- RNase T2 Digestion: The mRNA is digested with RNase T2, leaving only the cap tag (2 or 3 nucleotides) covalently attached to the cDNA.
- Adapter Ligation and Sequencing: A DNA adapter is ligated to the cap tag, and the chimeric molecule is sequenced. The resulting reads contain a "palindrome" that reveals the cap tag's sequence and the cDNA sequence, thereby mapping the cap status to its specific mRNA transcript [70].
Diagram 1: Cap2 methylation reduces RIG-I activation. Cap1 mRNA strongly activates RIG-I, triggering an interferon response. Cap2 mRNA shows markedly reduced activation, enabling immune evasion.
Modified Nucleosides: Rewriting the mRNA's Identity
Mechanism of Immune Suppression
A parallel and highly successful strategy for evading innate immunity involves incorporating modified nucleosides into the mRNA backbone. In vitro transcribed (IVT) mRNA is recognized as a pathogen-associated molecular pattern (PAMP) by various immune receptors, including Toll-like receptors 7 and 8 (TLR7/8) in endosomes and RIG-I in the cytosol. This recognition leads to a potent type I interferon response that inhibits translation and can induce apoptosis [71] [26].
The seminal discovery by Karikó and Weissman revealed that replacing uridine with its naturally occurring modified counterpart, pseudouridine (Ψ) or the more effective N1-methylpseudouridine (m1Ψ), abrogates this immune recognition. These modified nucleosides alter the secondary structure of the mRNA and mask its "foreign" signature, allowing it to be interpreted by the cell as "self" RNA. This dramatically reduces interferon signaling, increases mRNA stability, and enhances protein translation [71] [72] [73].
Impact on Vaccine Efficacy and Therapeutic Potential
The use of nucleoside-modified mRNA is a cornerstone of the successful COVID-19 vaccines (e.g., Comirnaty and Spikevax). Comparative studies have demonstrated its superiority over unmodified mRNA. For instance, in influenza mRNA vaccines, replacing uridine with m1Ψ led to a significant positive impact on the induction of functional antibody titers in mice and macaques by reducing innate immune sensing [72].
The choice of delivery system, specifically the ionizable lipid in lipid nanoparticles (LNPs), can influence the degree of benefit gained from nucleoside modification. The immunostimulatory nature of some LNPs can partially overshadow the effect of modification, highlighting the need for a holistic design approach [72].
Table 2: Impact of N1-methylpseudouridine (m1Ψ) Modification in an Influenza mRNA Vaccine Model
| LNP Ionizable Lipid | Impact of m1Ψ Modification on Immune Response | Notes |
|---|---|---|
| MC3 | Significant positive impact | Increased functional antibody titers; reduced innate cytokines [72] |
| KC2 | Significant positive impact | Similar beneficial effect as with MC3 LNP [72] |
| L319 | Minimal impact | LNP itself may be less immunostimulatory, reducing added benefit of modification [72] |
The experimental implementation of these strategies requires specific enzymatic and chemical reagents.
Table 3: Essential Research Reagents for mRNA Cap and Immune Modulation Studies
| Reagent / Tool | Function / Purpose | Key Feature |
|---|---|---|
| mRNA Cap 2´-O-Methyltransferase (e.g., M0366, NEB) | Enzymatically converts Cap-0 structures to Cap-1 in vitro. | Essential for producing immune-evasive cap structures for research mRNAs; requires m7G-capped RNA as substrate [74]. |
| Vaccinia Capping Enzyme (e.g., M2080, NEB) | Synthesizes the core m7G cap (Cap-0) on in vitro transcribed RNA. | Required to create a proper 5' end for subsequent Cap1 methylation [74]. |
| N1-methylpseudouridine-5'-Triphosphate | Modified nucleoside building block for IVT mRNA. | Directly replaces UTP in transcription reactions to produce immune-silenced mRNA; a key component of clinical-grade vaccines [71] [72]. |
| Ionizable Lipids (MC3, KC2, L319) | Key component of LNPs for in vivo mRNA delivery. | Different lipids have varying immunostimulatory properties, which can interact with cap and nucleoside modification strategies [72]. |
| CLAM-Cap-seq Protocol | Transcriptome-wide mapping of Cap2-modified mRNAs. | Provides a detailed methodology for identifying Cap2 targets and quantifying methylation status [70]. |
Application in Cell Fate Reprogramming and Cancer Immunotherapy
The principles of immune evasion are directly applicable to cell fate reprogramming. The field is leveraging iPSC technology to differentiate clinically useful immune cells (T-cells, NK cells, macrophages) at scale. The utilization of iPSCs allows for the introduction of genetic modifications to enhance anti-tumor properties. Using modified mRNA to deliver reprogramming factors (e.g., OCT4, SOX2, KLF4, c-MYC) or to engineer chimeric antigen receptors (CARs) minimizes the risk of activating detrimental innate immune pathways during the process, thereby improving efficiency and cell viability [75].
Furthermore, the immunomodulatory effects of mRNA can be harnessed therapeutically. A striking clinical discovery showed that cancer patients who received mRNA-based COVID-19 vaccines within 100 days of starting immunotherapy were twice as likely to be alive three years later. The proposed mechanism is that the mRNA vaccine acts as a systemic "alarm," putting the immune system on high alert. This trained immunity can help overcome resistance in "cold" tumors, making them more susceptible to subsequent checkpoint blockade therapy. This synergy between a non-specific mRNA vaccine and targeted immunotherapy opens new avenues for in vivo reprogramming of the tumor microenvironment [76].
Diagram 2: Immune recognition dictates mRNA therapeutic efficacy. Unmodified mRNA triggers innate immunity, inhibiting translation and therapeutic success. mRNA engineered with Cap2 and modified nucleosides evades detection, enabling efficient protein production and the desired cellular outcome.
Boosting Translation Efficiency and mRNA Stability through UTR and Poly-A Tail Engineering
In the rapidly advancing field of mRNA-based technology for cell fate reprogramming research, achieving precise control over protein expression is paramount. The transient nature of mRNA expression makes it particularly suitable for therapeutic applications, including the generation of induced pluripotent stem cells (iPSCs) and direct cell transdifferentiation, where persistent activity of reprogramming factors can be detrimental [32]. While much attention has historically focused on coding sequence optimization and delivery systems, the untranslated regions (UTRs) and poly-A tail of an mRNA transcript serve as critical regulatory hubs that profoundly influence translation efficiency and mRNA stability. These elements collectively determine the intracellular kinetics, bioavailability, and ultimately the therapeutic efficacy of synthetic mRNA [77].
Optimizing these non-coding regions presents a powerful strategy for enhancing the performance of mRNA therapeutics without altering the encoded protein. UTRs contain binding sites for RNA-binding proteins and microRNAs that modulate translational efficiency and decay rates, while the poly-A tail interacts with poly-A binding proteins to protect the transcript from degradation and facilitate translational initiation [78]. For cell fate reprogramming applications, where the precise timing and dosage of transcription factors like OCT4, SOX2, KLF4, and cMYC (OSKM) are critical, fine-tuning these elements can significantly improve reprogramming efficiency and kinetics while maintaining the non-integrating, transient nature of mRNA-based approaches [77].
This technical guide synthesizes current advances in UTR and poly-A tail engineering, providing researchers with evidence-based strategies to optimize mRNA constructs for enhanced stability and translational output, with particular emphasis on applications in regenerative medicine and cellular reprogramming.
UTR Engineering for Enhanced mRNA Performance
The Critical Role of 5' UTRs in Translation Initiation
The 5' UTR is a major determinant of translation initiation efficiency, serving as the landing platform for the ribosomal pre-initiation complex (PIC). Key considerations for 5' UTR design include avoiding upstream start codons (uAUGs) and associated open reading frames (uORFs) that can cap-trap ribosomes and reduce translation of the main ORF [79]. Additionally, minimizing stable secondary structures near the 5' cap is crucial, as these can impede scanning by the 43S PIC [79].
Table 1: Performance of Engineered 5' UTRs in mRNA Therapeutics
| 5' UTR Source/Design | Design Approach | Expression Improvement | Cell Type Validation | Key Findings |
|---|---|---|---|---|
| Deep Learning-Designed | Gradient descent optimization & generative neural networks | High editing efficiency with megaTAL mRNA | K562, HepG2, T cells | 24/29 designed UTRs showed high editing; correlation across cell types (r²=0.84-0.87) [79] |
| Human α-globin | Endogenous UTR | High potency | Multiple cell lines | Validated as strong performer compared to novel designs [80] |
| CYBA 5' UTR | Selected from long-lived mRNAs | Significantly increased protein levels | NIH3T3, A549, hAMSCs | Enhanced translation without affecting mRNA half-life [78] |
| Fully randomized screening | Massively parallel reporter assays | High mean ribosome load | HEK293T, HepG2, T cells | Identified UTRs with varying performance; high cross-celltype correlation [79] |
Innovative approaches to 5' UTR optimization now employ deep learning models such as Optimus 5-Prime, which predicts translation efficiency from sequence features. These models are trained on data from massively parallel reporter assays (MPRAs) that measure the Mean Ribosome Load (MRL) for thousands of 5' UTR variants [79]. Recent advances include Optimus 5-Prime(25), specifically trained on shorter, fully randomized 5' UTRs to minimize inclusion of unintended regulatory elements while maintaining high translational efficiency [79].
Experimental Protocol: Deep Learning-Guided 5' UTR Design
The following methodology outlines the process for designing and validating high-performance 5' UTRs:
Library Construction: Generate a library of 5' UTR sequences with either fully randomized 25-50nt regions or sequences generated by generative neural networks (DENs) or gradient descent optimization (Fast SeqProp) [79].
Massively Parallel Reporter Assays (MPRAs): Clone the 5' UTR library upstream of a reporter gene (e.g., EGFP) and downstream of a T7 promoter. Transcribe mRNA in vitro and transfert into relevant cell types (HEK293T, HepG2, or T cells) [79].
Polysome Profiling: After 8 hours incubation, treat cells with cycloheximide to freeze ribosomes, fractionate lysates through sucrose density gradients, and sequence RNA from each fraction [79].
Mean Ribosome Load Calculation: Calculate MRL for each 5' UTR by multiplying normalized read counts in each fraction by the corresponding number of ribosomes [79].
Model Training and Validation: Train convolutional neural networks on the MRL data, then use the models to design novel 5' UTRs with predicted high translation efficiency [79].
Functional Validation: Test top-performing designs in the context of therapeutic mRNAs, such as those encoding gene editing enzymes (megaTALs) or reprogramming factors, measuring functional outcomes (e.g., editing efficiency or reprogramming kinetics) [79].
3' UTR Engineering for mRNA Stability and Translational Control
The 3' UTR plays a pivotal role in determining mRNA stability through its influence on deadenylation rates and interaction with RNA-binding proteins. Unlike traditional approaches that primarily utilized globin 3' UTRs, recent screening efforts have identified novel 3' UTRs with superior stabilizing properties [77].
Table 2: Performance of Engineered 3' UTRs in mRNA Therapeutics
| 3' UTR Source/Design | Design Approach | Expression Improvement | Key Findings |
|---|---|---|---|
| FCGRT 3' UTR | Cell-based selection from ActD-treated hDCs | ~6-fold increase in half-life | Discovered via systematic evolution; augments protein expression [77] |
| CYBA 3' UTR | Selected from long-lived mRNAs | Significantly increased protein levels | Enhanced translational efficiency without affecting mRNA half-life [78] |
| VP6 3' UTR | Empirical screening | High potency | Validated as promising alternative to conventional UTRs [80] |
| SOD 3' UTR | Empirical screening | High potency | Identified as high-performing 3' UTR element [80] |
| Engineered AU-rich elements | Rational design with "AUUUA" repeats | Up to 5-fold increase in protein expression | Enhances stability via HuR protein binding; optimal repeats crucial [81] |
| LSP1 3' UTR | Cell-based selection | Enhanced stability | Identified through systematic enrichment process [77] |
A groundbreaking approach to 3' UTR discovery involves a cell-based selection process that mimics natural selection for mRNA stability. This method involves:
- Treating immature human dendritic cells (hDCs) with Actinomycin D to block new RNA synthesis, thereby enriching for stable mRNA species [77].
- Extracting remaining mRNAs after prolonged culture, fragmenting them, and cloning the fragments as 3' UTRs in a reporter construct.
- Conducting multiple rounds of selection with increasing stringency to identify 3' UTR elements that dramatically enhance mRNA half-life (up to 6-fold improvement compared to starting library) [77].
This approach has identified novel 3' UTRs from genes such as FCGRT, LSP1, and CCL22 that outperform conventional β-globin 3' UTRs in various therapeutic contexts, including cancer immunotherapy and cellular reprogramming [77].
Another innovative strategy involves the rational incorporation of AU-rich elements (AREs) in the 3' UTR. Contrary to traditional views that AREs primarily destablize mRNAs, engineered AREs containing specific "AUUUA" repeats can significantly enhance stability and translation by recruiting the RNA-binding protein HuR [81]. Knockdown experiments confirm that HuR is essential for this stabilizing effect, with optimized ARE repeats increasing protein expression up to 5-fold for various encoded proteins, including luciferase, EGFP, mCherry, and ovalbumin [81].
Poly-A Tail Engineering for Enhanced Stability and Expression
Structural Innovations in Poly-A Tail Design
The poly-A tail is critical for mRNA stability and efficient translation, primarily through its interaction with poly-A binding proteins (PABPs) that protect the transcript from degradation and facilitate circularization with the 5' end to enhance ribosome recycling [82]. While traditional optimization efforts have focused primarily on tail length, recent innovations explore structural modifications that profoundly influence stability.
Table 3: Performance of Engineered Poly-A Tails in mRNA Therapeutics
| Poly-A Tail Design | Structure | Expression Improvement | Key Findings |
|---|---|---|---|
| A50L50LO | Loop structure with complementary linker | Highest in vitro & in vivo expression | Small loop structure enhances stability; superior to conventional designs [82] |
| A30L70 | Linear heterologous tail | Strong expression, second to A50L50LO | Used in BioNTech COVID-19 vaccine; robust performance [82] |
| A120 | Homogenous adenosine tail | Moderate expression | Benchmark for traditional approaches; outperformed by structured tails [82] |
| A50L50LX | Linear linker without complementarity | Lower expression than looped version | Demonstrates importance of structure beyond sequence [82] |
| Heterologous A/G tail | Mixed adenosine/guanosine | High potency comparable to A30L70 | Novel tail design with potential for therapeutic applications [80] |
A breakthrough in poly-A tail engineering involves the introduction of loop structures within the tail region. The A50L50LO design, which incorporates complementary sequences that form small stable loops, demonstrated superior protein expression both in vitro and in vivo compared to conventional linear poly-A tails [82]. This design maintained high luciferase expression for extended periods (24+ hours) in mouse models and significantly increased human erythropoietin (hEPO) levels compared to other tail configurations [82].
The mechanism behind this enhancement appears to involve impeded deadenylation, as secondary structures in the poly-A tail region can slow the rate of deadenylation complex progression, thereby extending mRNA half-life [82]. This principle is observed in certain viral transcripts that incorporate non-adenosine ribonucleotides to inhibit deadenylation and enhance stability [82].
Experimental Protocol: Evaluating Poly-A Tail Designs
To systematically evaluate poly-A tail configurations:
Tail Design: Construct mRNA variants with different poly-A tail structures:
- A50L50LO: 50 adenosine bases + linker + 50 adenosine bases with complementary linker sequence forming a loop
- A50L50LX: 50 adenosine bases + linker + 50 adenosine bases without complementarity
- A30L70: 30 adenosine bases + linker + 70 adenosine bases (BioNTech design)
- A120: Homogenous 120 adenosine tail [82]
In Vitro Testing: Transfert constructed mRNAs (500 ng/well) into multiple cell lines (e.g., Nor10, HeLa, A549, HepG2). Measure luciferase activity at 6, 24, and 48 hours post-transfection [82].
In Vivo Validation: Formulate selected mRNAs into lipid nanoparticles (LNPs). Administer intramuscularly or intravenously to C57BL/6 mice (5 μg dose). Monitor luciferase expression using IVIS imaging at 6 and 24 hours, or measure serum hEPO levels by ELISA at 2 and 6 hours [82].
Immune Response Evaluation: For vaccine applications, immunize mice with mRNA encoding antigenic proteins (e.g., HPV E6/E7 or influenza HA). Analyze antigen-specific CD8+ T cell responses by flow cytometry and antibody titers by ELISA [82].
Integrated UTR and Poly-A Tail Optimization Strategies
Combinatorial Approaches for Maximum Expression
The most effective mRNA optimization strategies combine enhanced 5' UTRs, 3' UTRs, and poly-A tails in a synergistic manner. Research demonstrates that pairing the human α-globin 5' UTR with novel 3' UTRs like VP6 or SOD, along with advanced poly-A tails (A30L70 or heterologous A/G tails), produces superior results compared to any single modification alone [80].
For cell fate reprogramming applications, the combination of optimized UTRs has proven particularly valuable. In the context of generating induced pluripotent stem cells (iPSCs) from somatic cells, mRNA formulations incorporating stability-enhancing 3' UTRs significantly improve the efficiency and kinetics of reprogramming by maintaining appropriate levels of pluripotency factors like OCT4, SOX2, KLF4, and cMYC [77]. The transient nature of mRNA delivery is ideally suited for reprogramming applications, as it avoids the risk of genomic integration associated with viral methods while allowing precise control over factor expression timing and dosage [32].
The CYBA UTR system exemplifies the success of combinatorial approaches. When both 5' and 3' UTRs from the CYBA gene were combined, they significantly enhanced protein expression of reporter genes and therapeutic proteins like human bone morphogenetic protein 2 (hBMP2) without altering mRNA half-life, suggesting a pure translation-enhancing effect [78]. This combination induced successful osteogenic differentiation in human adipose mesenchymal stem cells (hAMSCs), demonstrating its therapeutic potential in regenerative medicine [78].
The Scientist's Toolkit: Essential Reagents for mRNA Engineering
Table 4: Key Research Reagents for mRNA UTR and Poly-A Tail Engineering
| Reagent/Tool | Function | Application Examples |
|---|---|---|
| Deep Learning Models (Optimus 5-Prime) | Predicts translation efficiency from 5' UTR sequence | Designing novel high-performance 5' UTRs [79] |
| Cell-based Selection System | Identifies naturally occurring stabilizing 3' UTR elements | Discovery of FCGRT, LSP1 3' UTRs with enhanced stability [77] |
| Modified Nucleotides (5-methyl CTP, 2-thio UTP) | Reduces immunogenicity, increases stability | Generating chemically-modified mRNA with decreased innate immune recognition [78] |
| Poly-A Tailing Kits | Adds defined poly-A tails of specific lengths and structures | Constructing A50L50LO, A30L70 tail variants [82] |
| UTR Database (http://utrdb.ba.itb.cnr.it/) | Repository of natural UTR sequences | Source of CYBA, DECR1, GMFG UTR sequences [78] |
| Lipid Nanoparticles (LNPs) | In vivo delivery of engineered mRNA | Formulating mRNA for animal studies and therapeutic applications [82] |
| Polysome Profiling Reagents | Separates mRNAs based on ribosome number | Measuring translation efficiency in MPRAs [79] |
UTR and poly-A tail engineering represents a powerful approach for enhancing the therapeutic potential of mRNA-based technologies, particularly in the context of cell fate reprogramming and regenerative medicine. Through strategic optimization of these non-coding regions, researchers can significantly improve both the stability and translational efficiency of synthetic mRNA transcripts without altering the encoded protein.
The most promising developments in this field include deep learning-guided 5' UTR design, cell-based selection of stabilizing 3' UTR elements, and structural innovations in poly-A tail engineering that go beyond simple length optimization. The integration of these approaches—combining enhanced 5' UTRs, novel 3' UTRs, and structured poly-A tails—provides synergistic improvements in protein expression that can dramatically enhance the efficacy of mRNA therapeutics.
For the field of cell fate reprogramming, these advances enable more precise control over the timing and dosage of reprogramming factors, leading to improved efficiency in generating iPSCs and transdifferentiated cells. As mRNA technology continues to evolve, further optimization of these regulatory elements will undoubtedly unlock new therapeutic possibilities and enhance our ability to manipulate cellular behavior for regenerative applications.
The therapeutic application of messenger RNA (mRNA) for cell fate reprogramming represents a frontier in biomedical science, offering potential solutions for regenerative medicine, cancer immunotherapy, and genetic disorders. The clinical success of lipid nanoparticles (LNPs) in mRNA COVID-19 vaccines has established this platform as the leading vehicle for nucleic acid delivery [83]. However, the transition from vaccination to complex therapeutic applications like cell reprogramming necessitates overcoming fundamental biological barriers, with endosomal escape standing as the most critical bottleneck.
The intracellular delivery pathway for mRNA-loaded LNPs follows a multi-step cascade: selective organ-targeting delivery, cellular uptake, endosomal escape, and cytosolic mRNA release (the SCER cascade) [84]. While each step presents challenges, endosomal escape is particularly limiting—estimates suggest that less than 2% of internalized LNPs successfully release their cargo into the cytosol [85]. For cell fate reprogramming, where precise temporal control of protein expression is crucial, this inefficiency can determine therapeutic success or failure. This technical guide examines the mechanisms underlying endosomal escape and provides evidence-based strategies for optimizing LNPs to overcome this critical barrier.
The Fundamental Challenge: Mechanisms and Barriers to Endosomal Escape
The Current Understanding of Escape Mechanisms
Ionizable lipids, the cornerstone of modern LNP formulations, are engineered to undergo protonation in the acidic environment of endosomes (pH ~6.0-6.5) [84] [83]. This pH-dependent ionization is believed to facilitate several interconnected processes:
- Membrane Interaction: Protonated lipids gain positive charges, enabling electrostatic interaction with negatively charged endosomal membrane lipids [86].
- Phase Transition: These interactions may promote a transition from the lamellar endosomal membrane to an inverted hexagonal (HII) phase, creating non-bilayer structures that facilitate membrane disruption [86].
- Lipid Exchange: The acidic environment triggers tight interaction between LNPs and endosomal membranes, potentially enabling lipid exchange and local membrane destabilization [86].
Recent research utilizing galectin-9 as a sensitive biomarker for membrane damage has provided direct visual evidence of LNP-induced endosomal disruption [87] [86]. Live-cell microscopy studies demonstrate that only a fraction of internalized LNPs trigger galectin-9 recruitment, and surprisingly, many damaged endosomes contain little or no RNA cargo, suggesting complex payload partitioning during endosomal maturation [86].
Quantifying the Efficiency Bottleneck
Advanced microscopy techniques have revealed multiple points of inefficiency in the cytosolic delivery pathway. The table below summarizes key quantitative findings from recent studies:
Table 1: Quantified Barriers to LNP-Mediated Cytosolic Delivery
| Barrier Point | Efficiency Metric | Experimental System | Reference |
|---|---|---|---|
| Galectin-9+ membrane damage induction | Only a fraction of internalized LNPs trigger detectable damage | Live-cell imaging of galectin-9 recruitment in cultured cells | [86] |
| RNA cargo presence in damaged endosomes | 67-74% for siRNA-LNPs; ~20% for mRNA-LNPs | Single vesicle tracking and fluorescence quantification | [86] |
| Functional cytosolic release | Estimated <2% of internalized RNA reaches cytosol | Correlation of uptake with protein expression | [85] |
| Ionizable lipid-RNA segregation | Observed in endosomal sorting compartments | Super-resolution microscopy of dual-fluorescent LNPs | [86] |
These findings highlight that endosomal escape inefficiency stems from multiple factors: not all LNPs damage their containing endosomes, not all damaged endosomes contain RNA cargo, and even when release occurs, it may be partial or incomplete.
Optimization Strategies for Enhanced Endosomal Escape
Rational Design of Ionizable Lipids
The chemical structure of ionizable lipids profoundly influences their endosomal escape capability through multiple mechanisms:
pKa Optimization: The apparent pKa of ionizable lipids in LNPs should ideally range between 6.0-7.0 to ensure sufficient protonation in endosomes while maintaining neutral charge in circulation [88] [84]. This precise pKa tuning balances efficient endosomal escape with reduced cytotoxicity and extended circulation time.
Molecular Architecture: Specific structural elements correlate with enhanced performance:
AI-Driven Design: Machine learning approaches now enable rational design of ionizable lipids by predicting both apparent pKa and mRNA delivery efficiency [88]. One recent study virtually screened nearly 20 million potential lipid structures, identifying several that matched or exceeded the performance of benchmark lipids like MC3 and SM-102 [88].
Table 2: Experimentally Validated Ionizable Lipids and Performance Characteristics
| Lipid Name/Class | Key Structural Features | Reported Performance | Reference |
|---|---|---|---|
| DLin-MC3-DMA (MC3) | Linoleyl tails, dimethylamino head | Benchmark for siRNA delivery; moderate mRNA delivery | [83] |
| SM-102 | Heptadecane chain, ether linkages | Superior to MC3 in mRNA vaccines | [88] |
| C12-200 | Ether linkages, cyclic amine head | Effective for intravenous mRNA delivery | [89] |
| L319 | Ester bonds for biodegradability | Improved liver clearance vs MC3 | [83] |
| 306-O12B | O-series with ester bond | Selective liver targeting | [84] |
| AI-designed lipids | Varied, often with benzene rings | Matched or exceeded MC3 performance | [88] |
Advanced Formulation Strategies
Helper Lipid Selection and Ratios
The composition of helper lipids significantly influences endosomal escape efficiency:
DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine): This cone-shaped phospholipid promotes the formation of inverted hexagonal phases that facilitate membrane fusion [89] [83]. Formulations combining cationic lipids like DOTAP with DOPE have demonstrated superior transfection efficiency compared to other helper lipids [89].
Cholesterol: At approximately 30-40 mol% of the formulation, cholesterol enhances LNP stability and facilitates membrane fusion through its effects on membrane fluidity [89] [84].
PEGylated Lipids: Typically comprising 1.5-2 mol% of formulations, PEG-lipids (e.g., DSPE-PEG2000) prevent particle aggregation and reduce macrophage clearance, but excessive PEG content can inhibit cellular uptake and endosomal escape [89] [84].
Buffer Composition and Lyophilization
The buffer environment significantly impacts LNP stability and function:
- Optimal Buffer Systems: Research has identified that sodium potassium magnesium calcium and glucose solution (SPMCG) better preserves LNP integrity and transfection efficiency compared to standard buffers like PBS [89].
- Lyophilization Considerations: Proper freeze-drying with appropriate cryoprotectants can maintain LNP activity after rehydration, enhancing storage stability without compromising endosomal escape capability [89].
Experimental Approaches for Evaluating Endosomal Escape
Direct Visualization Methods
Advanced imaging techniques provide direct assessment of endosomal escape efficiency:
Galectin Recruitment Assays: Transfection with fluorescently tagged galectin-9 (e.g., Gal9-YFP) enables real-time monitoring of endosomal membrane damage [87] [86]. Protocol: Seed cells in glass-bottom dishes, transfect with Gal9-YFP, treat with LNPs, and image using confocal microscopy with time-lapse capability. Damage quantification is based on counting Gal9-positive vesicles per cell.
Dual-Fluorescent LNPs: Co-labeling ionizable lipids (e.g., BODIPY-MC3) and RNA payloads with spectrally distinct fluorophores enables tracking of component segregation during endosomal processing [86].
Super-Resolution Microscopy: Techniques like STORM or STED microscopy resolve LNP-endosome interactions at nanoscale resolution, revealing localization patterns predictive of escape efficiency [86].
Functional Assessment Methods
Dose-Response Analysis: Comparing LNP uptake (measured via fluorescent lipid tags) with functional protein output (via luciferase or fluorescence assays) provides indirect quantification of escape efficiency [85].
ESCRT Machinery Monitoring: Tracking recruitment of ESCRT components (e.g., ALIX, TSG101) to damaged endosomes helps identify non-productive membrane repair pathways that limit escape [86].
The following diagram illustrates the key intracellular barriers and experimental assessment points in the LNP delivery pathway:
The Scientist's Toolkit: Essential Reagents and Methodologies
Table 3: Key Research Reagents for Studying and Optimizing Endosomal Escape
| Reagent/Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Ionizable Lipids | DLin-MC3-DMA, SM-102, C12-200, AI-designed lipids | Core functional component enabling pH-dependent endosomal escape | Optimize pKa (6.0-7.0 range); consider biodegradability via ester linkages |
| Helper Lipids | DOPE, cholesterol | Promote inverted hexagonal phase formation and membrane fusion | DOPE enhances escape; cholesterol optimizes stability (30-40 mol%) |
| PEGylated Lipids | DMG-PEG2000, DSPE-PEG2000 | Stabilize particles, prevent aggregation, modulate pharmacokinetics | Limit to 1.5-2 mol%; higher percentages inhibit cellular uptake |
| Membrane Damage Reporters | Galectin-9-YFP, Galectin-3-mCherry | Visualize and quantify endosomal disruption in live cells | Galectin-9 most sensitive; use time-lapse imaging for kinetics |
| Endosomal Markers | Rab5-GFP (early endosomes), Rab7-GFP (late endosomes) | Identify endosomal maturation stages | Co-localization with LNPs reveals trafficking routes |
| ESCRT Components | ALIX, TSG101 antibodies | Detect membrane repair responses that limit escape | Immunofluorescence or live-cell imaging of tagged proteins |
| Buffer Systems | SPMCG buffer, citrate buffers | Maintain LNP stability and function during formulation | Superior to PBS for preserving transfection efficiency |
| Cryoprotectants | Trehalose, sucrose | Enable lyophilization while maintaining LNP activity | Critical for long-term storage of optimized formulations |
Overcoming the endosomal escape bottleneck requires a multifaceted approach integrating rational lipid design, sophisticated formulation strategies, and rigorous assessment methodologies. The emerging toolkit—from AI-driven lipid discovery to advanced imaging techniques—provides unprecedented capability to optimize this critical delivery step. For cell fate reprogramming applications, where precise control of protein expression dynamics is essential, enhancing endosomal escape efficiency from the current <2% to even modestly higher levels could dramatically expand the therapeutic potential of mRNA-based technologies. Continued focus on understanding the fundamental mechanisms underlying LNP-endosome interactions will enable the design of next-generation delivery systems capable of meeting the exacting demands of cell reprogramming and other advanced therapeutic applications.
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using the Yamanaka factors (OCT4, SOX2, KLF4, and c-Myc, collectively known as OSKM) revolutionized regenerative medicine [90]. However, the clinical application of this technology faces a significant challenge: the fine balance between achieving effective reprogramming and mitigating oncogenic risk. The Yamanaka factors, particularly c-Myc and KLF4, possess inherent oncogenic potential, and prolonged expression can lead to genomic instability and tumor formation [91] [92]. Within the broader context of mRNA-based cell fate reprogramming, managing this risk is paramount. This whitepaper provides an in-depth technical guide to protocols designed to harness the rejuvenative power of OSKM while minimizing oncogenic consequences through transient expression strategies. These approaches are foundational for developing safe and effective therapies for age-related diseases, regenerative medicine, and progeroid syndromes [93] [90] [94].
Oncogenic Mechanisms of Yamanaka Factors
Understanding the specific oncogenic mechanisms of each factor is crucial for designing safer reprogramming protocols.
Table 1: Oncogenic Risk Profiles of Yamanaka Factors
| Factor | Primary Function in Reprogramming | Oncogenic Mechanism | Associated Risks |
|---|---|---|---|
| c-Myc | Master regulator of proliferation; amplifies transcriptional output [91]. | Opens chromatin; drives uncontrolled cell cycle progression; classic oncogene [91] [92]. | Significantly increases tumorigenesis risk; mice transplanted with c-Myc-induced iPSCs developed teratomas [91]. |
| KLF4 | Regulates pluripotency genes like NANOG; context-dependent transcriptional activator/repressor [90] [91]. | Functions as both a tumor suppressor and oncogene; overexpression found in squamous cell cancers [91]. | Contributes to tumorigenesis in specific cellular contexts [91]. |
| OCT4 | Pioneer transcription factor; essential for establishing and maintaining pluripotency [91]. | If persistently expressed, can force cells into a pluripotent state, the precursor to teratoma formation [90] [94]. | OCT4 overexpression is present in various cancers, including ovarian, cervical, and colorectal [91]. |
| SOX2 | Partners with OCT4 to regulate pluripotency genes; critical for neural development [91] [95]. | Forced expression in differentiated cells can destabilize cellular identity and promote oncogenesis. | Expressed in at least 25 cancer types, including breast, gastric, and brain tumors [91]. |
The diagram below illustrates the core-risk relationship in OSKM-mediated reprogramming.
Strategic Approaches for Risk Mitigation
Factor Modification: The OSK vs. OSKM Paradigm
A primary strategy to reduce oncogenic risk involves modifying the factor cocktail itself. The most prominent approach is the exclusion of c-Myc.
- Evidence for OSK Efficacy: Studies have demonstrated that OSK-only reprogramming is sufficient to induce significant epigenetic rejuvenation. In one key study, wild-type mice treated with OSK via an AAV9 delivery system exhibited a 109% extension in remaining lifespan and a reduced frailty index, all without reported teratoma formation [93].
- Trade-offs: While safer, OSK-only reprogramming is generally less efficient than OSKM and may require longer exposure or optimized delivery to achieve comparable rejuvenation effects [92]. The absence of c-Myc's potent chromatin-opening activity can slow the epigenetic resetting process.
Temporal Control: The Critical Role of Transient Expression
The core principle of safe reprogramming is to separate rejuvenation from full dedifferentiation. This is achieved through precise temporal control of factor expression.
Table 2: Quantitative Outcomes of Transient Expression Protocols In Vivo
| Induction Protocol | Model System | Key Outcomes | Tumorigenesis |
|---|---|---|---|
| Cyclic (2-day ON, 5-day OFF) [93] | Progeric LAKI mice | 33% median lifespan increase; restoration of mitochondrial ROS and H3K9me levels. | None reported after 35 cycles. |
| Cyclic (1-day ON, 6-day OFF) with OSK [93] | 124-week-old wild-type mice | 109% remaining lifespan extension; improved frailty index score. | No teratomas reported. |
| Single, short-term (2.5-week pulse) [95] | 2-month-old progeric mice | Increased maximum lifespan by 11 weeks; improved lean mass and motor skills in old age. | Not detected. |
| Continuous low-dose (0.2 mg/ml DOX) [95] | Heterozygous progeric mice | Increased median age of death from 42.6 to 55.6 weeks. | Not detected. |
The workflow for developing a transient protocol is outlined below.
Advanced Delivery Modalities: mRNA and Chemical Reprogramming
The delivery method is critical for ensuring transient expression and avoiding genomic integration.
mRNA-Based Delivery: Synthetic, chemically modified mRNA is a leading platform for safe, transient OSKM delivery [96] [97].
- Mechanism: mRNA is translated directly in the cytoplasm into functional proteins without entering the nucleus, eliminating the risk of insertional mutagenesis [96] [1].
- Key Advance: Incorporation of N1-methylpseudouridine suppresses the innate immune response to exogenous mRNA, enabling robust protein expression without excessive inflammation [1] [97].
- Delivery System: Lipid Nanoparticles (LNPs) protect mRNA from degradation and facilitate efficient cellular uptake [97].
Chemical Reprogramming: This non-genetic approach uses cocktails of small molecules to induce reprogramming, offering a promising alternative to factor-based methods [93].
- Evidence: A "7c" cocktail of small molecules has been shown to rejuvenate aged mouse fibroblasts, resetting epigenetic clocks and improving mitochondrial function [93].
- Safety Profile: Notably, chemical reprogramming with the 7c cocktail upregulated the p53 pathway, a key tumor suppressor, unlike OSKM-mediated reprogramming which often downregulates it to enhance efficiency [93].
The Scientist's Toolkit: Essential Reagents for mRNA-Based Reprogramming
Table 3: Key Research Reagent Solutions for mRNA-Based OSKM Delivery
| Reagent / Technology | Function | Technical Notes |
|---|---|---|
| N1-methylpseudouridine [1] [97] | Nucleoside modification that suppresses innate immune recognition of synthetic mRNA, enhancing stability and translation efficiency. | Critical for reducing immunogenicity and achieving high levels of protein expression. Replaces all uridine in the IVT reaction. |
| Lipid Nanoparticles (LNPs) [96] [97] | Delivery vector that encapsulates and protects mRNA, facilitating cellular uptake and endosomal escape. | Typically composed of ionizable cationic lipids, phospholipids, cholesterol, and PEG-lipids. Tissue-specific targeting remains a research focus. |
| PureCap Technology [97] | mRNA synthesis method that produces highly pure, completely 5'-capped mRNA, avoiding uncapped impurities that trigger immune responses. | Cap2 structures further reduce immunogenicity. Purified capped mRNA shows >10x higher translational activity than standard methods. |
| Non-Replicating mRNA Construct [1] | The therapeutic mRNA molecule, featuring a 5' cap, 5' UTR, OSKM ORF, 3' UTR, and poly(A) tail. | Optimized 5' and 3' UTRs are crucial for ribosome recruitment and translational control. The ORF can be a single polycistronic sequence or separate mRNAs. |
| Tissue Nanotransfection (TNT) [57] | A physical, non-viral delivery platform using nanoelectroporation to locally deliver genetic cargo (mRNA, plasmid) directly into tissues. | Enables in vivo reprogramming with high specificity and minimal off-target effects. Useful for localized rejuvenation therapies. |
Experimental Protocol: A Detailed Methodology for Transient mRNA Transfection
This protocol outlines the process for rejuvenating human dermal fibroblasts using transient, non-integrating mRNA delivery of OSKM factors.
Step 1: mRNA Preparation
- Source: Obtain in vitro transcribed (IVT) mRNA for OCT4, SOX2, KLF4, and c-Myc. Alternatively, use a single polycistronic construct. Ensure all mRNAs are fully capped (preferably Cap1 or Cap2) and incorporate N1-methylpseudouridine to minimize immune activation [1] [97].
- Quality Control: Verify mRNA integrity, purity, and concentration using analytical methods such as UV spectrophotometry and capillary electrophoresis.
Step 2: Cell Culture and Seeding
- Cell Line: Human dermal fibroblasts (HDFs) from a validated source (e.g., ATCC).
- Culture Conditions: Maintain HDFs in standard fibroblast growth medium (e.g., DMEM + 10% FBS) at 37°C with 5% CO₂.
- Seeding for Transfection: One day prior to transfection, seed HDFs at 50-70% confluence in a multi-well plate pre-coated with a suitable extracellular matrix (e.g., Matrigel or Fibronectin) to enhance cell adhesion and health.
Step 3: mRNA Transfection
- Complexation: Complex the OSKM mRNAs with a commercial LNP transfection reagent or a custom formulation optimized for fibroblasts, according to the manufacturer's instructions. A typical ratio of 1-2 µg mRNA per well of a 12-well plate can be used as a starting point.
- Transfection: Replace the cell culture medium with the mRNA-LNP complex. Incubate cells for 4-6 hours [96].
- Pulse Period: This constitutes one "pulse" of OSKM expression.
Step 4: Post-Transfection and Cycling
- Remove Complex: After the pulse period, carefully remove the transfection medium and replace it with fresh fibroblast growth medium.
- Chase Period: Allow the cells to recover for 5 days without any transfection [93]. This "chase" period lets the translated proteins degrade and allows the cells to re-establish their identity in a rejuvenated state.
- Cycle Repetition: For partial reprogramming protocols, repeat the pulse-chase cycle (1-day pulse, 5-day chase) for a predetermined number of cycles (e.g., 4-8 cycles). Monitor cells closely for any signs of morphological change indicative of dedifferentiation.
Step 5: Validation and Safety Assessment
- Efficacy Assays:
- Epigenetic Aging Clocks: Perform DNA methylation analysis (e.g., using Illumina EPIC arrays) and calculate biological age with established clocks (e.g., Horvath's clock) to confirm rejuvenation [93] [94].
- Functional Assays: Measure age-related markers such as mitochondrial respiration (Seahorse Analyzer), senescence-associated beta-galactosidase (SA-β-gal) activity, and transcriptomic analysis (RNA-seq) of age-related gene signatures [93] [94].
- Safety Assays:
- Identity Verification: Use immunocytochemistry and RT-qPCR to confirm the continued expression of key fibroblast markers (e.g., Vimentin, COL1A1) and the absence of pluripotency markers (e.g., NANOG, TRA-1-60) [94].
- Tumorigenicity Test: As a gold-standard safety test, inject ~1 million treated cells into immunodeficient mice and monitor for teratoma formation over several months [90] [94].
- Efficacy Assays:
The path to clinical translation of OSKM reprogramming hinges on the rigorous implementation of safety-first protocols. As detailed in this guide, the oncogenic risk associated with the Yamanaka factors can be effectively managed through a multi-pronged strategy: modifying the factor cocktail, exerting precise temporal control over their expression, and employing non-integrating delivery systems like modified mRNA. The evidence from in vivo models is promising, demonstrating that transient induction can yield significant rejuvenation and lifespan extension without tumor formation. Continued refinement of these protocols, particularly in achieving tissue-specific targeting and standardizing safety assays, will be crucial for advancing mRNA-based cell fate reprogramming from a powerful research tool to a transformative therapeutic modality.
The immense clinical potential of mRNA-based technology for cell fate reprogramming is fundamentally constrained by a single, critical challenge: the need for precision delivery. While mRNA therapeutics can instruct cells to produce therapeutic proteins, their application hinges on the ability to deliver these genetic instructions specifically to the target tissues or organs relevant to the disease. Without this specificity, mRNA nanomedicines face significant drug loss, potential off-target effects, and reduced therapeutic efficacy, as they predominantly accumulate in the liver following systemic administration due to inherent biological barriers [98]. The development of strategies to overcome this hepatic tropism and achieve targeted extrahepatic delivery represents the current frontier in advancing mRNA therapeutics beyond vaccines and liver-directed applications. This guide synthesizes the latest technological advances—including novel nanoparticle designs, targeting ligands, and formulation techniques—that are creating new pathways for tissue-specific mRNA delivery, thereby unlocking the full potential of mRNA for cell fate reprogramming research and therapeutic development.
Core Targeting Strategies and Mechanisms
Achieving tissue-specific delivery requires a multifaceted approach that manipulates the interactions between nanoparticles and biological systems. The following core strategies form the foundation of modern targeted mRNA nanomedicine.
Passive and Endogenous Targeting
Passive targeting leverages the natural physicochemical properties of nanoparticles—such as size, surface charge, and composition—to influence their biodistribution. Nanoparticles in a specific size range (typically 10-150 nm) can preferentially extravasate in tissues with leaky vasculature, such as tumors or sites of inflammation, through the Enhanced Permeability and Retention (EPR) effect [98]. Simultaneously, endogenous targeting exploits the body's own biological "homing" mechanisms. A prime example is the selective organ targeting (SORT) methodology, which deliberately incorporates supplementary "SORT molecules" into traditional lipid nanoparticle (LNP) formulations. By carefully tuning the percentage and type of these molecules (such as cationic, anionic, or ionizable lipids), researchers can systematically shift LNP accumulation from the liver to the lungs, spleen, or other tissues without the need for external targeting ligands [99] [100]. The biochemical basis for this lies in the fact that different lipid compositions recruit distinct arrays of apolipoproteins (Apo) from the blood onto the LNP surface. This formed "biomolecular corona" then determines which cellular receptors the particle engages, thereby dictating its final tissue destination [99].
Active Targeting
Active targeting represents a more direct approach by functionalizing the surface of delivery vehicles with targeting ligands that bind specifically to receptors highly expressed on the surface of target cells [53]. This strategy can be visualized as a key-and-lock mechanism, where the nanoparticle is precisely guided to its intended cellular lock.
Diagram 1: Active targeting utilizes surface ligands to bind cell-specific receptors.
These ligands include antibodies or their fragments, peptides, aptamers, glycans, and small molecules [53] [100]. The conjugation of these ligands can be achieved through various methods, including post-insertion techniques, click chemistry, or the incorporation of pH-sensitive lipids [53]. The primary advantage of active targeting is its potential for high cellular specificity, which can enhance uptake in the target tissue and reduce non-specific delivery.
Table 1: Comparison of Core Targeting Strategies for mRNA Delivery
| Strategy | Mechanism | Key Advantages | Inherent Limitations | Exemplary Technologies |
|---|---|---|---|---|
| Passive Targeting | Leverages NP size/charge for natural biodistribution patterns (e.g., EPR effect in tumors). | Simple formulation; No complex conjugation chemistry required. | Limited targeting precision; Highly dependent on pathophysiology. | Size-tuned LNPs (~100 nm) for tumor accumulation [98]. |
| Endogenous Targeting | Modulates the biomolecular corona (e.g., with apolipoproteins) to exploit natural trafficking pathways. | Does not require external ligands; High potential for clinical translation. | Requires extensive lipid screening; Organ selectivity can be species-dependent. | SORT LNPs [99]; LNP composition tuning (pKa, lipid ratios) [100]. |
| Active Targeting | Uses surface-bound ligands (Abs, peptides, etc.) for specific receptor binding on target cells. | Highest theoretical specificity; Can target specific cell subtypes. | Ligand conjugation can complicate manufacturing and affect LNP stability; Potential for immunogenicity. | Antibody-functionalized LNPs; Peptide-decorated NPs [53] [100]. |
Advanced Delivery Platforms and Formulation Methodologies
The SORT (Selective Organ Targeting) Platform
The SORT platform is a groundbreaking methodology that provides a systematic, rational framework for designing LNPs that target specific tissues. The core protocol involves adding a fifth component—the SORT molecule—to the standard four-component LNP formulation.
Detailed Experimental Protocol for SORT LNP Preparation (Pipette Mixing Method):
- Lipid Stock Solution Preparation: Dissolve the constituent lipids—ionizable lipid (e.g., DLin-MC3-DMA, ALC-0315), phospholipid (e.g., DOPE, DSPC), cholesterol, PEG-lipid (e.g., DMG-PEG2000), and the SORT molecule (e.g., 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) for positive charge, or 1,2-dioleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DOPG) for negative charge)—in pure ethanol. The molar ratios are critical; for example, a 30:30:38:2:10 ratio (ionizable lipid:phospholipid:cholesterol:PEG-lipid:SORT molecule) can be used as a starting point, with the SORT percentage being the key variable for tuning organ selectivity [99].
- Aqueous Phase Preparation: Dilute the mRNA in an aqueous buffer, typically a 50 mM sodium acetate buffer at pH 4.0. This acidic environment is crucial for the subsequent protonation of the ionizable lipid and efficient mRNA encapsulation.
- Nanoparticle Formation: In a tube, rapidly mix the ethanolic lipid solution with the aqueous mRNA solution using a pipette. The sudden change in polarity and pH leads to the spontaneous self-assembly of lipids into nanoparticles, entrapping the mRNA molecules.
- Buffer Exchange and Purification: Dialyze the resulting LNP suspension against a large volume of phosphate-buffered saline (PBS) at pH 7.4 for several hours to remove the ethanol and adjust the pH to physiological conditions. This step also ensures the ionizable lipids deprotonate, reducing systemic toxicity.
- Characterization: Determine the particle size and polydispersity (PDI) using dynamic light scattering (DLS). Measure the encapsulation efficiency of the mRNA using a Ribogreen assay, which quantifies the difference in fluorescence between intact LNPs (where RNA is shielded) and lysed LNPs (where all RNA is exposed) [99].
The biochemical logic of SORT is that incorporating a cationic lipid like DOTAP promotes targeting to the lungs, while anionic lipids shift distribution to the spleen. The specific SORT molecule and its molar ratio directly influence the surface charge and the composition of the protein corona that forms in the blood, thereby dictating organotropism [99] [100].
Novel Nanomaterial Systems
Beyond conventional LNPs, new nanostructural designs are emerging to address the limitations of current delivery systems.
Lipid Nanoparticle Spherical Nucleic Acids (LNP-SNAs): This novel architecture, developed at Northwestern University, involves a core LNP loaded with CRISPR machinery or mRNA, which is then surrounded by a dense shell of DNA. This spherical nucleic acid (SNA) structure is not just a protective coating; it actively promotes cellular uptake through its ability to engage with cell surface receptors more effectively than unadorned LNPs. In tests, LNP-SNAs demonstrated up to three times higher cellular uptake and a corresponding threefold boost in gene-editing efficiency compared to standard LNPs, while also showing reduced toxicity [101]. This highlights the principle that the structure of a nanomaterial, not just its chemical components, is a critical determinant of its delivery potency.
Poly(Beta-Amino Esters) and Hybrid Systems: As alternatives to lipid-based systems, polymeric vectors like poly(beta-amino esters) (PBAEs) are being explored for their tunable biodegradability and potential for enhanced endosomal escape. Furthermore, hybrid systems that combine the favorable properties of lipids and polymers (e.g., lipid-polymer nanoparticles) are under investigation to improve stability, delivery efficiency, and enable tissue-specific targeting [102].
Experimental Workflow for Targeted LNP Development
The journey from concept to a characterized targeted LNP follows a multi-stage pipeline, integrating formulation, characterization, and validation. The following diagram and table outline the key stages and essential reagents for this process.
Diagram 2: Key stages in developing and validating targeted LNPs.
Table 2: The Scientist's Toolkit: Essential Reagents for Targeted LNP Research
| Reagent / Material | Function / Role | Specific Examples |
|---|---|---|
| Ionizable Lipids | pH-dependent charge; core component for mRNA encapsulation and endosomal escape. | DLin-MC3-DMA, ALC-0315, SM-102, L319 [53] [100]. |
| SORT Molecules | Tunes organotropism by modulating LNP surface charge and protein corona. | Cationic: DOTAP; Anionic: DOPG [99]. |
| Helper Phospholipids | Stabilizes LNP bilayer structure; can promote membrane fusion. | DOPE, DSPC [99] [100]. |
| Cholesterol | Modulates membrane fluidity and enhances structural stability of LNPs. | Pharmaceutical grade cholesterol [100]. |
| PEG-Lipids | Reduces nonspecific protein adsorption, improves stability, and prolongs circulation half-life. | DMG-PEG2000, ALC-0159 [99] [53]. |
| Targeting Ligands | Confers active targeting to specific cell surface receptors. | Antibodies/fragments, peptides, aptamers, glycans [53]. |
| mRNA Constructs | The therapeutic cargo; reporter mRNAs are used for screening. | Firefly luciferase (FLuc) mRNA for biodistribution; GFP mRNA for cellular uptake [99]. |
Application in Cell Fate Reprogramming and Disease Modeling
The ability to achieve tissue-specific mRNA delivery is transformative for cell fate reprogramming research, as it enables precise manipulation of gene expression in vivo. This capability moves beyond the traditional paradigm of ex vivo cell engineering.
A powerful application is the restoration of tumor suppressor genes. For instance, in HER2-low breast cancer models, researchers have successfully co-delivered mRNAs encoding the tumor suppressor p53 and the HER2 receptor using LNPs. This dual approach simultaneously restored the function of a critical guardian of the genome and sensitized the tumor cells to existing HER2-targeted therapies like trastuzumab. The mRNA-mediated upregulation of HER2 on the cancer cell surface created a neoantigen-like effect, making the previously resistant tumors vulnerable to antibody-dependent cellular cytotoxicity [103]. This strategy demonstrates how targeted mRNA delivery can be used not just for protein replacement, but for dynamically reprogramming cell identity and therapeutic susceptibility.
Furthermore, the first personalized in vivo CRISPR treatment was successfully administered to an infant with a rare genetic liver disease (CPS1 deficiency). The therapy, delivered via LNP, was developed and approved in just six months, establishing a landmark precedent for rapid, on-demand gene editing. Notably, the use of LNPs allowed for multiple doses to be safely administered to increase the percentage of edited cells—a flexibility not typically possible with viral vectors due to immune concerns [104]. This case underscores the potential of targeted mRNA/LNP systems to enable complex cell fate reprogramming for personalized medicine, moving from "CRISPR for one to CRISPR for all" [104].
The frontier of next-generation delivery systems is defined by an increasing level of control over the in vivo journey of mRNA nanomedicines. The strategies outlined—from the rational design of SORT LNPs to the structural innovation of LNP-SNAs and the precision of active targeting ligands—collectively empower researchers to direct therapeutic cargo to specific organs, tissues, and even cell subtypes. This precision is the key that unlocks the full potential of mRNA technology for cell fate reprogramming, enabling the development of sophisticated therapies for cancer, genetic disorders, and regenerative medicine.
Future progress will be driven by several key factors: the continued development of novel, biodegradable ionizable lipids with improved tissue selectivity; the integration of artificial intelligence and machine learning to predict the performance of novel LNP formulations, thereby accelerating the design cycle [105] [100]; and a deeper understanding of the fundamental interactions between nanoparticles and biological systems, particularly the protein corona. As these areas advance, the vision of delivering bespoke genetic instructions to any cell type in the body to treat or cure disease moves closer to reality, firmly establishing targeted mRNA delivery as a cornerstone of next-generation therapeutics.
Benchmarking mRNA Technology: A Comparative Analysis with CRISPR, RNAi, and Viral Vectors
The advent of genetic vaccine technologies and advanced cell engineering tools has positioned mRNA-based platforms and viral vector systems at the forefront of biomedical innovation. While both platforms achieved remarkable success during the COVID-19 pandemic, their underlying biological mechanisms present distinct safety and immunogenicity profiles that significantly impact their application in cell fate reprogramming research. This technical analysis provides a comprehensive comparison of these platforms, focusing on three critical parameters: genomic integration potential, immunogenicity, and clinical safety. Understanding these differences is paramount for researchers designing therapeutic strategies, particularly in the context of cellular reprogramming where genomic integrity and precise immune modulation determine translational success.
Table 1: Fundamental Characteristics of mRNA and Viral Vector Platforms
| Parameter | mRNA Platforms | Viral Vector Platforms |
|---|---|---|
| Genetic Material | Messenger RNA (mRNA) [106] | DNA (Adenovirus, AAV, Lentivirus) [106] [107] |
| Mechanism of Action | Direct protein translation in the cytoplasm [106] | Host cell nuclear entry and transcription [106] |
| Intracellular Duration | Transient (hours to days) [57] | Prolonged (weeks to months, potentially persistent) [108] |
| Genomic Integration | Non-integrating [60] [109] | Possible, depending on vector (e.g., Lentivirus); Adenovirus typically non-integrating [107] |
Mechanisms of Action and Genomic Integration
mRNA Platform Mechanism
mRNA vaccines and therapeutics function by introducing messenger RNA into the cell's cytoplasm, where it is directly translated into the target protein by ribosomes without any nuclear entry [106]. This protein then triggers the desired immune response or therapeutic effect. The mRNA molecule is typically encapsulated within lipid nanoparticles (LNPs) that protect it from degradation and facilitate cellular uptake through endocytosis. Once inside the cell, the LNP must escape the endosome to release the mRNA into the cytoplasm for translation [109]. This process is inherently non-integrating, as the genetic material never enters the nucleus and is degraded by normal cellular processes within days [60] [109]. The transient nature of this expression is a key safety feature for cell reprogramming applications, minimizing the risk of permanent genetic alterations.
Viral Vector Platform Mechanism
Viral vector platforms utilize engineered viruses, such as adenoviruses or adeno-associated viruses (AAVs), to deliver genetic material encoding a target antigen. These vectors are rendered replication-incompetent by deleting essential viral genes. Unlike mRNA platforms, viral vectors deliver DNA sequences to the host cell [106]. Following cellular entry, the genetic material is transported to the nucleus where it exists as an episome (for adenoviruses) or, in the case of some vectors like lentiviruses, can potentially integrate into the host genome [107]. This nuclear entry and potential for integration enables more sustained antigen expression but introduces the risk of insertional mutagenesis, where the integration disrupts normal host genes or regulatory elements, potentially leading to oncogenesis [107].
Figure 1: Comparative intracellular pathways of mRNA and viral vector platforms, highlighting key differences in genomic integration risk.
Immunogenicity Profiles
The immune response elicited by vaccine platforms is a critical determinant of both their efficacy and safety. mRNA and viral vector platforms trigger distinct innate and adaptive immune responses, which can be leveraged for different applications in cell reprogramming and therapeutic development.
Innate Immune Recognition
mRNA platforms can be recognized by various pattern recognition receptors (PRRs), including endosomal Toll-like receptors (TLR3, TLR7, TLR8) and cytosolic receptors (RIG-I), which detect the foreign RNA and trigger interferon (IFN) responses [109]. While this can enhance immunogenicity, excessive IFN signaling can inhibit mRNA translation. To mitigate this, mRNA platforms incorporate nucleoside modifications (e.g., pseudouridine) that reduce PRR recognition while enhancing protein expression [109]. Alternatively, some platforms use sequence engineering to deplete uridines, thereby minimizing immune recognition [109].
Viral vector platforms, particularly those based on adenovirus, trigger robust innate immunity through recognition of the viral capsid and viral-associated molecular patterns. This can lead to significant inflammatory responses and the production of pro-inflammatory cytokines [108]. A major limitation of viral vectors is preexisting immunity, where individuals with prior exposure to the viral vector have neutralizing antibodies that can reduce vaccine efficacy [108]. This is a significant consideration for therapeutic applications requiring repeated administration.
Adaptive Immune Responses
Comparative studies reveal distinct patterns in the adaptive immune responses generated by each platform. A 2024 randomized trial comparing a self-amplifying mRNA vaccine (ARCT-154) with the ChAdOx1-S adenovirus-vector vaccine found that the mRNA vaccine induced higher neutralizing antibody responses that persisted for one year post-vaccination [110]. This enhanced immunogenicity was associated with generally higher efficacy against COVID-19 [110].
Table 2: Comparative Immunogenicity and Clinical Safety Data
| Parameter | mRNA Platform (Comirnaty) | Viral Vector Platform (Vaxzevria) | Inactivated Vaccine (CoronaVac) |
|---|---|---|---|
| Most Frequent Local AE (1st Dose) | Pain (87.4%) [111] | Pain (84.4%) [111] | Pain (69.1%) [111] |
| Most Frequent Systemic AEs (1st Dose) | Fatigue (56.9%), Myalgia (37.2%), Fever (17.5%) [111] | Fever (76.7%), Headache (58.9%), Myalgia (53.3%) [111] | Fatigue (49.1%) [111] |
| Average Number of AEs (1st Dose) | Not specified | 6 [111] | 3 [111] |
| Neutralizing Antibody Persistence | Higher responses persisting to one year [110] | Lower comparative responses [110] | Not specified in dataset |
| Impact of Preexisting Immunity | No significant reduction in efficacy [108] | Reduced efficacy in seropositive individuals [108] | Not applicable |
Clinical Safety and Adverse Event Profiles
Large-scale clinical trials and post-marketing surveillance have provided comprehensive safety data for both platform technologies, revealing distinct adverse event profiles that inform their risk-benefit assessment.
Local and Systemic Reactogenicity
A 2025 prospective longitudinal study in Malaysia directly compared adverse events following primary and booster doses of different COVID-19 vaccines. The study found that after the first dose, Vaxzevria (viral vector) recipients reported the highest number of average adverse events (n=6), including a high incidence of fever (76.7%) and headache (58.9%) [111]. In contrast, Comirnaty (mRNA) recipients reported fewer systemic events initially, though these increased after the booster dose [111]. Both platforms demonstrated predominantly mild-to-moderate adverse events that were transient and resolved within 1-2 days [111] [110].
Lymph Node Reactivity and Imaging Considerations
From a diagnostic perspective, a 2023 study investigating the impact of COVID-19 vaccination on 18F-FDG PET/CT imaging found that ipsilateral axillary lymph node reactivity was a common finding after both vaccine types [112]. However, this reaction was more pronounced after mRNA vaccination, with a prevalence of 61% on semiquantitative assessment compared to 47.6% after viral vector vaccination [112]. This heightened reactivity indicates a more robust immune activation in regional lymph nodes following mRNA vaccination and must be considered when interpreting diagnostic imaging to avoid false-positive cancer diagnoses [112].
Implications for Cell Fate Reprogramming Research
The distinct profiles of mRNA and viral vector technologies have significant implications for their application in cell reprogramming and regenerative medicine, where genomic integrity and controlled immunogenicity are paramount.
Safety Advantages of mRNA in Cellular Reprogramming
The non-integrative nature of mRNA platforms presents a substantial safety advantage for cell fate reprogramming applications. Research at Harvard has demonstrated that using synthetic mRNA to reprogram human adult skin cells into induced pluripotent stem cells (iPSCs) avoids the risk of insertional mutagenesis associated with viral vector methods [60]. This mRNA-based approach achieved reprogramming efficiencies of 1-4% of starting cells—orders of magnitude higher than conventional viral methods—while maintaining genomic integrity [60]. Furthermore, mRNA technology enables direct programming of iPSCs into specific cell types without compromising the host genome, providing a safer paradigm for regenerative medicine [60].
Delivery System Innovations
Both platforms require sophisticated delivery systems to achieve efficient cellular transfection. For mRNA platforms, lipid nanoparticles (LNPs) have emerged as the dominant delivery vehicle, with components including ionizable lipids, phospholipids, cholesterol, and PEG-lipids that work synergistically to protect mRNA and facilitate cellular uptake [109] [107]. Viral vectors continue to leverage the natural infectivity of engineered viruses but face challenges including immune recognition and manufacturing complexity [107]. Emerging non-viral delivery technologies such as exosome-based delivery and virus-like particles (VLPs) offer promising alternatives with potentially improved safety profiles [107].
Table 3: The Scientist's Toolkit: Key Research Reagents for mRNA and Viral Vector Studies
| Research Reagent | Platform | Function/Application | Key Characteristics |
|---|---|---|---|
| Lipid Nanoparticles (LNPs) | mRNA | mRNA encapsulation and delivery [109] [107] | Ionizable lipid, phospholipid, cholesterol, PEG-lipid components [107] |
| Nucleoside-Modified mRNA | mRNA | Enhanced translation and reduced immunogenicity [109] | Incorporation of pseudouridine or N1-methylpseudouridine [109] |
| Adeno-Associated Viruses (AAVs) | Viral Vector | In vitro and in vivo gene delivery [107] | Minimal immunogenicity; broad tissue targeting; sustained expression [107] |
| Lentiviral Vectors | Viral Vector | Delivery to dividing and non-dividing cells [107] | Potential for genomic integration; stable long-term expression [107] |
| Polyethyleneimine (PEI) | Non-Viral Transfection | mRNA complexation and endosomal escape [107] | High positive charge density; can be modified to reduce cytotoxicity [107] |
| Tissue Nanotransfection (TNT) | Non-Viral Delivery | In vivo cellular reprogramming via nanoelectroporation [57] | Non-viral, high specificity, minimal cytotoxicity [57] |
The head-to-head comparison of mRNA and viral vector technologies reveals a complex landscape where neither platform is universally superior. The choice between platforms must be guided by the specific application, target population, and therapeutic goals. mRNA platforms offer distinct advantages in safety profile due to their non-integrating mechanism and reduced risk of insertional mutagenesis, making them particularly suitable for cell fate reprogramming applications where genomic integrity is paramount. Their ability to avoid preexisting immunity issues further enhances their utility for therapeutic strategies requiring repeated administration. Viral vector platforms provide the benefit of sustained antigen expression from a single dose but face challenges related to preexisting immunity and potential inflammatory responses. The ongoing innovation in delivery systems, particularly in non-viral approaches such as LNPs and tissue nanotransfection, continues to enhance the safety and efficacy of both platforms. For researchers in cell reprogramming and drug development, these comparative insights provide a critical foundation for selecting appropriate platform technologies that balance immunogenicity, safety, and efficacy for specific therapeutic applications.
The precision control of gene expression is a cornerstone of modern biological research and therapeutic development. Within cell fate reprogramming, two dominant paradigms have emerged: transient mRNA-based knockdown and permanent CRISPR-Cas9-mediated knockout. This technical guide delineates the fundamental mechanisms, experimental parameters, and application landscapes of these technologies. While mRNA knockdown techniques offer reversible gene suppression at the transcriptional level, CRISPR-Cas9 knockout provides permanent gene disruption at the DNA level. Framed within the context of mRNA-based technology for regenerative medicine, this review provides researchers with a structured comparison to inform methodological selection for specific experimental and therapeutic objectives.
The ability to precisely manipulate gene expression has revolutionized functional genomics and therapeutic development, particularly in the field of cell fate reprogramming. Gene knockdown and knockout represent two fundamental approaches to studying gene function by reducing or eliminating gene expression [113]. Knockdown, typically achieved through mRNA-based technologies like RNA interference (RNAi), results in the temporary inactivation of a particular gene product by targeting its mRNA [114]. In contrast, knockout, facilitated by CRISPR-Cas9 gene editing, involves the complete and permanent removal or disruption of target genes [114] [115].
The emergence of mRNA as a versatile tool for directing cellular behavior has introduced new possibilities for regenerative medicine [32]. Unlike traditional gene therapy approaches, mRNA therapeutics offer a non-integrative and controllable strategy for expressing therapeutic proteins or modulating gene expression [32] [116]. When contrasted with the permanent editing capabilities of CRISPR-Cas9, these technologies represent complementary approaches with distinct advantages for different research and clinical scenarios, particularly in the context of cell reprogramming strategies aimed at tissue regeneration and organ function restoration.
Molecular Mechanisms and Technologies
mRNA-Based Knockdown (RNA Interference)
RNA interference (RNAi) is a natural cellular mechanism that has been harnessed for targeted gene knockdown. The process involves small RNA molecules, such as small interfering RNA (siRNA) or microRNA (miRNA), that mediate sequence-specific gene silencing at the post-transcriptional level [117].
Mechanism of Action: The RNAi pathway begins with the introduction of double-stranded RNA (dsRNA) into the cell. This dsRNA is recognized and cleaved by the endonuclease Dicer into smaller fragments of approximately 21 nucleotides in length. These small RNAs then associate with the RNA-induced silencing complex (RISC), where the antisense strand is separated and used to guide the complex to complementary mRNA sequences. Once bound, the Argonaute protein within RISC cleaves the target mRNA, preventing translation of the encoded protein. If the match between the siRNA/miRNA and the target mRNA is imperfect, translation may be stalled without mRNA degradation, as the RISC complex physically blocks ribosomal progression [117].
Gene knockdown is considered a form of post-transcriptional regulation of gene expression that results in abortive protein translation and degradation of the target mRNA [114]. The effect is transient and reversible, making it particularly valuable for studying essential genes where permanent disruption would be lethal [113].
CRISPR-Cas9-Mediated Gene Knockout
CRISPR-Cas9 is an adaptive immune system from prokaryotes that has been repurposed as a highly precise genome editing tool. It enables permanent gene knockout through direct modification of the DNA sequence [115].
Mechanism of Action: The CRISPR-Cas9 system consists of two core components: a guide RNA (gRNA) and the Cas9 nuclease protein. The gRNA is a synthetic RNA composed of a CRISPR RNA (crRNA) region that specifies the target DNA sequence through complementary base pairing, and a trans-activating CRISPR RNA (tracrRNA) that serves as a binding scaffold for the Cas9 nuclease [115]. The Cas9 protein remains inactive without gRNA association [115].
The genome editing mechanism involves three sequential steps: recognition, cleavage, and repair. The gRNA directs Cas9 to the target DNA sequence, which must be adjacent to a Protospacer Adjacent Motif (PAM) sequence (5'-NGG-3' for the commonly used SpCas9). Upon PAM recognition and target sequence verification, Cas9 undergoes conformational changes that activate its nuclease domains. The HNH domain cleaves the complementary DNA strand, while the RuvC domain cleaves the non-complementary strand, resulting in a double-strand break (DSB) 3 base pairs upstream of the PAM site [115].
Cellular repair mechanisms then address this DSB primarily through the error-prone non-homologous end joining (NHEJ) pathway. NHEJ frequently introduces small insertions or deletions (indels) at the cleavage site. When these indels are not multiples of three nucleotides, they cause frameshift mutations that disrupt the reading frame, leading to premature termination codons and complete gene disruption [118]. The resulting mutant mRNA may also be degraded by nonsense-mediated decay, preventing production of functional protein [119].
Table 1: Core Components and Mechanisms
| Feature | mRNA Knockdown (RNAi) | CRISPR-Cas9 Knockout |
|---|---|---|
| Target Molecule | mRNA | DNA |
| Core Components | siRNA/miRNA, Dicer, RISC complex | gRNA, Cas9 nuclease |
| Mechanism | mRNA degradation or translational inhibition | DNA double-strand break and error-prone repair |
| Genetic Alteration | None | Permanent sequence modification |
| Key Enzymes | Dicer, Argonaute | Cas9 nuclease |
| Cellular Process | Post-transcriptional regulation | DNA damage repair |
Experimental Workflows and Methodologies
RNAi Knockdown Experimental Protocol
The implementation of an RNAi knockdown experiment follows a systematic workflow:
Design of RNAi Triggers: The first critical step involves designing highly specific siRNAs or miRNAs that target only the intended genes. Bioinformatics tools are used to select sequences with maximal target specificity and minimal off-target potential. Optimal siRNA designs typically include 19-21 nucleotide duplexes with specific thermodynamic properties and sequence characteristics to enhance RISC loading and target engagement [117].
Delivery into Cells: Designed RNAi triggers are introduced into cells using various methods. Options include:
- Synthetic siRNA duplexes transfected using lipid-based reagents
- Plasmid vectors expressing short hairpin RNAs (shRNAs)
- PCR products or in vitro transcribed siRNAs
- Viral vectors for stable expression in hard-to-transfect cells The relatively small size and RNA nature of these components simplify delivery, as animal cells possess the endogenous Dicer and RISC machinery necessary for the process [117].
Efficiency Validation: Knockdown efficiency is typically assessed 24-72 hours post-transfection using:
- Quantitative RT-PCR to measure reduction in target mRNA levels
- Immunoblotting to detect decreases in protein expression
- Immunofluorescence for spatial assessment of protein reduction
- Phenotypic monitoring for expected functional consequences [117]
The entire process from design to validation typically requires 3-7 days, with effects lasting from several days to a week, depending on the cell type and delivery method.
CRISPR-Cas9 Knockout Experimental Protocol
CRISPR-Cas9 knockout generation follows a distinct workflow with permanent outcomes:
gRNA Design and Selection: This crucial step involves designing guide RNAs that target specific genomic loci with high efficiency and minimal off-target effects. Computational tools identify target sequences matching the 5'-N(20)NGG-3' pattern near the 5' end of the coding sequence to maximize disruption. Multiple gRNAs are often designed and screened to identify the most effective ones [115] [117].
Component Delivery: CRISPR components are delivered to cells in various formats:
- Plasmid vectors encoding both gRNA and Cas9 nuclease
- In vitro transcribed (IVT) RNAs for gRNA and Cas9
- Ribonucleoprotein (RNP) complexes of synthetic gRNA and purified Cas9 protein
- Viral vectors (lentivirus, AAV) for specific applications The RNP format is increasingly preferred due to high editing efficiency, reduced off-target effects, and immediate nuclease activity without transcription/translation requirements [117].
Editing and Validation: Following delivery (24-72 hours), editing efficiency is analyzed using:
- Tracking of Indels by Decomposition (ICE) or T7E1 assays for preliminary efficiency assessment
- Sanger sequencing of cloned PCR products to characterize specific mutations
- Next-generation sequencing for comprehensive editing profile
- Western blotting and functional assays to confirm protein loss and phenotypic consequences [117]
Clonal Selection: For stable cell lines, single-cell cloning is performed after CRISPR treatment, followed by expansion and genotyping to identify homozygous knockout clones. This process typically requires 4-8 weeks from initial transfection to validated clonal lines.
Table 2: Experimental Workflow Comparison
| Experimental Stage | RNAi Knockdown | CRISPR-Cas9 Knockout |
|---|---|---|
| Design Phase | siRNA/miRNA design (3-5 days) | gRNA design and selection (5-7 days) |
| Delivery Format | Synthetic siRNA, shRNA vectors | Plasmids, IVT RNA, RNP complexes |
| Time to Effect | 24-48 hours | 24-72 hours (initial editing) |
| Validation Methods | qRT-PCR, Western blot | ICE assay, Sequencing, Western blot |
| Clonal Isolation | Not typically required | 4-8 weeks for stable lines |
| Effect Duration | Transient (3-7 days) | Permanent |
Technical Considerations and Applications
Advantages and Limitations
RNAi Knockdown Advantages:
- Reversibility: Temporary effects allow study of essential genes without lethal consequences [113]
- Rapid Implementation: Faster from design to results compared to CRISPR
- Safety Profile: No permanent genomic alterations, making it suitable for therapeutic applications where transient modulation is desired
- Dose Titration: Ability to achieve partial knockdown for studying gene dosage effects [117]
RNAi Knockdown Limitations:
- Incomplete Silencing: Residual protein expression may persist, potentially confounding phenotypic interpretation [113]
- Off-Target Effects: Sequence-dependent off-targeting remains challenging, with siRNAs potentially regulating non-target mRNAs with partial complementarity [117]
- Transient Nature: Requires repeated administration for long-term studies
- Interferon Response: Possible activation of innate immune responses in certain cell types [117]
CRISPR-Cas9 Knockout Advantages:
- Complete and Permanent: Ensures full gene disruption, eliminating confounding effects from residual protein expression [117]
- High Specificity: Advanced design tools and modified gRNAs have substantially reduced off-target effects compared to RNAi [117]
- Versatility: Platform can be adapted for knockin, base editing, and transcriptional regulation beyond simple knockout
- Stable Cell Lines: Once established, knockout lines provide consistent models for long-term studies
CRISPR-Cas9 Limitations:
- Knockout Escape: Functional residual proteins can be generated through alternative splicing or translation reinitiation, occurring in up to one-third of knockout attempts [119]
- Essential Gene Lethality: Complete knockout of essential genes may preclude functional study
- Technical Complexity: More labor-intensive protocol requiring specialized expertise
- Compensatory Mechanisms: Permanent knockout may trigger adaptive responses that mask true phenotypic consequences [113]
Applications in Cell Reprogramming and Regenerative Medicine
Both technologies serve distinct roles in cell fate reprogramming and regenerative medicine applications:
mRNA Reprogramming Applications:
- Induced Pluripotent Stem Cell (iPSC) Generation: mRNA-based delivery of reprogramming factors (Oct4, Sox2, Klf4, c-Myc) avoids genomic integration, enhancing safety profile [120] [116]
- Direct Lineage Conversion: mRNA-mediated expression of transcription factors enables transdifferentiation without pluripotent intermediate [57]
- Partial Reprogramming: Transient expression of reprogramming factors can reverse aging-related changes without altering cell identity [57]
- Therapeutic Protein Expression: mRNA delivery of regenerative factors (e.g., VEGF-A) promotes tissue repair in cardiac, liver, and epithelial regeneration [32] [116]
CRISPR-Cas9 Applications:
- Functional Genomics: Genome-wide knockout screens identify genes essential for reprogramming efficiency [115]
- Disease Modeling: Introduction of pathogenic mutations or correction of disease-causing variants in patient-derived cells [115] [118]
- Barrier Elimination: Knockout of developmental checkpoints that constrain cell fate transitions
- Safe Harbor Integration: Precise insertion of reprogramming cassettes into genomic safe harbor sites [118]
Table 3: Application in Reprogramming and Regenerative Medicine
| Application | mRNA Approach | CRISPR-Cas9 Approach |
|---|---|---|
| iPSC Generation | Non-integrating, transient factor expression | Permanent modification of endogenous pluripotency genes |
| Direct Reprogramming | Transient expression of lineage-specific factors | Stable activation of endogenous master regulators |
| Disease Modeling | Limited to gain-of-function or dominant-negative effects | Precise introduction or correction of disease mutations |
| Therapeutic Development | Protein replacement, vaccination, transient modulation | Gene therapy for monogenic disorders, engineered cell therapies |
| Study of Essential Genes | Partial knockdown to study dosage effects | Conditional knockout systems for spatial/temporal control |
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagent Solutions
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| RNAi Triggers | Synthetic siRNA, shRNA vectors, miRNA mimics | Induce sequence-specific mRNA degradation for gene knockdown |
| CRISPR Components | Synthetic gRNA, Cas9 expression vectors, RNP complexes | Facilitate targeted DNA cleavage for permanent gene knockout |
| Delivery Vehicles | Lipid nanoparticles, Electroporation systems, Viral vectors | Enable intracellular delivery of nucleic acids and proteins |
| Validation Tools | qPCR assays, Western blot antibodies, ICE analysis kits | Confirm editing efficiency and target modulation |
| Reprogramming Factors | mRNA encoding OSKM factors, CRISPRa systems | Enable cell fate conversion and pluripotency induction |
| Cell Culture Supplements | Reprogramming-enhancing small molecules, Culture matrices | Support cell survival and efficiency during reprogramming |
The strategic selection between mRNA-based knockdown and CRISPR-Cas9-mediated knockout technologies depends fundamentally on the specific research question, desired persistence of effect, and safety considerations. For regenerative medicine applications requiring transient modulation and high safety profiles, mRNA technologies offer distinct advantages. Conversely, for fundamental investigations of gene function requiring complete and permanent disruption, CRISPR-Cas9 remains the superior choice.
Future directions will likely see increased integration of both technologies, leveraging the precision of CRISPR for genomic modifications combined with the transient controllability of mRNA for fine-tuning cellular behavior. As both platforms continue to evolve—with advances in mRNA chemistry delivery and CRISPR specificity—their synergistic application will undoubtedly accelerate both basic research and clinical translation in cell fate reprogramming and regenerative medicine.
The programming of cellular identity is a cornerstone of modern regenerative medicine and drug development. Within this field, two RNA-based technologies—mRNA reprogramming and RNA interference (RNAi)—serve distinct but complementary roles. mRNA reprogramming involves the delivery of in vitro transcribed messenger RNA (mRNA) that encodes for transcription factors or other proteins to directly alter a cell's gene expression network and fate [96]. In contrast, RNAi is a powerful gene silencing mechanism that utilizes small RNA molecules like small interfering RNA (siRNA) to selectively degrade target messenger RNAs, thereby preventing the synthesis of specific proteins [121] [122]. While mRNA technology aims to add new functions by expressing proteins, RNAi aims to subtract existing functions by knocking down gene expression.
The specificity of these tools—their ability to target only the intended genetic elements—and their potential for off-target effects—unintended modulation of non-target genes—are critical parameters that dictate their experimental utility and therapeutic safety. For researchers engineering cell fates, understanding this balance is essential for robust experimental design and accurate data interpretation. This review provides a technical comparison of these platforms, focusing on their molecular mechanisms, specificity profiles, and practical implementation for cell reprogramming applications.
Molecular Mechanisms and Specificity Profiles
mRNA Reprogramming: A Gain-of-Function Approach
mRNA reprogramming operates on a straightforward principle: the delivery of synthetic mRNA into the cytoplasm, where it is translated into protein using the host cell's native machinery. This process bypasses the need for nuclear entry or genomic integration, leading to transient but high-level protein expression. The core workflow involves transcribing mRNA in vitro from a DNA template encoding the reprogramming factor, modifying the nucleosides to reduce innate immune recognition, and then delivering the purified mRNA into target cells, often using lipid nanoparticles (LNPs) [96].
The specificity of mRNA reprogramming is primarily determined by the protein it encodes. For instance, delivering mRNA for the transcription factor MyoD directs cells toward a myogenic lineage, while mRNA for Ngn2, Pdx1, and MafA can reprogram cells into insulin-producing β-like cells [96]. The off-target effects associated with this method are less about mistargeting other RNA sequences and more related to the broad regulatory networks that the translated transcription factors influence. A key advantage is the minimal risk of genomic integration, as mRNA does not enter the nucleus, thus avoiding insertional mutagenesis [96]. However, a significant challenge is the potential to activate the cell's innate immune system. Unmodified mRNA can be recognized by pattern recognition receptors, triggering an antiviral response and leading to reduced translation and cell death [96]. This is commonly mitigated through nucleoside modifications (e.g., pseudouridine, N1-methylpseudouridine) and sophisticated mRNA sequence engineering [96].
RNAi: A Loss-of-Function Approach with Precision Silencing
RNAi, particularly via siRNA, mediates post-transcriptional gene silencing with high sequence specificity. The mechanism begins when a synthetic, double-stranded siRNA duplex is introduced into the cell cytoplasm. The RNA-induced silencing complex (RISC) loads the siRNA and discards the passenger strand. The guide strand then directs RISC to a perfectly complementary messenger RNA target. The Argonaute-2 (Ago-2) protein within RISC cleaves the target mRNA, preventing its translation and leading to its degradation [123] [124]. This catalytic cycle allows a single RISC complex to degrade multiple mRNA molecules.
The specificity of RNAi is conferred by the Watson-Crick base pairing between the siRNA guide strand and its target mRNA. However, a major challenge is off-target effects. These can occur if the guide strand exhibits partial complementarity, especially in the "seed region" (nucleotides 2-7), to non-target mRNAs. This can lead to their degradation or translational repression, mimicking the effect of endogenous microRNAs [123] [124]. Strategies to minimize this include careful siRNA design using computational algorithms (e.g., BLAST searches, machine learning models), chemical modifications like 2'-O-methylation, and creating asymmetric duplexes that favor loading of the correct guide strand [123].
Table 1: Core Mechanisms and Specificity Determinants of mRNA Reprogramming and RNAi
| Feature | mRNA Reprogramming | RNAi (siRNA) |
|---|---|---|
| Primary Goal | Induce new cellular function/identity | Knock down existing gene expression |
| Molecular Action | Protein translation in the cytoplasm | mRNA degradation in the cytoplasm |
| Key Specificity Determinant | Functional domain of the translated protein | Nucleotide sequence of the guide strand |
| Primary Off-Target Risks | • Ectopic activity of translated factor• Activation of innate immune receptors | • Seed-based off-targeting (miRNA-like effects)• Saturation of endogenous RNAi machinery |
| Common Mitigation Strategies | • Nucleoside modification (e.g., pseudouridine)• Codon optimization• Controlled, pulsed delivery | • Advanced siRNA design algorithms• Chemical modifications (2'-OMe, LNA)• Asymmetric strand design |
Diagram 1: Core mechanistic workflows of mRNA reprogramming and RNAi (siRNA).
A Side-by-Side Comparison of Off-Target Effects
A systematic comparison of off-target effects is crucial for selecting the appropriate technology for a reprogramming application. The nature, consequences, and mitigation strategies for these effects differ significantly between the two platforms.
mRNA Reprogramming Off-Target Effects: The primary risk stems from the potent, pleiotropic actions of the translated proteins. For example, transcription factors used for reprogramming can bind to thousands of genomic sites, potentially activating or repressing non-target genes and leading to incomplete reprogramming or the emergence of aberrant cell types [96]. Furthermore, the intrinsic immunogenicity of exogenous RNA can trigger a Type I interferon response, causing global translational shutdown and apoptosis, which is counterproductive to reprogramming [96]. Mitigation relies on suppressing this immune recognition through nucleoside modifications and optimized purification.
RNAi Off-Target Effects: The main issue is sequence-based cross-talk. Due to the central role of the seed region, a single siRNA can inadvertently regulate hundreds of non-target transcripts, creating a "miRNA-like" signature that can confound phenotypic interpretation [123] [124]. At high concentrations, siRNAs can also saturate the endogenous RNAi machinery, particularly the Exportin-5 pathway, disrupting the processing and function of natural microRNAs and leading to widespread cellular dysfunction [123].
Table 2: Comprehensive Analysis of Off-Target Effects and Mitigation
| Parameter | mRNA Reprogramming | RNAi (siRNA) |
|---|---|---|
| Molecular Origin of Off-Targets | Protein-DNA/protein-protein interactions of the translated factor | Partial complementarity of siRNA guide strand to non-target mRNAs |
| Primary Consequence | • Aberrant differentiation• Uncontrolled cell fate changes | • False-positive/negative phenotypic data• Misinterpretation of gene function |
| Immune Activation | High risk (without modification) due to recognition by TLRs and RIG-I | Lower risk, but possible with certain sequences or delivery vehicles |
| Kinetics of Effect | Off-target protein activity is transient, ceasing as mRNA degrades | Off-target silencing can be long-lasting due to the catalytic nature of RISC |
| Key Mitigation Strategies | • Use of modified nucleosides (pseudouridine)• Codon/UTR optimization• Pulsed, low-dose delivery regimens | • Seed region optimization in design• Chemical modifications (2'-OMe)• Pooled siRNA or deconvolution screens |
Experimental Protocols for Assessing Specificity in Reprogramming Studies
To ensure the validity of reprogramming experiments, rigorous controls and validation assays are non-negotiable. Below are detailed protocols for profiling the specificity of each technology.
Protocol for mRNA Reprogramming Specificity Assessment
This protocol is designed to confirm on-target activity and identify immune activation or aberrant differentiation.
Cell Transfection & Sampling:
- Transfert cells with the modified mRNA using a preferred method (e.g., Lipid Nanoparticles, electroporation). Include a control group transfected with a non-reprogramming mRNA (e.g., GFP mRNA).
- Collect cell pellets and supernatant at 6h, 24h, 48h, and 96h post-transfection.
On-Target Efficiency Analysis:
- Quantitative PCR (qPCR): At each time point, extract total RNA and perform reverse transcription. Use qPCR to measure the expression of canonical markers of the target cell lineage (e.g., INS for β-cells, TNNT2 for cardiomyocytes). Calculate fold-change relative to the GFP control.
- Immunocytochemistry/Western Blot: Confirm the presence and intracellular localization of the translated reprogramming protein using specific antibodies. Quantify the percentage of positively staining cells.
Immune Response Profiling:
- Using the same cDNA, perform qPCR for interferon-stimulated genes (ISGs) such as IFIT1, OAS1, and MX1. Compare Ct values to the GFP control to assess immune activation.
- Analyze the cell culture supernatant for secreted interferon-beta using a commercial ELISA kit.
Off-Target Cell Fate Analysis:
- At day 7-10, perform high-content imaging or flow cytometry to analyze the expression of markers for unintended lineages. For example, if reprogramming toward neurons, also check for mesodermal or endodermal markers.
Protocol for RNAi Specificity Assessment in a Reprogramming Context
This protocol focuses on confirming specific gene knockdown and identifying transcriptome-wide off-targets.
Cell Transfection & Experimental Design:
- Transfert cells with the target siRNA, a non-targeting siRNA (scrambled control), and if possible, a second, independent siRNA targeting the same gene (to control for seed-based off-targets).
- Use a validated transfection reagent (e.g., RNAiMAX, Lipofectamine 2000) at a concentration that achieves >70% knockdown efficiency with minimal cytotoxicity.
On-Target Knockdown Validation:
- qPCR: 48 hours post-transfection, extract RNA and perform qPCR to measure the mRNA level of the target gene. Normalize to a housekeeping gene (e.g., GAPDH, ACTB) and calculate % knockdown relative to the scrambled control. Efficiency should be >70%.
- Western Blot: 72-96 hours post-transfection, analyze protein lysates to confirm reduction of the target protein.
Genome-Wide Off-Target Screening:
- RNA Sequencing (RNA-seq): Perform total RNA-seq on triplicate samples of cells treated with the target siRNA and the scrambled control.
- Bioinformatic Analysis:
- Map sequencing reads to the reference genome and quantify gene expression.
- Perform differential gene expression analysis (e.g., using DESeq2). Genes significantly downregulated in the target siRNA sample versus control are candidate off-targets.
- Cross-reference the list of significantly downregulated genes with in silico predictions of off-targets for the siRNA guide strand using tools like BLAST or specialized siRNA design software.
Successful implementation of these technologies requires a suite of reliable reagents and tools. The following table details key solutions for conducting mRNA reprogramming and RNAi experiments.
Table 3: Research Reagent Solutions for mRNA and RNAi Studies
| Reagent / Solution | Function | Example Applications & Notes |
|---|---|---|
| N1-methylpseudouridine | Chemically modified nucleoside that suppresses innate immune recognition of in vitro transcribed mRNA. | Critical for enhancing mRNA stability and translational efficiency in reprogramming protocols [96]. |
| Lipid Nanoparticles (LNPs) | A non-viral delivery system for encapsulating and delivering RNA molecules into cells with high efficiency. | Used for both siRNA and mRNA delivery; composition (ionizable lipid, PEG, cholesterol, phospholipid) can be tuned [121] [96]. |
| Triantennary GalNAc (N-acetylgalactosamine) | A targeting ligand conjugated to siRNA for receptor-mediated uptake by hepatocytes. | Enables highly efficient siRNA delivery to the liver, reducing the required dose and minimizing systemic exposure [123] [122]. |
| Locked Nucleic Acid (LNA) | A high-affinity RNA analogue where the ribose is locked in a C3'-endo conformation, increasing duplex stability. | Used in "gapmer" antisense oligonucleotides and to enhance the potency and duration of siRNA activity [123] [125]. |
| Cholesterol-conjugated siRNA | A simple conjugate that promotes association with lipid membranes and facilitates cellular uptake. | Useful for in vivo delivery to certain tissues, including the central nervous system, without complex formulations [121]. |
| Support Vector Machine (SVM) Algorithms | A class of machine learning algorithms used to predict highly active and specific siRNA sequences. | Integrated into modern siRNA design software to minimize off-target effects and maximize on-target knockdown during the design phase [123]. |
Diagram 2: A visual summary of the primary off-target risks and corresponding mitigation strategies for mRNA reprogramming and RNAi technologies.
mRNA reprogramming and RNAi are powerful but fundamentally different tools in the cell engineer's arsenal. The choice between them is not a matter of which is superior, but which is more appropriate for the specific biological question. mRNA reprogramming excels as a gain-of-function tool for directly instilling new cellular identities, though its off-target profile is linked to the broad regulatory nature of transcription factors and RNA immunogenicity. RNAi is a premier loss-of-function tool for dissecting the mechanistic role of specific genes in the reprogramming process, with its main off-target challenge being unintended, sequence-based silencing.
For researchers, this dictates a strategic approach: use mRNA to express master regulators and drive fate changes, and employ RNAi to knock down barriers to reprogramming or to validate the necessity of specific pathways. The future of precise cell engineering lies in their intelligent combination. Using RNAi to silence fibroblast identity genes alongside mRNA to activate neuronal programs, for example, could yield more pure and efficient reprogramming outcomes. As both technologies continue to evolve—with advances in self-amplifying mRNA, novel nanoparticle formulations for extrahepatic delivery, and more sophisticated AI-driven sequence design—their specificity will only improve, further solidifying RNA as a central pillar of next-generation regenerative medicine and therapeutic development.
The pursuit of reversing cellular aging has been fundamentally reshaped by the advent of partial reprogramming technologies, particularly those utilizing mRNA-based delivery of reprogramming factors. This approach enables transient, controlled expression of rejuvenating factors without genomic integration, offering a promising pathway for therapeutic applications [60] [96]. Within the broader context of mRNA technology for cell fate reprogramming, this whitepaper examines the rigorous validation frameworks necessary to demonstrate authentic age reversal.
Partial reprogramming distinguishes itself from full reprogramming by applying reprogramming factors—such as the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC, or OSKM)—for short, controlled durations. This transient exposure aims to reset epigenetic aging signatures while maintaining cellular identity, thereby avoiding complete dedifferentiation into pluripotent stem cells [93] [126]. The ultimate goal is not to create pluripotent cells but to restore function to aged somatic cells by reversing age-associated epigenetic and transcriptional alterations. Advanced delivery systems, including tissue nanotransfection (TNT) and lipid nanoparticles (LNPs), are being developed to enable precise, in vivo delivery of mRNA-encoded factors, forming the technological foundation for future clinical translation [57] [96].
Scientific Foundations of Partial Reprogramming
Core Mechanisms: From Epigenetic Reset to Functional Restoration
Partial reprogramming for rejuvenation operates primarily through epigenetic remodeling, reversing the cumulative changes that disrupt gene expression patterns in aged cells. Key molecular mechanisms include:
- DNA Methylation Reset: Age-related changes in DNA methylation patterns, as measured by epigenetic clocks, are reversed toward a more youthful configuration without erasing cellular identity [93] [127].
- Histone Modification Restoration: Treatment reduces age-associated heterochromatin loss and restores youthful patterns of histone modifications, such as H3K9me3 and H3K27me3 [93] [128].
- Transcriptomic Rejuvenation: The overall gene expression profile shifts to resemble that of younger cells, with downregulation of inflammatory pathways and restoration of metabolic function [93] [128].
- Mitochondrial Rejuvenation: Enhanced oxidative phosphorylation and reduced mitochondrial ROS production contribute to restored cellular energy metabolism [93] [128].
The discovery that biological age is surprisingly fluid has accelerated interest in reprogramming approaches. Research indicates that biological age can increase rapidly in response to severe stress (e.g., major surgery, pregnancy, COVID-19) but can also be reversed following recovery, demonstrating the plasticity of the aging process and the potential for therapeutic intervention [127].
Critical Distinction: Partial vs. Full Reprogramming
A critical consideration in rejuvenation research is maintaining the delicate balance between reversing aging hallmarks and preserving cellular identity. Full reprogramming to pluripotency, while effective for erasing all aging signatures, presents significant safety risks including teratoma formation and loss of tissue-specific function [126]. In contrast, partial reprogramming utilizes shortened exposure times or modified factor cocktails to achieve epigenetic rejuvenation while lineage-specific genes remain expressed, thus preserving cellular identity and function [93] [126].
Validation Frameworks for Age Reversal
Epigenetic Clocks: Measurement and Interpretation
Epigenetic clocks have emerged as the primary quantitative tool for assessing biological age reversal, though their interpretation requires sophisticated understanding.
Table 1: Generations of Epigenetic Clocks for Age Reversal Validation
| Generation | Representative Clocks | Training Basis | Utility in Reprogramming Studies |
|---|---|---|---|
| First Generation | Horvath, Hannum | Chronological age | Baseline age estimation; limited correlation with functional rejuvenation |
| Second Generation | PhenoAge, GrimAge, GrimAge2 | Multiple biomarkers & mortality risk | Better predicts health outcomes; used in TRIIM trial showing 2-year decrease [127] |
| Third Generation | DunedinPACE, DunedinPoAm | Pace of aging | Measures rate of aging change; sensitive to short-term interventions |
| Fourth Generation | Causal Clocks | Mendelian randomization | Aims to target causally implicated sites; emerging experimental tool [127] |
However, a crucial limitation was highlighted in a recent analysis suggesting that current epigenetic clocks may not distinguish between different types of age-related methylation changes [129]. Dr. Josh Mitteldorf's perspective paper identifies Type 1 methylation (potentially causative of damage) and Type 2 methylation (part of repair responses) as functionally distinct categories. Clocks that fail to differentiate these may produce misleading results, potentially interpreting suppression of repair mechanisms as successful rejuvenation [129]. This underscores the necessity of complementing epigenetic clock data with functional assessments.
Functional Assays for Rejuvenation Validation
Beyond epigenetic metrics, comprehensive validation requires demonstration of functional improvement across multiple cellular and physiological domains.
Table 2: Functional Assays for Validating Rejuvenation
| Aging Hallmark | Key Assays | Expected Outcome with Rejuvenation |
|---|---|---|
| Genomic Instability | γH2AX foci quantification, COMET assay | Significant reduction in DNA damage markers [128] |
| Cellular Senescence | SA-β-Gal staining, p16/p21 expression | Decreased senescent cell burden [128] |
| Mitochondrial Function | ROS measurement, OCR analysis, ATP production | Reduced oxidative stress, enhanced oxidative phosphorylation [93] [128] |
| Nuclear Architecture | Lamin B1 expression, heterochromatin markers | Restoration of nuclear envelope integrity, reduced heterochromatin loss [128] |
| Stem Cell Function | Transplantation assays, differentiation capacity | Improved regenerative capacity in muscle, neural, and other stem cells [93] |
| Transcriptomic Age | RNA sequencing, aging signature analysis | Shift toward youthful expression patterns [93] |
In Vivo Healthspan and Lifespan Outcomes
Ultimately, therapeutic rejuvenation must demonstrate translation to organism-level benefits. Several studies have shown promising results:
- Lifespan Extension: Partial reprogramming with OSK factors delivered via AAV9 extended remaining lifespan in 124-week-old wild-type mice by 109% compared to untreated controls [93].
- Healthspan Improvement: Treated mice exhibited significantly improved frailty index scores (6.0 vs. 7.5 in controls) and enhanced tissue function [93].
- Chemical Reprogramming Benefits: A reduced two-compound cocktail (2c) extended median lifespan in C. elegans by over 42% and improved multiple healthspan markers, including stress resistance and thermotolerance [128].
Experimental Protocols for Rejuvenation Research
mRNA-Based Partial Reprogramming Protocol
The following protocol outlines a standardized approach for mRNA-mediated partial reprogramming of human fibroblasts, adaptable to other cell types:
Day 0: Cell Plating
- Plate primary human dermal fibroblasts (aged donors preferred) in 6-well plates at 50,000 cells/well in standard fibroblast growth medium.
- Incubate at 37°C, 5% CO₂ for 24 hours to ensure 70-80% confluence at time of transfection.
Days 1-6: Daily mRNA Transfection
- Prepare mRNA-LNP complexes containing modified mRNAs encoding OCT4, SOX2, KLF4, and c-MYC (OSKM) with 1-methylpseudouridine incorporation to reduce immunogenicity [60] [96].
- Replace medium with fresh growth medium 30 minutes before transfection.
- Transfect cells using optimized lipid nanoparticles (LNPs) at 0.5-1.0 μg mRNA per well.
- Incubate for 4-6 hours, then replace with fresh growth medium.
- Repeat transfection daily for 6 consecutive days [128].
Day 7: Analysis and Validation
- Harvest cells for downstream analysis including epigenetic clock assessment, RNA sequencing, functional assays, and senescence markers.
- Include appropriate controls: untreated aged cells, young reference cells, and vehicle-treated aged cells.
Chemical Partial Reprogramming Protocol
For non-genetic approaches, chemical reprogramming offers an alternative strategy:
Seven-Compound (7c) Cocktail Treatment
- Prepare 7c cocktail: CHIR99021 (GSK-3β inhibitor), DZNep (EZH2 inhibitor), Forskolin (adenylyl cyclase activator), TTNPB (RAR agonist), Valproic acid (HDAC inhibitor), Repsox (TGF-β inhibitor), and Tranylcypromine (LSD1 inhibitor) [128].
- Treat aged human fibroblasts continuously for 6 days with refreshed medium and compounds daily.
- For reduced cocktail approach, use only CHIR99021 and VPA (2c cocktail) for focused epigenetic remodeling [128].
- Assess rejuvenation markers post-treatment as described in Section 4.1.
In Vivo Delivery Methods
For translational applications, several delivery platforms have been developed:
- AAV-Mediated Delivery: Using AAV9 capsid for broad tissue distribution with doxycycline-inducible OSK expression (1-day pulse, 6-day chase) in cyclic regimens [93].
- Tissue Nanotransfection (TNT): Non-viral nanoelectroporation platform for localized delivery of plasmid DNA or mRNA reprogramming factors directly to tissues [57].
- Chemical Cocktail Delivery: Systemic administration of small molecule cocktails (7c or 2c) without genetic material [128].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Partial Reprogramming Studies
| Reagent Category | Specific Examples | Function in Reprogramming |
|---|---|---|
| Reprogramming Factors | Modified mRNA encoding OSKM, OSK, or alternative factors | Directly induce epigenetic remodeling; modified bases reduce immune recognition [60] [96] |
| Delivery Vehicles | Lipid nanoparticles (LNPs), Tissue Nanotransfection (TNT) chips | Enable efficient cellular uptake while protecting mRNA payload [57] [96] |
| Chemical Inducers | CHIR99021, VPA, TTNPB, Forskolin, DZNep, Repsox, Tranylcypromine | Small molecules modulating signaling pathways to mimic transcription factor effects [128] |
| Epigenetic Age Assays | Illumina EPIC arrays, targeted bisulfite sequencing panels | Quantify DNA methylation changes for epigenetic clock calculation [127] |
| Senescence Detectors | SA-β-Gal staining kits, p16/p21 antibodies, lamin B1 antibodies | Assess cellular senescence burden before and after treatment [128] |
| DNA Damage Kits | γH2AX antibodies, COMET assay kits | Quantify genomic instability reduction following rejuvenation [128] |
Critical Considerations and Future Directions
Safety Assessment and Risk Mitigation
The transition of partial reprogramming toward clinical applications requires careful safety evaluation:
- Tumorigenicity Screening: Comprehensive teratoma formation assays in immunodeficient mice over extended periods (minimum 6 months) [93] [126].
- Identity Preservation Validation: Single-cell RNA sequencing to confirm maintenance of cell-type-specific gene expression patterns following reprogramming [93].
- Genomic Integrity Checks: Karyotyping and whole-genome sequencing to detect any chromosomal abnormalities or mutations introduced during the process [60].
- Integration-Free Confirmation: PCR-based assays to verify absence of vector integration when using mRNA or episomal approaches [60] [96].
Technical Challenges and Optimization Opportunities
Current limitations present opportunities for methodological advancement:
- Delivery Precision: Developing tissue-specific targeting systems using ligand-decorated LNPs or improved TNT configurations [57] [96].
- Dosing Optimization: Establishing minimum effective exposure times and factor concentrations to achieve rejuvenation while avoiding dedifferentiation [93].
- Personalization Approaches: Adapting protocols based on initial epigenetic age, cell type, and donor characteristics [126].
- Novel Factor Discovery: Screening for new reprogramming molecules that separate rejuvenation effects from pluripotency induction [128].
The field of partial reprogramming for rejuvenation has progressed from fundamental discovery to preliminary validation, with mRNA-based technologies offering a particularly promising pathway toward clinical translation. The comprehensive validation framework outlined in this whitepaper—integrating epigenetic clocks with functional assays and in vivo outcomes—provides researchers with a rigorous methodology for distinguishing authentic age reversal from mere epigenetic cosmetic changes. As safety and delivery technologies continue to advance, particularly in mRNA delivery and chemical reprogramming, the potential for developing genuine rejuvenation therapies targeting age-related diseases comes increasingly into focus. The ongoing refinement of both reprogramming protocols and validation standards will be essential for translating these promising laboratory findings into meaningful clinical applications.
The field of regenerative medicine has been transformed by the emergence of messenger RNA (mRNA) as a powerful tool for directing cellular behavior. Unlike traditional gene therapy approaches, mRNA therapeutics offer a non-integrative and controllable strategy for expressing therapeutic proteins, providing precision, safety, and transience in directing cellular behavior [32]. This technology enables efficient protein supplementation, cell reprogramming, and cell transdifferentiation, allowing for the precise modulation of cell fate and function [32]. The application of mRNA-based technology for cell fate reprogramming represents a paradigm shift in how researchers approach degenerative diseases and injury-related conditions, offering potential solutions for cardiac repair, liver regeneration, pulmonary recovery, and epithelial healing [32].
The fundamental advantage of mRNA-based platforms lies in their high programmability, significantly improved stability, and immunogenicity achieved through nucleotide modifications and advanced delivery systems such as lipid nanoparticles (LNPs) [130]. As a synthetic immunization strategy, mRNA vaccines deliver mRNA encoding specific antigens to host cells, enabling them to express proteins from external sources and thereby eliciting targeted immune responses within the body [130]. This mechanism not only mimics the immune response induced by natural viral infection but also offers substantial flexibility and safety, making it ideal for therapeutic applications beyond infectious diseases [130].
Fundamental Principles of mRNA Technology
Molecular Structure and Design
The design of synthetic mRNA molecules represents a critical foundation for effective cell fate reprogramming. These molecules typically comprise several key components: a 5′ cap structure, untranslated regions (UTRs), open reading frames (ORFs) encoding the antigen, and a poly(A) tail at the 3′ end [130]. Each component serves a specific function in enhancing mRNA stability, translational efficiency, and ultimately, the potency of the resulting therapeutic. The 5′ cap protects the mRNA from degradation and facilitates ribosomal binding, while the optimized UTRs regulate stability and translation rates. The ORF contains the precise genetic sequence for the target protein, and the poly(A) tail further stabilizes the transcript and promotes translation initiation [130].
Recent advances in mRNA chemistry have focused on nucleotide modifications to reduce innate immunogenicity and enhance protein expression [130]. These modifications include the incorporation of pseudouridine and other nucleoside analogs that decrease recognition by pattern recognition receptors, thereby minimizing inflammatory responses while maintaining translational fidelity. Additionally, codon optimization strategies have been employed to match the codon usage frequency of the target organism, further increasing translational efficiency and protein yield [130]. The continuous refinement of these molecular design elements has substantially improved the therapeutic potential of mRNA platforms for cell fate reprogramming applications.
Delivery Platforms and Formulations
The effective delivery of mRNA molecules into target cells represents a significant challenge that has been addressed through innovations in nanocarrier systems. Lipid nanoparticles (LNPs) have emerged as the leading delivery platform, providing protection for the fragile mRNA molecules and facilitating cellular uptake through endocytic pathways [130]. These LNPs typically consist of ionizable lipids, phospholipids, cholesterol, and PEG-lipids that self-assemble into stable nanoparticles around the mRNA core. The ionizable lipids are particularly crucial as they enable endosomal escape through their pH-dependent structural changes, releasing the mRNA payload into the cytoplasm where translation can occur [130].
The development of these delivery systems has been instrumental in overcoming biological barriers such as enzymatic degradation, renal clearance, and immune recognition. Beyond LNPs, researchers are exploring alternative delivery platforms including polymeric carriers, peptide-based systems, and hybrid nanoparticles to further improve tissue targeting and intracellular delivery efficiency [130]. Each platform offers distinct advantages for specific applications, with ongoing research focused on optimizing biodistribution, cellular uptake, and endosomal escape capabilities to enhance the efficacy of mRNA-based cell fate reprogramming strategies.
Quantitative Analysis of Pre-clinical and Clinical Data
Key Pre-clinical Studies
Pre-clinical investigations have established the foundational principles of mRNA-based cell fate reprogramming, with several landmark studies providing critical insights into the mechanisms and potential applications of this technology.
Table 1: Summary of Key Pre-clinical Studies in mRNA-Based Cell Fate Reprogramming
| Study Focus | Experimental Model | Key Findings | Implications |
|---|---|---|---|
| Landscape of Cell Fate Decisions [131] [132] | 52-gene human stem cell regulatory network | Quantified potential landscape with two basins of attraction representing stem and differentiated cell states; identified irreversible differentiation and reprogramming paths | Provides theoretical framework for understanding cellular reprogramming dynamics; predicts critical gene targets |
| mRNA Mechanism in Immuno-activation [76] | Preclinical models in combination with clinical correlation | mRNA vaccines function as immune alarms, increasing PD-L1 expression on cancer cells; creates ideal environment for checkpoint inhibitors | Explains synergistic effect of mRNA vaccines with immunotherapy; supports combination treatment approaches |
| Cellular Reprogramming Pathways [131] [132] | Computational analysis of gene regulatory networks | Both potential landscape and non-equilibrium curl flux determine cell differentiation dynamics; kinetic paths deviate from steepest descent gradient | Suggests optimal reprogramming pathways can be engineered through targeted manipulation of key network nodes |
The groundbreaking work on quantifying cell fate decisions revealed that the underlying landscape for stem cell networks contains two basins of attractions representing stem and differentiated cell states [131] [132]. Through sophisticated computational modeling of a 52-gene regulatory network, researchers demonstrated that the differentiation and reprogramming paths are irreversible and determined by both the potential landscape and probabilistic flux [131] [132]. This fundamental understanding of cellular dynamics provides a theoretical framework for designing more efficient mRNA-based reprogramming strategies by targeting critical nodes in the regulatory network.
Complementing this theoretical work, mechanistic studies have revealed that mRNA vaccines function as powerful immune activators, essentially serving as immune system alarms that put the body's immune system on high alert [76]. In preclinical models, this effect was shown to trigger adaptive responses in cancer cells, including increased expression of the immune checkpoint protein PD-L1 as a defense mechanism [76]. This discovery created the rationale for combining mRNA vaccines with immune checkpoint inhibitors, establishing a foundational principle that has translated into clinical applications beyond infectious diseases.
Early-Phase Clinical Trial Evidence
The transition from pre-clinical models to human trials has yielded compelling evidence supporting the potential of mRNA-based approaches in therapeutic applications, particularly in oncology and regenerative medicine.
Table 2: Key Clinical Trial Findings for mRNA-Based Therapies
| Trial/Study Description | Patient Population | Intervention | Key Outcomes | Significance |
|---|---|---|---|---|
| Retrospective Analysis of mRNA COVID Vaccines + Immunotherapy [76] | 1,000+ cancer patients (2019-2023) | mRNA COVID vaccine within 100 days of immune checkpoint inhibitors | 2x higher 3-year survival in vaccinated patients; advanced NSCLC: 37.33 vs 20.6 months median survival | First large-scale evidence of mRNA vaccine enhancing immunotherapy efficacy |
| mRNA Cancer Vaccine Trials Growth [133] | Global clinical trial analysis | Various mRNA cancer vaccines | 42% CAGR in trial initiations since 2020; 88% of mRNA therapies in preclinical/early clinical phases | Demonstrates rapid expansion and investment in mRNA therapeutic platform |
| mRNA Deal-Making Trends [133] | Industry-wide analysis | mRNA platform technology investments | $5B total deal value in H1 2025; $2B upfront cash; 57% of deals focus on platform technologies | Reflects strong industry confidence and financial investment in mRNA future |
A remarkable retrospective study analyzing over 1,000 cancer patients treated between August 2019 and August 2023 revealed that patients who received mRNA-based COVID vaccines within 100 days of starting immune checkpoint therapy were twice as likely to be alive three years after treatment compared to those who never received a vaccine [76]. This striking survival benefit was observed across multiple cancer types, with particularly impressive results in advanced non-small cell lung cancer, where vaccinated patients demonstrated a median survival of 37.33 months compared to 20.6 months in unvaccinated patients [76]. These findings have prompted the design of a randomized Phase III trial to determine if mRNA COVID vaccines should become part of the standard of care for patients receiving immune checkpoint inhibition.
The broader clinical landscape for mRNA therapies shows explosive growth, with the number of mRNA cancer vaccine trials initiated each year steadily increasing at a compound annual growth rate (CAGR) of 42% since 2020 [133]. Current data indicates that 88% of mRNA therapies are still in preclinical or early-stage clinical phases, highlighting the extensive pipeline of potential applications awaiting clinical validation [133]. This growth is supported by substantial financial investments, with H1 2025 alone generating $5 billion in total deal value and $2 billion in upfront cash for mRNA-based therapeutic development [133].
Experimental Protocols and Methodologies
Protocol for Analyzing Cell Fate Decisions
The investigation of cell fate decisions requires sophisticated computational approaches to model complex gene regulatory networks. The following protocol outlines the key methodology used in landmark studies to quantify landscape and biological paths for differentiation and reprogramming:
Network Construction and Modeling
- Assemble a comprehensive gene regulatory network incorporating 52 genes critical for stem cell fate decisions [131] [132]
- Define regulatory relationships (activation/repression) between network nodes based on experimental literature
- Formulate chemical reaction rate equations representing the dynamics of gene expression for each node
Landscape Reconstruction
- Apply self-consistent mean field approximation to solve the high-dimensional system [131] [132]
- Calculate steady-state probability distribution P using moment equations with Gaussian approximation
- Compute potential landscape U using the relationship U = -lnP, where P represents the probability distribution
- Project the 52-dimensional landscape onto 2-dimensional state space using key marker genes (NANOG and GATA6) for visualization
Kinetic Path Quantification
- Employ path integral methods to identify the most probable differentiation and reprogramming paths [131] [132]
- Calculate probabilistic flux to understand non-equilibrium dynamics
- Analyze path irreversibility by comparing differentiation and reprogramming trajectories
This methodology allows researchers to quantify the topography of the potential landscape through barrier heights and transition rates, which determine the global stability and kinetic speed of cell fate decision processes [131] [132].
Protocol for Evaluating mRNA Vaccine Efficacy in Oncology Models
The evaluation of mRNA vaccine efficacy in enhancing responses to immunotherapy involves both retrospective clinical analysis and mechanistic preclinical studies:
Retrospective Clinical Analysis
- Identify cancer patients treated with immune checkpoint inhibitors between specified date ranges [76]
- Stratify patients based on receipt of mRNA COVID vaccine within 100 days of starting immunotherapy
- Collect comprehensive clinical data including cancer type, stage, biomarker status, and treatment history
- Assess overall survival from immunotherapy initiation using Kaplan-Meier methods
- Perform multivariate Cox regression to adjust for potential confounding variables
Mechanistic Preclinical Studies
- Administer mRNA vaccines to appropriate animal models of cancer [76]
- Analyze immune activation through flow cytometry of tumor-infiltrating lymphocytes
- Measure PD-L1 expression on tumor cells following vaccination using immunohistochemistry
- Evaluate tumor growth kinetics and survival in vaccinated versus control animals
- Conduct immune cell depletion studies to identify critical effector populations
This combined clinical and preclinical approach provides both correlative evidence of efficacy and mechanistic insights into how mRNA vaccines enhance anti-tumor immunity [76].
Signaling Pathways and Molecular Mechanisms
The molecular mechanisms underlying mRNA-based cell fate reprogramming involve complex signaling networks and regulatory pathways. The following diagram illustrates the key signaling pathways involved in mRNA-mediated cell fate reprogramming:
Diagram 1: Signaling Pathways in mRNA-Mediated Cell Fate Reprogramming. This diagram illustrates the key molecular mechanisms through which mRNA-based interventions direct cellular reprogramming, including direct transcription factor expression and immune-mediated pathway modulation.
The diagram illustrates two primary mechanisms through which mRNA-based interventions direct cell fate reprogramming. The first involves direct expression of reprogramming transcription factors that alter gene regulatory networks, while the second leverages immune activation through antigen presentation and cytokine release to create microenvironments conducive to cellular reprogramming [76] [130]. The convergence of these pathways on the gene regulatory network ultimately drives transitions between cellular states.
The dynamics of cell fate decisions can be conceptually understood through the landscape and flux framework illustrated below:
Diagram 2: Landscape and Flux Model of Cell Fate Transitions. This diagram conceptualizes cellular states as basins of attraction separated by energy barriers, with probabilistic flux driving irreversible transitions between states.
This landscape model visualizes cellular states as basins of attraction separated by energy barriers, with the relative depth and size of these basins determining state stability [131] [132]. The probabilistic flux represents non-equilibrium dynamics that make differentiation and reprogramming paths irreversible, explaining why these processes follow distinct biological trajectories [131] [132]. This framework provides researchers with a quantitative approach to identifying key genes and regulatory connections that control the stability of stem cell and differentiated states.
Research Reagent Solutions
The implementation of mRNA-based cell fate reprogramming protocols requires specialized reagents and materials designed to address the unique challenges of mRNA manipulation and delivery. The following table catalogues essential research reagents and their applications in this rapidly advancing field.
Table 3: Essential Research Reagents for mRNA-Based Cell Fate Reprogramming
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Nucleotide Modifications (Pseudouridine, N1-methylpseudouridine) | Reduce innate immune recognition; enhance translational efficiency | Critical for minimizing inflammatory responses while maintaining protein expression levels [130] |
| Lipid Nanoparticles (LNPs) | mRNA encapsulation and delivery; facilitate cellular uptake and endosomal escape | Formulations typically include ionizable lipids, phospholipids, cholesterol, and PEG-lipids [130] |
| In Vitro Transcription (IVT) Kits | Synthetic mRNA production | Must include capping enzymes and poly(A) polymerase for generating functional mRNA constructs [130] |
| Codon Optimization Software | Enhance translational efficiency through sequence optimization | Algorithms designed to match codon usage frequency of target species while maintaining protein structure [130] |
| Stem Cell Marker Antibodies (Anti-NANOG, Anti-OCT4) | Identification and characterization of pluripotent stem cells | Essential for validating reprogramming efficiency and characterizing resulting cell phenotypes [131] [132] |
| Differentiation Marker Antibodies (Anti-GATA6, Anti-CDX2) | Detection of differentiated cell states | Used to confirm successful differentiation and quantify population heterogeneity [131] [132] |
| Immune Profiling Panels | Characterization of immune responses to mRNA therapies | Include cytokines (IFN-α, TNF-α) and cell surface markers for comprehensive immune monitoring [76] [130] |
The selection of appropriate reagents is critical for successful implementation of mRNA-based reprogramming protocols. Nucleotide modifications represent a particularly crucial category, as they significantly impact both the immunogenicity and translational efficiency of synthetic mRNA constructs [130]. Similarly, the formulation of lipid nanoparticles must be carefully optimized for specific applications, as variations in lipid composition can dramatically alter tissue tropism, cellular uptake efficiency, and endosomal escape capabilities [130].
For researchers validating cell fate changes, comprehensive sets of validated antibodies against key stem cell and differentiation markers are indispensable tools [131] [132]. These reagents enable quantitative assessment of reprogramming efficiency through techniques such as flow cytometry, immunocytochemistry, and Western blotting. Additionally, immune profiling reagents are essential for characterizing the innate and adaptive immune responses triggered by mRNA delivery, which can have significant impacts on both safety and efficacy outcomes [76] [130].
The accumulating evidence from both pre-clinical studies and early-phase clinical trials strongly supports the transformative potential of mRNA-based technology for cell fate reprogramming. The quantitative landscape models have provided unprecedented insights into the dynamics of cellular differentiation and reprogramming, revealing the fundamental principles that govern cell fate decisions [131] [132]. Meanwhile, clinical evidence has demonstrated that mRNA platforms can produce powerful biological effects, such as enhancing responses to immunotherapy in cancer patients [76]. These findings collectively establish mRNA technology as a versatile and powerful platform for therapeutic development.
Looking forward, several key challenges and opportunities will shape the future trajectory of this field. Current research remains limited regarding the in vivo biodistribution, long-term safety, and precise immune response mechanisms of mRNA-based interventions, particularly for complex applications like cell fate reprogramming [130]. Additionally, the immunosuppressive nature of microenvironments in damaged or diseased tissues presents a significant barrier that must be addressed through combinatorial approaches [130]. The identification and validation of ideal target antigens for specific applications, optimization of delivery systems for precise tissue targeting, and development of cost-effective manufacturing processes for personalized therapies represent additional frontiers for innovation [130].
The remarkable growth in mRNA technology development—evidenced by the 42% CAGR in mRNA cancer vaccine trials since 2020 and the $5 billion in deal value in H1 2025 alone—suggests that these challenges are being aggressively addressed by the research community [133]. As the field continues to mature, mRNA-based cell fate reprogramming is poised to become an increasingly powerful approach for addressing degenerative diseases, injury conditions, and cancer, ultimately fulfilling the promise of regenerative medicine through precise, safe, and transient manipulation of cellular behavior [32]. The continued integration of computational modeling, innovative delivery technologies, and sophisticated molecular design will enable researchers to overcome current limitations and unlock the full therapeutic potential of this revolutionary technology.
Conclusion
mRNA-based cell fate reprogramming represents a paradigm shift in regenerative medicine, offering a uniquely safe and transient method for generating iPSCs and engineering cell lineages without altering the host genome. The convergence of advancements in mRNA design—such as nucleoside modifications and high-purity capping—with sophisticated delivery platforms like LNPs and TNT, has positioned this technology for broad clinical impact. While challenges in delivery efficiency and precise temporal control of expression remain, the ongoing optimization of these systems continues to enhance their therapeutic potential. Future research must focus on developing tissue-specific delivery vectors, refining protocols for in vivo reprogramming, and advancing clinical trials. The integration of mRNA reprogramming with other technologies, such as next-generation CRISPR base editing, promises to unlock powerful new strategies for treating degenerative diseases, genetic disorders, and age-related decline, ultimately paving the way for a new era in personalized regenerative therapeutics.
References
- 1. Progress and prospects of mRNA-based drugs in pre- ... [https://www.nature.com/articles/s41392-024-02002-z]
- 2. Cell Reprogramming and Differentiation Utilizing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10971469/]
- 3. Therapeutic applications of cell engineering using mRNA ... [https://www.sciencedirect.com/science/article/pii/S0167779924001914]
- 4. Therapeutic applications of cell engineering using mRNA ... [https://pubmed.ncbi.nlm.nih.gov/39153909/]
- 5. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7918810/]
- 6. Clinical development of therapeutic mRNA applications [https://www.sciencedirect.com/science/article/pii/S1525001625002084]
- 7. Understanding COVID-19 mRNA Vaccines [https://www.genome.gov/about-genomics/fact-sheets/Understanding-COVID-19-mRNA-Vaccines]
- 8. Clinical translation of human iPSC technologies [https://pmc.ncbi.nlm.nih.gov/articles/PMC12498231/]
- 9. Application of Modified mRNA in Somatic Reprogramming ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8348611/]
- 10. Efficient mRNA delivery with graphene oxide ... [https://www.sciencedirect.com/science/article/abs/pii/S016836591630373X]
- 11. Feeder-Free Derivation of Human Induced Pluripotent ... [https://www.nature.com/articles/srep00657]
- 12. Opportunities and Challenges in the Delivery of mRNA-Based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7076378/]
- 13. Advances in Induced Pluripotent Stem Cell Reprogramming ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12577567/]
- 14. iPSC-based Platforms Market 2025 Report and Trends [https://www.towardshealthcare.com/insights/ipsc-based-platforms-market-sizing]
- 15. Pluripotent Stem Cells: Recent Advances and Emerging ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12025069/]
- 16. conversion by Cell | Stem fate & Therapy mRNA Cell Research [https://stemcellres.biomedcentral.com/articles/10.1186/scrt46]
- 17. mRNA Therapeutic Modalities Design, Formulation and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8773365/]
- 18. Modifications of mRNA vaccine structural elements for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8450916/]
- 19. mRNA-based vaccines and therapeutics: an in-depth survey ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-023-00977-5]
- 20. Unlocking the promise of mRNA therapeutics [https://www.nature.com/articles/s41587-022-01491-z]
- 21. Accelerating life sciences research | OpenAI [https://openai.com/index/accelerating-life-sciences-research-with-retro-biosciences/]
- 22. Roles of mRNA poly(A) tails in regulation of eukaryotic gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7614307/]
- 23. Enhancing mRNA Therapeutics in Two Shakes of a Poly(A) ... [https://en.vectorbuilder.com/resources/vector-academy/troubleshooting/mRNA-polyA-tail.html]
- 24. Press release: The Nobel Prize in Physiology or Medicine ... [https://www.nobelprize.org/prizes/medicine/2023/press-release/]
- 25. From rejection to the Nobel Prize: Karikó and Weissman's ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10663363/]
- 26. mRNA vaccines — a new era in vaccinology [https://www.nature.com/articles/nrd.2017.243]
- 27. Innate immune mechanisms of mRNA vaccines - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9641982/]
- 28. mRNA medicine: Recent progresses in chemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12311907/]
- 29. Highly efficient reprogramming to pluripotency and directed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3656821/]
- 30. Cell Reprogramming and Differentiation Utilizing ... [https://www.preprints.org/manuscript/202311.0233]
- 31. RNA-Based Tools for Nuclear Reprogramming and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3838600/]
- 32. mRNA-based technology for engineered regenerative ... [https://www.sciencedirect.com/science/article/pii/S305056232500176X]
- 33. Programming human cell fate: overcoming challenges and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10753584/]
- 34. Understanding mRNA technology as platform for biopharma [https://custombiotech.roche.com/global/en/post-listing/understanding-mrna-technology-as-platform-for-biopharma.html]
- 35. Messenger RNA [https://en.wikipedia.org/wiki/Messenger_RNA]
- 36. mRNA Reprogramming [https://allelebiotech.com/technologies/mrna-reprogramming/]
- 37. mRNA technology simply explained [https://www.susupport.com/blogs/knowledge/mrna-technology-simply-explained]
- 38. Mapping, modeling, and reprogramming cell-fate decision ... [https://arxiv.org/html/2412.00667v1]
- 39. mRNA Capping: Process, Mechanisms, Methods & Applications [https://lifesciences.danaher.com/us/en/library/mrna-capping.html]
- 40. Co-transcriptional capping [https://www.takarabio.com/learning-centers/mrna-and-cdna-synthesis/mrna-synthesis/5-prime-capping-of-mrna/co-transcriptional-capping?srsltid=AfmBOor8BW5dQij7r-q2uSPrF2z7VOXu0whsXJ7ryL5JYWPzGgigcjOO]
- 41. mRNA Capping Methods Explained [https://rna.bocsci.com/support/mrna-capping-techniques-cap-0-cap-1-and-co-transcriptional-methods.html]
- 42. Clean Capped mRNA: The Future of Vaccine Development [https://www.trilinkbiotech.com/cleancap-vaccine-development]
- 43. RNA Capping [https://www.neb.com/en-ca/products/rna-synthesis-and-modification/rna-capping?srsltid=AfmBOorF28cooQ_uvNUoGrcqg_kImk_esgurFblovm-38R0zMEjoOzpI]
- 44. A comparative exploration of mRNA capping enzymes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11625350/]
- 45. Cap analogs with a hydrophobic photocleavable tag ... [https://www.nature.com/articles/s41467-023-38244-8]
- 46. Methods of IVT mRNA capping - Behind the Bench [https://www.thermofisher.com/blog/behindthebench/methods-of-ivt-mrna-capping/]
- 47. Acuitas Therapeutics Unveils Next-Generation Lipid ... [https://crisprmedicinenews.com/press-release-service/card/acuitas-therapeutics-unveils-next-generation-lipid-nanoparticle-advancements-at-the-2025-mrna-health/]
- 48. MRNA Vaccine Drugs Development Services Market - Global [https://www.globenewswire.com/news-release/2025/11/17/3189330/0/en/MRNA-Vaccine-Drugs-Development-Services-Market-Global-Forecast-2025-2030-Opportunities-Through-Increased-Investment-Diverse-Therapeutic-Applications-and-Scalable-Production-Demand.html]
- 49. A complete guide to understanding Lipid nanoparticles (LNP) [https://insidetx.com/resources/reviews/complete-guide-to-understanding-lipid-nanoparticles-lnp/]
- 50. Introduction to Lipid Nanoparticle: LNP Formulation ... [https://www.precigenome.com/lipid-nanoparticles-lnp/introduction-lnp-formulation-design-preparation?srsltid=AfmBOooyLkiWlB8gIZ5cl9m2VGLV1IoHdF3agOlAMFG1os3GEYIY4TUi]
- 51. Exploring the Challenges of Lipid Nanoparticle Development [https://pmc.ncbi.nlm.nih.gov/articles/PMC12031360/]
- 52. Recent Advances in Lipid Nanoparticles and Their Safety ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11510967/]
- 53. Current landscape of mRNA technologies and delivery ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-024-01080-z]
- 54. Five Challenges Of Applying Lipid Nanoparticles As Delivery ... [https://us.progen.com/post/Five-Challenges-Of-Applying-Lipid-Nanoparticles-As-Delivery-Tools-For-Therapeutic-Agents]
- 55. Endosomal escape and current obstacles in ionizable lipid ... [https://www.sciencedirect.com/science/article/pii/S0378517325011007]
- 56. Deciphering the biological fate of mRNA-LNP-based ... [https://www.sciencedirect.com/science/article/pii/S2211383525007737]
- 57. Tissue nanotransfection and cellular reprogramming in ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1558735/full]
- 58. Tissue nanotransfection and cellular reprogramming in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12213499/]
- 59. Tissue Nanotransfection Silicon Chip and Related ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10820803/]
- 60. HSCI researchers achieve major breakthrough in cell ... [https://www.hsci.harvard.edu/hsci-researchers-achieve-major-breakthrough-cell-reprogramming]
- 61. Tissue Nanotransfection Silicon Chip and Related ... [https://www.mdpi.com/2079-4991/14/2/217]
- 62. Lipid nanoparticle delivery of the CRISPR/Cas9 system ... [https://www.nature.com/articles/s41598-025-03671-8]
- 63. Tissue nano-transfection of antimicrobial genes drives ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365924007442]
- 64. Current Progress and Future Perspectives of RNA-Based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12153701/]
- 65. Induced Pluripotent Stem Cells (iPSC) and Their Use in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590075/]
- 66. Modeling Human Neural Development and Disease with ... [https://www.isscr.org/upcoming-programs/poster-award-webinar]
- 67. Editorial: Studying rare diseases using induced pluripotent ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1686438/full]
- 68. Cell-engineered technologies for wound healing and ... [https://www.nature.com/articles/s44385-025-00042-w]
- 69. Induced Pluripotent Stem Cell (iPSC) Industry Trends for ... [https://bioinformant.com/ipsc-industry-trends/]
- 70. mRNA ageing shapes the Cap2 methylome in mammalian ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9891201/]
- 71. Nucleoside-modified messenger RNA [https://en.wikipedia.org/wiki/Nucleoside-modified_messenger_RNA]
- 72. The impact of nucleoside base modification in mRNA vaccine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10280092/]
- 73. Engineering of the current nucleoside-modified mRNA- ... [https://www.sciencedirect.com/science/article/pii/S0753332221007356]
- 74. mRNA Cap 2 O Methyltransferase [https://www.neb.com/en-us/products/m0366-mrna-cap2-o-methyltransferase?srsltid=AfmBOooqa7Q14syce2KFD3yEQCRPDVkWgzjMtLmGZLN5ZF3qHXuEwl3X]
- 75. Cell Fate Reprogramming in the Era of Cancer ... [https://pubmed.ncbi.nlm.nih.gov/34367185/]
- 76. ESMO 2025: mRNA-based COVID vaccines generate ... [https://www.mdanderson.org/newsroom/research-newsroom/-esmo-2025--mrna-based-covid-vaccines-generate-improved-response.h00-159780390.html]
- 77. Improving mRNA-Based Therapeutic Gene Delivery by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6453560/]
- 78. Human cellular CYBA UTR sequences increase mRNA ... [https://www.nature.com/articles/srep39149]
- 79. Optimizing 5'UTRs for mRNA-delivered gene editing using ... [https://www.nature.com/articles/s41467-024-49508-2]
- 80. Evaluation of synthetic mRNA with selected UTR ... [https://pubmed.ncbi.nlm.nih.gov/40822034/]
- 81. Enhancing mRNA translation efficiency by introducing ... [https://pubmed.ncbi.nlm.nih.gov/40125272/]
- 82. Loop structure in poly(A) tail of mRNA vaccine enhances ... [https://www.nature.com/articles/s41541-025-01287-7]
- 83. Lipid nanoparticles for mRNA delivery [https://www.nature.com/articles/s41578-021-00358-0]
- 84. Rational Design of Lipid Nanoparticles [https://pmc.ncbi.nlm.nih.gov/articles/PMC11323194/]
- 85. Protein corona formed on lipid nanoparticles compromises ... [https://www.nature.com/articles/s41467-025-63726-2]
- 86. Cellular and biophysical barriers to lipid nanoparticle ... [https://www.nature.com/articles/s41467-025-60959-z]
- 87. The kinetics of endosomal disruption reveal differences in ... [https://pubmed.ncbi.nlm.nih.gov/40684993/]
- 88. Artificial intelligence-driven rational design of ionizable ... [https://www.nature.com/articles/s41467-024-55072-6]
- 89. Optimization of Lipid Nanoformulations for Effective mRNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9259059/]
- 90. Current advances and future prospects of cell ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1546423/full]
- 91. Application of the Yamanaka Transcription Factors Oct4 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10531188/]
- 92. Resetting the Biological Clock: OKSM and the Reversal of ... [https://www.pharmasalmanac.com/articles/resetting-the-biological-clock-oksm-and-the-reversal-of-aging]
- 93. The long and winding road of reprogramming-induced ... [https://www.nature.com/articles/s41467-024-46020-5]
- 94. Cellular reprogramming and epigenetic rejuvenation [https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-021-01158-7]
- 95. A single short reprogramming early in life initiates and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9649606/]
- 96. Therapeutic applications of cell engineering using mRNA ... [https://www.sciencedirect.com/science/article/abs/pii/S0167779924001914]
- 97. Cell Reprogramming and Differentiation Utilizing ... [https://www.mdpi.com/2221-3759/12/1/1]
- 98. Developing mRNA Nanomedicines with Advanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11842722/]
- 99. Preparation of selective organ- targeting ... | Nature Protocols [https://www.nature.com/articles/s41596-022-00755-x?error=cookies_not_supported]
- 100. Lipid nanoparticle-mediated targeted and its... mRNA delivery [https://pubs.rsc.org/en/content/articlehtml/2025/tb/d5tb01556a]
- 101. CRISPR's Efficiency Triples With Spherical Nucleic Acid ... [https://news.feinberg.northwestern.edu/2025/09/05/crisprs-efficiency-triples-with-spherical-nucleic-acid-delivery-system/]
- 102. Exploring the future of mRNA delivery: Beyond lipid ... [https://pubmed.ncbi.nlm.nih.gov/40683021/]
- 103. mRNA nanomedicine reprograms the cell cycle to increase ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365925009976]
- 104. CRISPR Clinical Trials: A 2025 Update [https://innovativegenomics.org/news/crispr-clinical-trials-2025/]
- 105. mRNA therapeutics: Transforming medicine through ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590236/]
- 106. Viral vector vaccines vs. mRNA vaccines: The differences [https://www.susupport.com/blogs/knowledge/viral-vector-vaccines-vs-mrna-vaccines-the-differences]
- 107. Viral vs Non-Viral mRNA Delivery Methods [https://www.bocsci.com/research-area/comparing-viral-and-non-viral-vectors-for-mrna-delivery.html?srsltid=AfmBOoqZfdU1IyFiqU9GSa1riR7oGIGJMb1yTKt6lCC28Hf-ToVRwKWG]
- 108. Comparative analysis of adenovirus, mRNA, and protein ... [https://insight.jci.org/articles/view/198069]
- 109. mRNA vaccines for infectious diseases: principles ... - Nature [https://www.nature.com/articles/s41573-021-00283-5]
- 110. A randomized trial comparing safety, immunogenicity and ... [https://www.nature.com/articles/s41541-024-01017-5]
- 111. Safety comparison of mRNA, viral vector, and inactivated ... [https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-025-11254-1]
- 112. Comparison between viral vector and mRNA based COVID ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9803623/]
- 113. Gene Knockdown vs. Knockout: RNAi vs. CRISPR ... [https://synapse.patsnap.com/article/gene-knockdown-vs-knockout-rnai-vs-crispr-approaches]
- 114. Knockout vs. Knockdown [https://www.huabio.com/blogs/news/knockout-vs-knockdown?srsltid=AfmBOopROZt1MEtCKxOGnkzsUX5j8WCGiWn48_KtrCPNhGt0HdAgcoAJ]
- 115. Mechanism and Applications of CRISPR/Cas-9-Mediated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8388126/]
- 116. mRNA: A Game Changer in Drug Development [https://www.trilinkbiotech.com/mrna-game-changer-in-drug-development]
- 117. RNAi vs. CRISPR: Guide to Selecting the Best Gene ... [https://www.synthego.com/blog/rnai-vs-crispr-guide/]
- 118. CRISPR Knockouts vs. Knockins: Key Differences [https://www.zeclinics.com/blog/what-are-knockouts-and-knockins]
- 119. Escaping from CRISPR–Cas-mediated knockout [https://cmbl.biomedcentral.com/articles/10.1186/s11658-024-00565-x]
- 120. Why mRNA Reprogramming Is the Key to Safer and Faster ... [https://www.reprocell.com/blog/why-mrna-reprogramming-is-the-key-to-safer-and-faster-ipsc-generation]
- 121. The new era of siRNA therapy: Advances in cancer treatment [https://pmc.ncbi.nlm.nih.gov/articles/PMC12148947/]
- 122. siRNA-based delivery systems: Technologies, carriers ... [https://www.sciencedirect.com/science/article/abs/pii/S0014299925001955]
- 123. RNA Therapeutics: Delivery Problems and Solutions—A Review [https://pmc.ncbi.nlm.nih.gov/articles/PMC12567017/]
- 124. Small interfering RNA (siRNA) Therapeutics [https://www.delveinsight.com/blog/small-interfering-rna-sirna-therapeutics-competitive-landscape-technology-and-pipeline-insights-2015-a-delveinsights-report]
- 125. RNA-Targeting Techniques: A Comparative Analysis of ... [https://www.mdpi.com/2073-4425/16/10/1168]
- 126. Epigenetic Reprogramming: Reverse Aging at the Cellular ... [https://www.hubmeded.com/blog/epigenetic-reprogramming]
- 127. Turning back the epigenetic clock: Can we reverse ageing? [https://www.ddw-online.com/turning-back-the-epigenetic-clock-can-we-reverse-ageing-38601-202511/]
- 128. Chemical reprogramming ameliorates cellular hallmarks of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12340157/]
- 129. Why Epigenetic Clocks May Fail to Measure Anti- ... [https://www.aging-us.com/news-room/why-epigenetic-clocks-may-fail-to-measure-anti-aging-effects]
- 130. mRNA Vaccines: Current Applications and Future Directions [https://pmc.ncbi.nlm.nih.gov/articles/PMC12572956/]
- 131. Quantifying Cell Fate Decisions for Differentiation and ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003165]
- 132. Quantifying Cell Fate Decisions for Differentiation and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3731225/]
- 133. Beacon RNA: mRNA Summit 2025 Presentation [https://www.beacon-intelligence.com/conference-reports/mrna-summit-2025-post-conference-report/]