MSC Exosomes as Immunomodulatory Agents: Mechanisms and Applications in M2 Macrophage Polarization
This article synthesizes current research on how mesenchymal stem/stromal cell (MSC)-derived exosomes orchestrate macrophage polarization toward the anti-inflammatory M2 phenotype.
MSC Exosomes as Immunomodulatory Agents: Mechanisms and Applications in M2 Macrophage Polarization
Abstract
This article synthesizes current research on how mesenchymal stem/stromal cell (MSC)-derived exosomes orchestrate macrophage polarization toward the anti-inflammatory M2 phenotype. We explore foundational mechanisms including CD73/adenosine signaling, exosomal miRNA delivery, and protein-mediated pathways. For researchers and drug development professionals, we detail methodological approaches for exosome characterization and therapeutic application across disease models, discuss strategies for optimizing exosome potency through preconditioning and engineering, and evaluate validation techniques and comparative efficacy of exosomes from different MSC sources. This comprehensive analysis highlights MSC exosomes as a promising cell-free therapeutic platform for modulating immune responses in inflammatory and autoimmune diseases.
The Biological Framework: Understanding Macrophage Plasticity and MSC Exosome Cargo
Macrophages, key components of the innate immune system, exhibit remarkable functional plasticity that allows them to respond dynamically to microenvironmental cues. This adaptability, referred to as macrophage polarization, enables these cells to adopt distinct functional phenotypes—primarily the pro-inflammatory M1 and anti-inflammatory M2 states—in response to various stimuli [1]. Rather than existing as discrete populations, macrophages occupy a dynamic continuum with M1 and M2 phenotypes representing the extremes of this functional spectrum [2]. The balance between these polarization states plays a critical role in immune homeostasis, tissue repair, and disease pathogenesis, making the understanding of this process fundamental to immunology research and therapeutic development.
Within the tumor microenvironment (TME), macrophage polarization becomes particularly significant. Tumor-associated macrophages (TAMs) demonstrate a profound impact on tumor progression, with their functional shift representing a core mechanism in cancer immunobiology [1]. The emerging role of mesenchymal stem cell (MSC) derived exosomes in modulating this polarization, specifically toward the M2 phenotype, presents a promising therapeutic avenue for immune-mediated diseases and cancer treatment [3] [4] [5].
The Macrophage Polarization Spectrum
Classically Activated M1 Macrophages
M1 macrophages are predominantly polarized through classical activation pathways in response to pro-inflammatory stimuli. Key inducers include interferon-gamma (IFN-γ), lipopolysaccharide (LPS), and other pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) [6]. The signaling pathways involved in M1 polarization are complex and multifaceted:
- IFN-γ activation: Binds to its receptor and activates the JAK1/2-STAT1 pathway, driving expression of pro-inflammatory genes [6]
- LPS signaling: Binds to TLR4-MD2 receptor, activating both the IRAK1/TRAF6/NF-κB and MyD88/NF-κB pathways, and phosphorylating IRF3 to activate interferon-sensitive response elements (ISRE) [6]
- TNF-α signaling: Activates the JNK cascade, stimulating AP-1 pathway activation [6]
- GM-CSF signaling: Exerts effects through JAK2/STAT5 pathway activation [6]
The metabolic profile of M1 macrophages is characterized by preferential utilization of glycolysis, generating substantial amounts of nitric oxide (NO) and pro-inflammatory cytokines [1]. This metabolic reprogramming supports their inflammatory functions and distinguishes them metabolically from their M2 counterparts.
Alternatively Activated M2 Macrophages
M2 macrophages undergo alternative activation primarily in response to type 2 T helper (Th2) cell-derived factors including interleukin-4 (IL-4), IL-6, IL-10, and IL-13, as well as other mediators such as transforming growth factor-β (TGF-β), glucocorticoids, and immune complexes [1]. The molecular mechanisms governing M2 polarization include:
- IL-4/IL-13 signaling: Activates STAT6 and IRF4 through JAK-STAT pathways, driving expression of characteristic M2 markers [1] [2]
- Metabolic programming: Preferential reliance on oxidative phosphorylation and fatty acid oxidation to produce anti-inflammatory cytokines such as IL-10 [1]
- Transcriptional regulation: Core regulatory factors STAT6 and IRF4 promote secretion of anti-inflammatory factors while enhancing lipid metabolism through PPARγ/LXR signaling networks [1]
M2 macrophages contribute significantly to tissue repair, inflammation resolution, and immunoregulation, making them attractive targets for therapeutic manipulation in inflammatory and autoimmune conditions.
Comparative Analysis of M1 and M2 Phenotypes
Table 1: Characteristic Features of M1 and M2 Macrophage Phenotypes
| Feature | M1 Macrophages | M2 Macrophages |
|---|---|---|
| Activation Stimuli | IFN-γ, LPS, TNF-α, GM-CSF [6] | IL-4, IL-13, IL-10, TGF-β, glucocorticoids [1] |
| Key Signaling Pathways | JAK/STAT1, NF-κB, IRF3/5 [1] [6] | STAT6, IRF4, PI3K/AKT, PPARγ/LXR [1] |
| Metabolic Preference | Glycolysis [1] | Oxidative phosphorylation, Fatty acid oxidation [1] |
| Characteristic Markers | CD80, CD86, iNOS, MHC-II, TNF-α, IL-12 [1] [6] | CD206, CD163, Arg-1, IL-10, TGF-β [1] [5] |
| Major Functions | Pro-inflammatory responses, Pathogen clearance, Anti-tumor immunity [1] | Anti-inflammatory responses, Tissue repair, Angiogenesis, Immunosuppression [1] |
| Secreted Factors | TNF-α, IL-1β, IL-6, IL-12, IL-23, CXCL chemokines [1] [6] | IL-10, TGF-β, VEGF, CCL17, CCL22 [1] |
Table 2: M1 and M2 Cytokine Profiles and Functional Outcomes
| Phenotype | Key Cytokines/Chemokines | Functional Outcomes |
|---|---|---|
| M1 | TNF-α, IL-1α, IL-1β, IL-6, IL-12, IL-23, CXCL9, CXCL10 [1] [6] | Activation of cytotoxic T lymphocytes, Induction of tumor cell apoptosis, Enhanced immune surveillance [1] |
| M2 | IL-10, TGF-β, IL-1RA, CCL17, CCL22 [1] [4] | Promotion of tumor angiogenesis, Immune evasion, Tumor cell proliferation and metastasis [1] |
MSC Exosomes as Regulators of Macrophage Polarization
Mechanisms of MSC Exosome-Mediated M2 Polarization
Mesenchymal stem cell-derived exosomes have emerged as potent mediators of macrophage polarization toward the M2 phenotype. These nano-sized extracellular vesicles (50-150 nm in diameter) facilitate cell-to-cell communication by transferring bioactive molecules, including proteins, lipids, and RNA species [4]. Multiple mechanistic studies have elucidated how MSC exosomes promote M2 polarization:
CD73-mediated adenosine signaling: MSC exosomes express CD73 (ecto-5'-nucleotidase), which catalyzes the conversion of AMP to adenosine. Adenosine then binds to A2A and A2B receptors on macrophages, activating AKT/ERK-dependent signaling pathways that drive M2 polarization [3]. Inhibition of CD73 activity or adenosine receptors abolishes this polarizing effect, confirming the critical nature of this pathway [3].
EDA-FN activated TLR signaling: Extra domain A-fibronectin (EDA-FN) present in MSC exosomes activates MyD88-dependent Toll-like receptor signaling, promoting M2-like macrophage polarization [3]. This mechanism works synergistically with CD73-mediated pathways to enhance the immunomodulatory effects.
MicroRNA-mediated regulation: MSC exosomes contain numerous immunomodulatory microRNAs that can be transferred to macrophages, altering gene expression and promoting M2 polarization. For instance, exosomal miR-let7 suppresses pro-inflammatory pathways and promotes M2 marker expression [4].
Metabolic reprogramming: MSC exosomes influence macrophage metabolism, shifting them toward oxidative phosphorylation and fatty acid oxidation—metabolic states characteristic of M2 macrophages [1] [4].
Diagram 1: MSC exosome mechanisms in M2 macrophage polarization
Experimental Evidence and Functional Outcomes
Multiple experimental approaches have demonstrated the efficacy of MSC exosomes in promoting M2 macrophage polarization with subsequent functional consequences:
In vitro polarization assays: Treatment of primary macrophages or THP-1-derived macrophages with MSC exosomes consistently increases expression of M2 markers (CD206, Arg-1, IL-10, TGF-β) while decreasing M1 markers (CD80, CD86, TNF-α, IL-12) [3] [5]. This effect is comparable to the polarization induced by dexamethasone, a potent anti-inflammatory steroid [5].
Disease models: In preclinical models of hyperoxia-induced lung injury, MSC exosomes promoted infiltration of M2-like macrophages while reducing M1-like macrophages and pro-inflammatory cytokines such as TNF-α [3]. Similar effects were observed in models of atherosclerosis, where MSC exosomes attenuated disease progression via miR-let7 mediated infiltration and polarization of M2 macrophages [4].
Therapeutic outcomes: The shift toward M2 polarization mediated by MSC exosomes correlates with improved tissue repair, reduced inflammation, and functional recovery in various disease contexts, including skeletal muscle injury, cardiovascular diseases, autoimmune conditions, and central nervous system disorders [4].
Research Methodologies and Experimental Protocols
Standardized Macrophage Polarization Protocols
Table 3: Experimental Conditions for Macrophage Polarization Studies
| Macrophage Type | Polarization Stimuli | Culture Duration | Key Markers for Validation |
|---|---|---|---|
| M0 (Naive) | M-CSF (40 ng/mL) [3] | 7-9 days [3] | CD14, CD68 [5] |
| M1 | IFN-γ (20-100 ng/mL) + LPS (10-100 ng/mL) [2] [6] | 24-48 hours [2] | CD80, CD86, iNOS, TNF-α, IL-12 [1] [5] |
| M2 | IL-4 (20-40 ng/mL) or IL-13 (20-40 ng/mL) [2] | 24-48 hours [2] | CD206, CD163, Arg-1, IL-10, TGF-β [1] [5] |
MSC Exosome Isolation and Characterization
A standardized protocol for MSC exosome isolation and application includes the following critical steps:
MSC Culture and Serum Adaptation: Culture MSC lines (e.g., E1-MYC 16.3 human ESC-derived MSCs) in chemically defined medium. Gradually adapt cells to serum-free conditions by reducing FBS concentration from 10% to elimination over several steps (5% → 2.5% → 1% → 0%) at 48-hour intervals [5].
Exosome Isolation: Collect conditioned medium after 48 hours in serum-free conditions. Perform sequential centrifugation: initial centrifugation at 3,000 rpm for 10 minutes at 4°C to remove dead cells and debris, followed by addition of exosome isolation solution (e.g., Exocib), overnight incubation at 4°C, and final centrifugation at 3,000 rpm for 40 minutes at 4°C to pellet exosomes [5].
Exosome Quantification and Characterization:
- Protein quantification: Use Nanodrop spectrophotometry (280 nm) or micro-Bradford assay [5]
- Size distribution analysis: Dynamic Light Scattering (DLS) with Zetasizer instruments [5]
- Morphological validation: Field Emission Scanning Electron Microscopy (FESEM) or Transmission Electron Microscopy (TEM) [5]
- Surface marker confirmation: Flow cytometry for tetraspanins (CD9, CD63, CD81) and CD73/NT5E activity assays [3] [5]
Exosome-Macrophage Co-culture: Treat macrophages with 10 μg/mL MSC exosomes for 24-48 hours. For inhibition studies, co-treat with CD73 inhibitors (e.g., PSB12379 at 10 nM) or adenosine receptor antagonists to confirm mechanism-specific effects [3].
Diagram 2: Experimental workflow for MSC exosome isolation and macrophage treatment
Assessment of Polarization Status
Comprehensive evaluation of macrophage polarization status following exosome treatment involves multiple methodological approaches:
- Surface marker analysis: Flow cytometry for M1 (CD80, CD86) and M2 (CD206, CD163) markers [5]
- Cytokine profiling: ELISA for pro-inflammatory (TNF-α, IL-12, IL-6) and anti-inflammatory (IL-10, TGF-β) cytokines in culture supernatants [5]
- Gene expression analysis: Real-time PCR for M1 (iNOS, IL-1β) and M2 (Arg-1, IL-10) marker genes [5]
- Metabolic assessment: Spectrophotometric assays for oxidative stress markers (MDA, NO) and antioxidant capacity (TAC, CAT, SOD) [5]
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Studying MSC Exosome-Mediated Macrophage Polarization
| Reagent Category | Specific Examples | Function/Application |
|---|---|---|
| Polarization Inducers | IFN-γ (M1), LPS (M1), IL-4 (M2), IL-13 (M2) [2] [6] | Direct polarization of macrophages to specific phenotypes |
| MSC Exosome Markers | CD9, CD63, CD81 antibodies [4] [5] | Identification and validation of exosome isolates |
| Functional Antibodies | CD80-FITC (M1), CD86-PE (M1), CD206-APC (M2) [5] | Flow cytometry analysis of macrophage surface markers |
| Pathway Inhibitors | PSB12379 (CD73 inhibitor), A2A/A2B receptor antagonists [3] | Mechanistic studies of exosome-mediated polarization |
| Cytokine Detection | TNF-α, IL-12, IL-10, TGF-β ELISA kits [5] | Quantification of inflammatory and anti-inflammatory mediators |
| Cell Lines | THP-1 monocyte cell line, Primary human or rat macrophages [3] [5] | Consistent cellular models for polarization studies |
| Exosome Isolation Kits | Exocib kit, Tangential flow filtration systems [3] [5] | Standardized isolation of exosomes from conditioned media |
Technical Considerations and Limitations
While the M1/M2 dichotomy provides a valuable framework for studying macrophage functions, several technical and conceptual limitations must be considered:
Phenotypic continuum: Macrophages exist along a dynamic continuum rather than in discrete categories, exhibiting remarkable plasticity shaped by local microenvironmental cues including metabolic signaling and extracellular matrix composition [1]. Single-cell transcriptomics has revealed that conventional binary classification systems are biologically inadequate for capturing the true complexity of macrophage functional states [1].
Marker co-expression: Recent studies have identified macrophage populations that co-express both classical M1 markers (e.g., iNOS) and alternative M2 markers (e.g., Arg-1), demonstrating capacity for rapid functional switching that challenges traditional classification systems [1].
Methodological limitations: Current surface marker-based detection methods (e.g., CD86 for M1-like or CD206 for M2-like phenotypes) may fail to comprehensively characterize macrophage heterogeneity, particularly when analyzing at single-cell resolution [1]. Future investigations should integrate single-cell multi-omics with spatial profiling technologies to achieve higher-resolution characterization of macrophage subsets [1].
These considerations highlight the need for multi-dimensional profiling approaches and functionally defined classification frameworks that transcend conventional surface marker-based paradigms in macrophage polarization research.
Mesenchymal stem cell-derived exosomes (MSC-Exos) have emerged as a paradigm-shifting therapeutic modality in regenerative medicine and immunomodulation. These nano-sized extracellular vesicles (30-150 nm in diameter) mediate intercellular communication by transferring functional proteins, lipids, and nucleic acids to recipient cells, thereby orchestrating diverse biological processes [7] [8]. The therapeutic potential of MSC-Exos mirrors that of their parent cells but with significant advantages, including lower immunogenicity, enhanced stability, and the ability to cross biological barriers [9] [8]. This technical guide provides a comprehensive examination of MSC-Exos, with a specific focus on their biogenesis, molecular composition, and characterization, framed within the context of their crucial role in modulating macrophage polarization toward the anti-inflammatory M2 phenotype—a key mechanism underpinning their therapeutic efficacy in inflammatory and degenerative diseases [10] [11].
Biogenesis and Composition of MSC Exosomes
Biogenesis Pathways
The formation of MSC-Exos is a meticulously regulated process originating from the endosomal system [9]:
- Early Endosome Formation: The process initiates with the invagination of the plasma membrane, forming early endosomes that serve as the primary vesicular compartments for exosome production [9].
- Multivesicular Body (MVB) Development: Early endosomes mature into late endosomes, which subsequently evolve into multivesicular bodies (MVBs) through inward budding of the endosomal membrane. During this process, cytosolic components are sequestered into intraluminal vesicles (ILVs) within the MVBs [12] [8].
- Exosome Release: MVBs follow one of two pathways: degradation through fusion with lysosomes or exocytosis through fusion with the plasma membrane, releasing ILVs into the extracellular space as exosomes [13] [14].
This biogenesis is governed by both ESCRT (Endosomal Sorting Complex Required for Transport)-dependent and ESCRT-independent mechanisms, with key regulatory proteins including Rab GTPases (Rab27a, Rab27b), TSG101, and Alix [14] [9].
Molecular Composition
MSC-Exos encapsulate a diverse array of biomolecules that reflect their biological functions:
- Proteins: MSC-Exos contain transmembrane proteins (CD9, CD63, CD81), heat shock proteins (HSP60, HSP70, HSP90), biogenesis-related proteins (Alix, TSG101), and adhesion molecules (CD29, CD44, CD73) [10] [14].
- Nucleic Acids: Exosomes carry various genetic materials, including mRNAs, microRNAs (miRNAs), and other non-coding RNAs, which can regulate gene expression in recipient cells [10] [7].
- Lipids: The exosomal membrane is enriched in cholesterol, ceramides, and sphingolipids, which provide structural integrity and facilitate membrane fusion [14].
Table 1: Essential Markers for MSC-Exos Characterization
| Marker Category | Specific Markers | Function and Significance |
|---|---|---|
| Transmembrane/Tetraspanin Proteins | CD9, CD63, CD81 | Form oligomeric complexes; participate in membrane fusion and cargo sorting; universal exosome markers [13] [8] [14] |
| ESCRT-Associated Proteins | TSG101, Alix | Involved in the biogenesis and cargo sorting during MVB formation [8] [14] |
| Heat Shock Proteins | HSP70, HSP90 | Involved in protein folding and antigen presentation [14] |
| MSC-Surface Markers | CD29, CD44, CD73, CD105 | Reflect mesenchymal origin; CD73 has immunomodulatory function via adenosine production [10] [3] |
| Lipid Raft Proteins | Flotillin-1, Flotillin-2 | Contribute to the structure of specialized membrane microdomains [14] |
MSC Exosomes in Macrophage Polarization to M2 Phenotype
Macrophage Plasticity and Polarization
Macrophages are highly plastic immune cells that differentiate into distinct functional phenotypes in response to microenvironmental cues [10]. The classically activated M1 phenotype is induced by IFN-γ and LPS, promoting inflammation through pro-inflammatory cytokines (TNF-α, IL-1, IL-6). Conversely, the alternatively activated M2 phenotype is induced by IL-4 and IL-13, exerting anti-inflammatory effects and facilitating tissue repair [10] [3]. In pathological conditions such as spinal cord injury, infiltrating macrophages predominantly adopt the M1 phenotype, exacerbating tissue damage. MSC-Exos have demonstrated the capacity to shift this balance toward the M2 phenotype, thereby promoting resolution of inflammation and tissue regeneration [10] [11].
Mechanisms of M2 Polarization by MSC Exosomes
MSC-Exos employ multiple sophisticated mechanisms to promote M2 macrophage polarization, with two particularly well-characterized pathways highlighted below.
CD73/Adenosine Pathway
MSC-Exos express CD73 (ecto-5'-nucleotidase) on their surface, which catalyzes the conversion of extracellular adenosine monophosphate (AMP) to adenosine [3]. Adenosine then binds to A2A and A2B receptors on macrophages, activating AKT/ERK-dependent signaling pathways that drive M2 polarization [3]. This pathway has been experimentally validated using specific inhibitors: PSB12379 (CD73 inhibitor) and antagonists of A2A/A2B receptors, which abolish the polarizing effect of MSC-Exos [3].
EDA-Fibronectin/TLR Pathway
MSC-Exos contain extra domain A-fibronectin (EDA-FN), which activates the MyD88-dependent Toll-like receptor (TLR) signaling pathway in macrophages, promoting their differentiation toward the M2 phenotype [3]. This mechanism has been demonstrated in both mouse and human monocytes [3].
In addition to these pathways, exosomal miRNAs play a significant role. MSC-Exos deliver specific miRNAs to macrophages, modulating signaling pathways that influence polarization. For instance, exosomal miR-223 can downregulate pro-inflammatory genes in target cells [11].
Experimental Characterization of MSC Exosomes
Isolation and Purification Methods
The reliability of experimental data on MSC-Exos is contingent upon rigorous isolation and characterization. The primary methods include:
- Ultracentrifugation (Gold Standard): Involves differential centrifugation steps, culminating in high-speed ultracentrifugation (100,000-120,000 × g) to pellet exosomes. While cost-effective, it is time-consuming and may compromise vesicle integrity [14] [15].
- Tangential Flow Filtration (TFF): Utilizes a hollow fiber membrane to concentrate and purify exosomes based on size. This method is scalable for clinical-grade production and offers higher efficiency and better preservation of exosome integrity [3] [15].
- Size Exclusion Chromatography (SEC): Separates exosomes from contaminants based on hydrodynamic volume, yielding high-purity preparations suitable for functional studies [14].
- Commercial Kits: Polymer-based precipitation kits offer convenience for small-scale studies but may co-precipitate non-exosomal contaminants [14].
Table 2: Key Experimental Protocols for MSC-Exos Macrophage Polarization Studies
| Experimental Area | Protocol Description | Key Reagents and Parameters |
|---|---|---|
| Exosome Isolation | Ultracentrifugation: Sequential centrifugation to eliminate cells, debris, and microvesicles, followed by ultracentrifugation at 100,000-120,000 × g [14] [15]. | PBS for washing/resuspension; sucrose buffer for density gradient [15]. |
| Macrophage Differentiation | Isolation of PBMCs via Ficoll-Paque density gradient centrifugation; differentiation with macrophage colony-stimulating factor (M-CSF, 40 ng/mL) for 7-9 days [3]. | Ficoll-Paque; M-CSF; culture media (RPMI with 10% FBS) [3]. |
| Polarization Assay | Treatment of primary macrophages with MSC-Exos (e.g., 10 μg/mL) for 24-48 hours; assessment of phenotype via marker analysis [3]. | MSC-Exos characterized by protein/particle concentration; LPS/IFN-γ for M1 control; IL-4/IL-13 for M2 control [3]. |
| Mechanistic Inhibition Studies | Co-treatment of macrophages with MSC-Exos and specific pathway inhibitors to confirm mechanism [3]. | PSB12379 (CD73 inhibitor, 10 nM); A2A/A2B adenosine receptor antagonists; inhibitors of AKT/ERK phosphorylation [3]. |
Characterization Techniques
Comprehensive characterization is essential per MISEV (Minimal Information for Studies of Extracellular Vesicles) guidelines:
- Physical Characterization: Nanoparticle Tracking Analysis (NTA) and Dynamic Light Scattering (DLS) determine particle size distribution and concentration. Transmission Electron Microscopy (TEM) confirms spherical morphology and bilayer structure [13] [14].
- Biochemical Characterization: Western blot, flow cytometry, or ELISA are used to detect positive markers (CD9, CD63, CD81, TSG101) and negative markers (calnexin, apolipoproteins) [13] [15].
- Functional Assays: Uptake studies using fluorescently labeled exosomes, along with in vitro and in vivo models to assess specific biological functions such as M2 macrophage polarization [3] [11].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for MSC-Exos and Macrophage Polarization Research
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| MSC-Exos Sources | Human bone marrow, adipose tissue, umbilical cord, dental pulp, placenta [13] [7] [15]. | Provide biologically active exosomes; source impacts exosomal cargo and functional efficacy [13] [15]. |
| Characterization Antibodies | Anti-CD9, Anti-CD63, Anti-CD81, Anti-TSG101, Anti-Alix, Anti-HSP70 [13] [14] [15]. | Essential for Western blot, flow cytometry, and immunoaffinity capture to confirm exosome identity. |
| Pathway Inhibitors | PSB12379 (CD73 inhibitor), A2A/A2B adenosine receptor antagonists, AKT/ERK pathway inhibitors [3]. | Tool compounds for mechanistic studies to dissect specific signaling pathways in macrophage polarization. |
| Macrophage Polarization Cytokines | IFN-γ, LPS (for M1 polarization); IL-4, IL-13 (for M2 polarization) [10] [3]. | Used as positive and negative controls in polarization assays to validate the experimental system. |
| Cell Culture Reagents | M-CSF (for macrophage differentiation), Ficoll-Paque (for PBMC isolation), defined exosome-production media [3] [15]. | Enable the differentiation and maintenance of primary macrophages and the production of contaminant-free exosomes. |
MSC exosomes represent a sophisticated biological system for intercellular communication, with a defined biogenesis pathway and a characteristic composition of proteins, lipids, and nucleic acids. The precise combination of tetraspanins (CD9, CD63, CD81) and ESCRT-related proteins (TSG101) serves as a fundamental signature for their identification. Their profound capacity to polarize macrophages toward an M2 phenotype via mechanisms such as the CD73/adenosine and EDA-FN/TLR pathways underscores their therapeutic potential in immune-mediated and degenerative diseases. As research progresses, the translation of MSC-Exos into clinical applications will heavily rely on standardized, reproducible experimental protocols—from isolation and characterization to functional validation—enabling their development as a consistent and effective cell-free therapeutic biologic.
The tumor microenvironment (TME) is a landscape of dynamic cellular crosstalk, often co-opting physiological pathways to foster immunosuppression and support tumor growth. Among these, the purinergic signaling pathway, specifically the CD73/adenosine axis, has emerged as a master regulator of immune evasion. The core of this mechanism involves the metabolic conversion of extracellular ATP (eATP), a damage-associated molecular pattern (DAMP) that stimulates immunity, into immunosuppressive extracellular adenosine (eADO). This adenosine primarily activates two G-protein-coupled receptors, A2A and A2B, on immune cells, triggering intracellular signaling cascades that promote the polarization of macrophages towards an M2, pro-tumorigenic phenotype [16] [17]. This review details the core mechanism of this axis and frames it within the context of a burgeoning field of research: the modulation of this pathway by Mesenchymal Stem Cell (MSC)-derived exosomes. Understanding this interaction is critical for developing novel immunotherapies that can reprogram the TME and enhance anti-tumor immunity.
The Core Mechanism: From ATP to Adenosine and M2 Polarization
The CD39-CD73 Ectoenzyme Cascade: Generating Immunosuppressive Adenosine
The adenosinergic pathway initiates with the release of eATP from stressed, damaged, or dying cells within the TME. The conversion of this immunostimulatory eATP into immunosuppressive eADO is a two-step process catalyzed by the ectoenzymes CD39 and CD73.
- Step 1: CD39 (ENTPD1) catalyzes the rate-limiting hydrolysis of eATP (or ADP) to AMP [16] [18].
- Step 2: CD73 (NT5E), an ecto-5'-nucleotidase, then performs the final dephosphorylation of AMP to adenosine [19] [16] [18].
This pathway is significantly amplified in the TME, particularly under hypoxic conditions. Hypoxia-inducible factor-1α (HIF-1α), upregulated in low oxygen, acts as a key transcription factor driving the expression of CD73, thereby enhancing adenosine production [19]. Furthermore, a critical feedforward circuit exists in areas like colorectal cancer, where adenosine signaling through the A2B receptor on cancer-associated fibroblasts (CAFs) further upregulates their CD73 expression, creating a potent, self-reinforcing immunosuppressive niche [20].
A2A and A2B Receptor Signaling: Divergent Affinity, Convergent Outcome on Macrophages
Once generated, extracellular adenosine exerts its effects by binding to P1 purinergic receptors. The A2A and A2B receptors are the primary mediators of adenosine-induced immunosuppression, both coupling to Gs proteins and activating adenylate cyclase to increase intracellular cyclic AMP (cAMP) levels [21] [16]. However, they exhibit distinct affinities and contextual roles.
- A2A Receptor (High Affinity): Activated under physiological and moderately elevated adenosine concentrations. Its signaling robustly elevates cAMP, which suppresses the function of effector immune cells like T cells and NK cells [16].
- A2B Receptor (Low Affinity): Requires the high adenosine concentrations typically found in pathological sites like the TME for activation. It is crucial for sustaining the immunosuppressive network, notably by promoting the M2 polarization of macrophages and driving collagen production and fibrosis [21] [22] [20].
The elevated intracellular cAMP activates Protein Kinase A (PKA) and the exchange protein directly activated by cAMP (Epac). These downstream effectors, in turn, inhibit pro-inflammatory pathways and activate transcription factors that drive the expression of M2-associated genes [21]. The table below summarizes the key features of these receptors.
Table 1: Characteristics of A2A and A2B Adenosine Receptors in the TME
| Feature | A2A Receptor (A2AR) | A2B Receptor (A2BR) |
|---|---|---|
| Affinity for Adenosine | High (nM range) [16] | Low (μM range) [16] |
| Primary Signaling | Gs → ↑ cAMP → PKA [21] [16] | Gs → ↑ cAMP → PKA / Epac [21] |
| Key Context of Action | Physiological & pathological adenosine levels | Pathologically high adenosine (e.g., hypoxic TME) [22] |
| Major Role in Macrophages | Inhibition of pro-inflammatory (M1) responses; promotion of M2 polarization [17] | Direct driving of M2 polarization; enhancement of pro-fibrotic responses [22] |
| Cross-talk | Synergizes with PD-1/CTLA-4 signaling [22] | Forms feedforward circuit with CAF-CD73 [20] |
The MSC Exosome Connection: Modulators of the Adenosine Axis
Emerging evidence positions MSC-derived exosomes as powerful, nanoscale mediators of intercellular communication that can directly influence the CD73/adenosine pathway and macrophage polarization. These extracellular vesicles transfer bioactive cargo—including microRNAs (miRNAs), proteins, and lipids—to recipient cells in the TME.
Documented Effects of MSC Exosomes on Macrophage Polarization
Studies consistently show that MSC-derived exosomes can drive a shift from a pro-inflammatory M1 phenotype to an anti-inflammatory, pro-repair M2 phenotype.
- SHED-MSC Exosomes: Exosomes from stem cells of human exfoliated deciduous teeth (SHED) were shown to skew M0/M1 macrophages to the M2 phenotype. Treatment led to a significant increase in M2 markers (CD206, Arg-1, IL-10, TGF-β) and a decrease in M1 markers (CD80, IL-12, TNF-α) [23].
- BMSC Exosomes in Osteomyelitis: Bone marrow MSC (BMSC)-derived exosomes carrying miR-223 were found to mitigate LPS-induced macrophage pyroptosis and inflammation, creating a less hostile environment that favors M2-like polarization [24].
- General Mechanism: The immunomodulatory effect is akin to that of the anti-inflammatory drug dexamethasone, highlighting the potent regulatory capacity of these exosomes [23].
Potential Mechanisms of Cross-talk with the Adenosine Pathway
The precise interaction between MSC exosomes and the adenosine pathway is an active area of research, with several plausible mechanistic links:
- Direct Cargo Delivery: MSC exosomes may carry and deliver functional CD39 or CD73 enzyme themselves, or miRNAs that regulate the expression of these ectoenzymes or adenosine receptors in recipient macrophages or CAFs.
- Reprogramming Recipient Cells: By promoting a generalized anti-inflammatory, M2-polarized state in macrophages via transferred miRNAs (e.g., miR-223 [24]) or cytokines (e.g., TGF-β [23]), exosomes could indirectly create a milieu that favors the stability and activity of the CD73/adenosine axis.
- Feedback Loop: Adenosine signaling through A2A/A2B receptors on MSCs could influence the cargo and release of their exosomes, establishing a feedback loop that fine-tunes the immunosuppressive landscape.
Diagram: Proposed Crosstalk Between MSC Exosomes and the Adenosine Axis in Macrophage Polarization
Experimental Protocols for Investigating the Axis
To empirically validate the relationships described, researchers can employ the following detailed methodologies.
Protocol 1: In Vitro Macrophage Polarization and Phenotyping
This protocol assesses the direct impact of adenosine receptor agonism/antagonism or MSC exosomes on macrophage polarization.
1. Macrophage Culture and Stimulation:
- Cells: Use human monocytic cell lines (e.g., THP-1) differentiated into M0 macrophages with PMA (e.g., 100 nM for 48 hours), or primary human monocyte-derived macrophages.
- Polarization Induction: Polarize M0 macrophages to M1 using LPS (e.g., 100 ng/ml) and IFN-γ (e.g., 20 ng/ml) [23].
- Experimental Treatment:
- Adenosine Pathway Modulation: Treat M1 or M0 macrophages with:
- A2A agonist (e.g., CGS-21680) and/or A2B agonist (e.g., BAY 60-6583).
- A2A antagonist (e.g., SCH-58261) and/or A2B antagonist (e.g., PSB-1115).
- Recombinant CD73 enzyme to increase ambient adenosine.
- CD73 inhibitor (e.g., APCP, AB680).
- MSC Exosome Treatment: Introduce purified MSC exosomes (e.g., 50-100 μg/mL) to the macrophage culture [23] [24].
- Adenosine Pathway Modulation: Treat M1 or M0 macrophages with:
2. Flow Cytometry Analysis:
- Harvest: Harvest macrophages after 24-48 hours of treatment.
- Staining: Stain cells with fluorochrome-conjugated antibodies against surface markers.
- Analysis: Analyze by flow cytometry. The M1 phenotype is characterized by high CD80, CD86, while the M2 phenotype is characterized by high CD206, CD163 [23] [25].
3. Cytokine Profiling:
- Collection: Collect cell culture supernatants after treatment.
- Quantification: Use ELISA or multiplex cytokine arrays to measure cytokine concentrations.
- Expected Shift: A successful M2 polarization will show increased IL-10 and TGF-β and decreased TNF-α, IL-12, and IL-6 [23] [25].
Protocol 2: Validating the cAMP/PKA/Snail Pathway in EMT and Invasion
This protocol, adaptable from gastric cancer studies, demonstrates a key downstream pathway of Adora2b activation relevant to cancer progression [22].
1. Cell Treatment:
- Use gastric cancer cell lines (e.g., MKN-45, MGC-803) or other relevant adenocarcinoma cells.
- Treat cells with an A2B receptor agonist (BAY 60-6583) or antagonist (PSB-1115) for specified durations (e.g., 1-6 hours for signaling, 24-48 hours for invasion).
2. Western Blot Analysis:
- Lysate Preparation: Prepare RIPA cell lysates.
- Electrophoresis and Transfer: Separate proteins by SDS-PAGE and transfer to PVDF membranes.
- Antibody Probing: Probe membranes with specific primary and secondary antibodies.
- Key Targets:
- Phospho-PKA Substrates: To confirm PKA activation.
- Total and Phospho-Snail: To assess Snail activation and stability.
- EMT Markers: E-cadherin (expected decrease), Vimentin (expected increase) [22].
3. Functional Invasion Assay:
- Setup: Use Matrigel-coated Transwell inserts.
- Seeding and Treatment: Seed serum-starved cells in the upper chamber with A2B agonist/antagonist. Place complete growth medium in the lower chamber as a chemoattractant.
- Incubation and Quantification: Incubate for 24-48 hours. Fix, stain (e.g., with crystal violet), and count cells that have invaded through the Matrigel. A2B activation is expected to increase invasive capacity [22].
Table 2: Key Quantitative Findings from Preclinical Studies of the Adenosine Axis
| Experimental Model | Intervention | Key Quantitative Outcome | Citation |
|---|---|---|---|
| GC Lung Metastasis Model | CD73 knockout | ↓ Metastatic nodules by 60%; ↑ CD8+ T cells 2.3-fold; ↓ Treg infiltration by 40% | [22] |
| GC Cell Line (MKN-45) | Adora2b knockdown | ↓ Snail/Vimentin expression by 30% | [22] |
| SHED-MSC Exosome on Macrophages | Exosome treatment | Significant (P<0.05) ↑ CD206, IL-10, TGF-β; ↓ CD80, IL-12, TNF-α | [23] |
| Dermal Fibrosis Model | A2AR stimulation | ↑ Collagen production; effect abrogated in A2AR knockout mice | [21] |
The Scientist's Toolkit: Essential Research Reagents
The following table compiles key reagents and tools essential for experimental investigation of the CD73/adenosine axis.
Table 3: Essential Reagents for Research on the CD73/Adenosine Axis
| Reagent / Tool | Function / Specificity | Example Use Case | |
|---|---|---|---|
| CD73 Inhibitor (e.g., AB680, APCP) | Small molecule inhibitor of CD73 enzymatic activity. | Blocking adenosine production in co-culture assays to assess its necessity for M2 polarization. | [16] [18] |
| A2A Receptor Antagonist (e.g., SCH-58261) | Selective antagonist for the A2A receptor. | Determining the specific contribution of A2A vs. A2B signaling in macrophage polarization. | [21] [20] |
| A2B Receptor Antagonist (e.g., PSB-1115) | Selective antagonist for the A2B receptor. | Investigating the role of A2B in fibrosis, CAF activation, and M2 polarization in hypoxic TME. | [22] [20] |
| Recombinant CD73 Protein | Active ecto-5'-nucleotidase enzyme. | Exogenously increasing adenosine levels in vitro to model the TME. | [18] |
| Anti-CD73 Neutralizing Antibody | Antibody that blocks CD73 enzyme function. | In vivo studies to inhibit the adenosine pathway and evaluate therapeutic efficacy. | [20] [18] |
| Flow Cytometry Antibodies (CD80, CD86, CD206, CD163) | Cell surface markers for M1 (CD80/86) and M2 (CD206/163) macrophages. | Phenotyping macrophage populations after experimental treatments. | [23] [25] |
| cAMP ELISA Kit | Quantifies intracellular cyclic AMP levels. | Directly measuring the activation of adenosine receptor downstream signaling. | [21] |
| MSC Exosome Isolation Kit | Isulates and purifies exosomes from MSC conditioned media. | Obtaining the exosomal fraction for functional studies on macrophage polarization. | [23] [24] |
The CD73/adenosine axis, operating through the A2A and A2B receptors, constitutes a powerful, metabolism-driven mechanism of immunosuppression by enforcing M2 macrophage polarization. The emerging role of MSC-derived exosomes as modulators of this axis introduces a new layer of complexity and a potential therapeutic avenue. Future research must focus on elucidating the specific cargo within MSC exosomes that interfaces with the purinergic pathway. Combining adenosine pathway inhibitors (targeting CD73, A2A, or A2B) with strategies that leverage or engineer MSC exosomes to deliver anti-tumor cargo represents a promising frontier. Such multi-targeted approaches are crucial for dismantling the robust immunosuppressive networks in the TME and enhancing the efficacy of next-generation cancer immunotherapies.
Exosomes are small extracellular vesicles (30-150 nm in diameter) that serve as crucial mediators of intercellular communication by transferring functional proteins, lipids, and nucleic acids between cells [26] [4]. These vesicles are formed through the endosomal pathway, originating from the inward budding of endosomal membranes to create intraluminal vesicles within multivesicular bodies (MVBs), which subsequently fuse with the plasma membrane to release exosomes into the extracellular space [26] [27]. The biogenesis process involves sophisticated machinery including the Endosomal Sorting Complex Required for Transport (ESCRT) and various associated proteins (Alix, TSG101), while Rab GTPases and SNARE proteins mediate MVB trafficking and fusion with the plasma membrane [26].
The therapeutic potential of mesenchymal stem cell (MSC)-derived exosomes has garnered significant scientific interest, particularly their ability to modulate immune responses by promoting the polarization of macrophages toward the anti-inflammatory M2 phenotype [3] [4]. This polarization is a critical mechanism in tissue repair, resolution of inflammation, and regenerative processes [28]. The diverse cargo loaded into MSC exosomes—including microRNAs, proteins, and cytokines—orchestrates this immunomodulatory effect through multiple synergistic pathways, making them promising candidates for therapeutic development in inflammatory diseases, cancer, and regenerative medicine [29] [4].
Exosomal Cargo Components and Their Mechanisms in M2 Macrophage Polarization
MicroRNAs (miRNAs) as Key Regulatory Molecules
Exosomal microRNAs are short non-coding RNA molecules that post-transcriptionally regulate gene expression in recipient cells [26]. The sorting of miRNAs into exosomes is a selective process mediated by RNA-binding proteins and specific sequence motifs [26]. Once delivered to target cells, these miRNAs can profoundly alter macrophage function and polarization state.
Table 1: Exosomal microRNAs in Macrophage Polarization
| microRNA | Biological Effect | Proposed Mechanism/Target | Experimental Context |
|---|---|---|---|
| miR-let7 | Promotes M2 polarization; Reduces atherosclerosis | Mediates infiltration and polarization of M2 macrophages | MSC exosomes in ApoE−/- mouse model [4] |
| miR-182 | Attenuates myocardial ischemia-reperfusion injury | Regulates macrophage polarization | MSC exosomes in cardiac injury model [4] |
| miR-21 | Anti-inflammatory effects | Not fully elucidated | MSC exosomes in immunomodulation [27] |
| miR-16 | Anti-inflammatory effects | Not fully elucidated | MSC exosomes in immunomodulation [27] |
The diversity of exosomal miRNAs allows for precise regulation of multiple targets within macrophage signaling networks. For instance, MSC-derived exosomes enriched in miR-let7 have been shown to promote M2 macrophage polarization and attenuate atherosclerosis progression in preclinical models [4]. Similarly, exosomal miR-182 contributes to cardioprotection following myocardial ischemia-reperfusion injury by modulating macrophage polarization [4]. The stability of miRNAs within the exosomal lipid bilayer enhances their potential as therapeutic agents, protecting them from degradation by extracellular RNases [26].
Proteins as Effectors of Immunomodulation
Exosomal proteins constitute another essential component of the MSC exosome cargo, with diverse functions ranging from enzymatic activity to signal transduction. Among these, CD73 and MFGE8 have emerged as critical mediators of M2 macrophage polarization.
Table 2: Exosomal Proteins in Macrophage Polarization
| Protein | Biological Effect | Mechanism of Action | Experimental Evidence |
|---|---|---|---|
| CD73 (NT5E) | Promotes M2-like macrophage polarization | Catalyzes AMP→adenosine; Binds A2A/A2B receptors; Activates AKT/ERK pathways | Human ESC-derived MSC exosomes; CD73 inhibition blocks polarization [3] |
| MFGE8 | Reduces fibrosis; Promotes M2b polarization; Enhances efferocytosis | Activates STAT3/Arg1 axis; Binds αvβ3 integrin; Activates Src-FAK-STAT3 signaling | ADMSC exosomes in porcine esophageal stricture model; CRC-derived EVs [30] [31] |
| Extra Domain A-Fibronectin (EDA-FN) | Induces M2 phenotype | Activates MyD88-dependent TLR signaling pathway | Mouse and human monocyte studies [3] |
CD73, a surface ecto-5'-nucleotidase, mediates its effects through the production of adenosine, which then binds to adenosine receptors A2A and A2B on macrophages, subsequently activating AKT/ERK-dependent signaling pathways [3]. The critical role of CD73 has been demonstrated through inhibition experiments where polarization of M2-like macrophages by MSC exosomes was abolished in the presence of CD73 inhibitors [3].
MFGE8 (lactadherin) plays a dual role in immunomodulation, both by promoting the polarization of M2b macrophages through the STAT3/Arg1 axis and by enhancing efferocytosis—the clearance of apoptotic cells—through interaction with αvβ3 integrins on macrophages [30] [31]. In a porcine model of esophageal stricture, MSC exosomes containing MFGE8 significantly reduced fibrosis and collagen deposition by modulating macrophage phenotype [30].
Cytokines and Signaling Molecules
While the search results provide less specific detail on individual cytokines in exosomal cargo compared to miRNAs and proteins, cytokines collectively contribute to the exosome-mediated immunomodulatory environment. The exosomal membrane contains various cytokines and surface proteins that can directly interact with receptors on target cells, while the luminal cargo includes additional signaling molecules that can be released into the target cell upon fusion [4] [27].
The collaborative action of these diverse cargo components enables MSC exosomes to precisely orchestrate macrophage polarization, making them potent regulators of the immune response in various pathological conditions.
Signaling Pathways in Exosome-Mediated M2 Polarization
The promotion of M2 macrophage polarization by MSC exosomes occurs through multiple interconnected signaling pathways, activated by the diverse exosomal cargo components.
Diagram 1: Signaling pathways in exosome-mediated M2 macrophage polarization. MSC exosomes deliver diverse cargo that activates multiple signaling pathways in macrophages, collectively promoting the anti-inflammatory M2 phenotype.
The CD73/adenosine pathway represents a crucial mechanism where exosomal CD73 catalyzes the conversion of AMP to adenosine, which then binds to A2A and A2B adenosine receptors on macrophages, subsequently activating AKT/ERK signaling [3]. Simultaneously, the MFGE8/integrin pathway involves exosomal MFGE8 binding to αvβ3 integrins, leading to activation of Src-FAK signaling and subsequent STAT3 phosphorylation and nuclear translocation, ultimately driving expression of M2-related genes like Arg1 [30] [31]. Additional pathways include EDA-FN-mediated activation of MyD88-dependent TLR signaling [3], while various exosomal miRNAs modulate multiple targets within these networks to reinforce M2 polarization.
Experimental Approaches and Methodologies
Exosome Isolation and Characterization
Standardized protocols for exosome isolation and characterization are critical for research reproducibility. The most common isolation method is multi-step ultracentrifugation, which involves sequential centrifugation steps to remove cells and debris (300-2000 × g), followed by ultracentrifugation at 100,000 × g to pellet exosomes [29] [31]. Alternative methods include size-exclusion chromatography, polymer-based precipitation, and immunoaffinity capture, with combinations of methods often providing superior purity [29].
Characterization of isolated exosomes typically involves:
- Nanoparticle Tracking Analysis (NTA): Determines particle size distribution and concentration [3] [30]
- Transmission Electron Microscopy (TEM): Visualizes exosome morphology and structure [30]
- Western Blotting: Detects exosomal markers (CD9, CD63, CD81, TSG101, Alix) and absence of negative markers (calnexin, albumin) [3] [30]
- Protein Quantification: Measures total exosomal protein content [3]
Functional characterization includes enzymatic assays for specific exosomal components, such as CD73/NT5E activity measured using phosphate detection systems [3].
Macrophage Polarization Assays
Experimental evaluation of macrophage polarization typically involves in vitro differentiation of macrophages from primary sources (e.g., peripheral blood mononuclear cells or bone marrow-derived cells) using macrophage colony-stimulating factor (M-CSF) [3] [31]. Following differentiation, macrophages are treated with MSC exosomes and assessed for polarization markers:
- Flow Cytometry: Surface expression of M2 markers (CD206, CD163) [30] [32]
- qRT-PCR: Gene expression of M2-associated markers (Arg1, YM1, IL-10) [30]
- ELISA/Immunoassay: Secretion of M2-related cytokines (IL-10, TGF-β) [30]
- Immunofluorescence: Cellular localization of polarization markers [30]
Inhibition studies using specific pharmacological inhibitors (e.g., PSB12379 for CD73) help establish the functional contribution of specific exosomal components to macrophage polarization [3].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Exosome-Macrophage Studies
| Reagent/Category | Specific Examples | Function/Application | Experimental Context |
|---|---|---|---|
| CD73 Inhibitors | PSB12379 | Inhibits CD73 enzymatic activity; Blocks adenosine production | Validates CD73-mediated polarization [3] |
| Adenosine Receptor Antagonists | A2A and A2B receptor inhibitors | Blocks adenosine receptor signaling | Confirms receptor involvement in polarization [3] |
| Signaling Inhibitors | AKT/ERK phosphorylation inhibitors | Blocks downstream signaling pathways | Validates AKT/ERK role in polarization [3] |
| Exosome Isolation Reagents | Ultracentrifugation equipment; Size-exclusion chromatography columns | Isolates and purifies exosomes from conditioned media | Standard exosome preparation [29] [31] |
| Macrophage Differentiation Factors | M-CSF (Macrophage Colony-Stimulating Factor) | Differentiates monocytes to macrophages | Primary macrophage culture [3] [31] |
| M2 Macrophage Detection Antibodies | Anti-CD206, Anti-CD163, Anti-Arg1 | Identifies and quantifies M2 polarization | Flow cytometry, immunofluorescence [30] [32] |
| Exosome Labeling Dyes | PKH-26, PKH-67 | Tracks exosome uptake by recipient cells | Cellular uptake studies [30] |
The diverse cargo of MSC exosomes—including specific microRNAs, proteins like CD73 and MFGE8, and cytokines—collectively orchestrates macrophage polarization toward the anti-inflammatory M2 phenotype through multiple synergistic signaling pathways. This mechanistic understanding provides a robust foundation for developing exosome-based therapeutics for inflammatory diseases, tissue repair, and immune-related disorders.
Future research directions should focus on standardizing exosome isolation protocols to ensure reproducibility, engineering exosomes to enhance their targeting and immunomodulatory potency, and conducting rigorous preclinical studies to validate efficacy and safety. The ability to harness and potentially enhance the natural immunomodulatory properties of MSC exosomes represents a promising frontier in regenerative medicine and immunotherapy, offering new avenues for treating conditions characterized by excessive inflammation or impaired tissue repair.
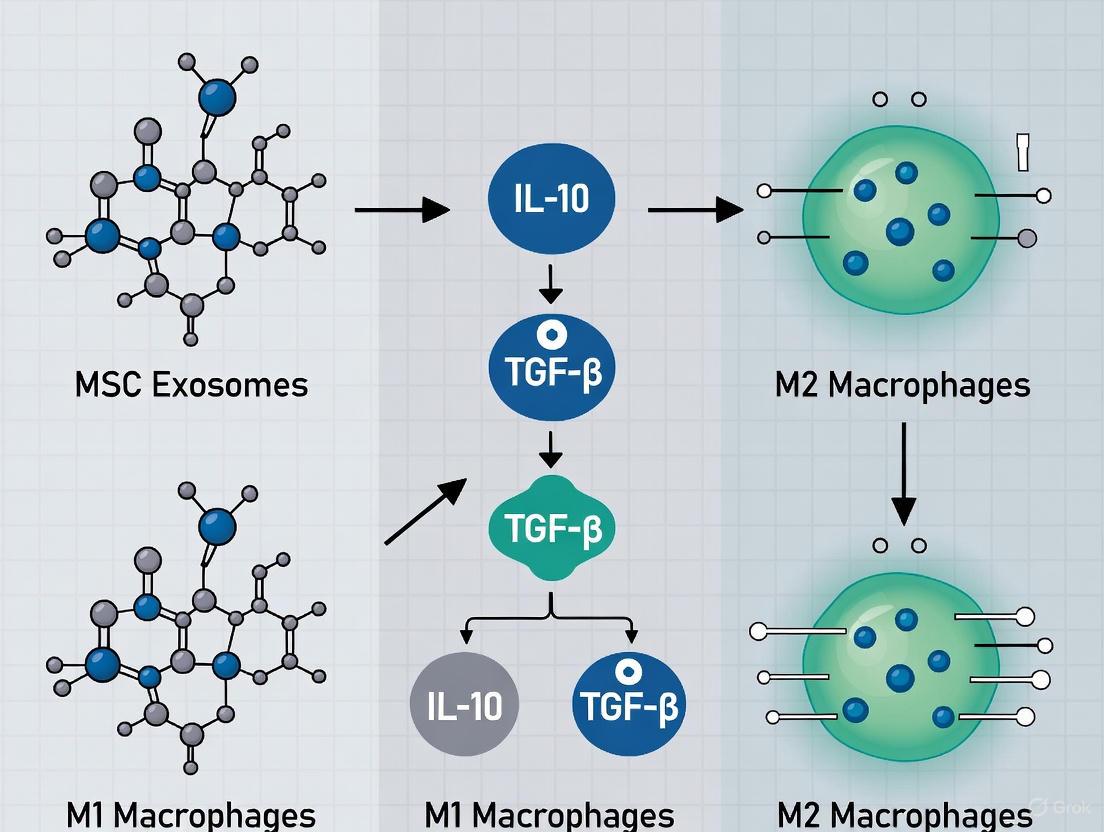
Mesenchymal stromal cell-derived exosomes (MSC-exosomes) have emerged as potent mediators of immunomodulation, with their ability to polarize macrophages toward an anti-inflammatory M2 phenotype being a key therapeutic mechanism. This whitepaper delineates the sophisticated synergistic signaling network through which MSC-exosomes coordinate this polarization, integrating the CD73/ecto-5'-nucleotidase activity with the Extra Domain A-Fibronectin (EDA-FN)/Toll-like Receptor (TLR) pathway and downstream AKT/ERK signaling cascades. We present a mechanistic model wherein exosomal CD73 catalyzes the production of adenosine, activating adenosine receptors A2A and A2B, while concurrent EDA-FN engagement of TLR4 initiates a MyD88-dependent signaling pathway. These inputs converge to activate AKT and ERK phosphorylation, driving the transcriptional reprogramming necessary for M2 macrophage polarization. This document provides a comprehensive technical guide, including summarized quantitative data, detailed experimental protocols, and pathway visualizations, to support researchers and drug development professionals in leveraging this synergistic signaling for therapeutic innovation.
Macrophages are innate immune cells with remarkable plasticity, capable of polarizing into pro-inflammatory M1 or anti-inflammatory M2 phenotypes in response to microenvironmental cues [3] [33]. The M2 phenotype, characterized by the expression of markers such as CD206 and CD163 and the secretion of anti-inflammatory mediators like IL-10 and TGF-β, plays a pivotal role in resolving inflammation, promoting tissue repair, and restoring homeostasis [3] [4]. Mesenchymal Stem/Stromal Cells (MSCs) and, more recently, their secreted exosomes, have been identified as powerful inducers of this beneficial M2 polarization [34] [4] [35].
Exosomes are nano-sized extracellular vesicles that serve as critical messengers for cell-to-cell communication, carrying a functional cargo of proteins, lipids, and nucleic acids [36]. The therapeutic efficacy of MSC-exosomes in a spectrum of preclinical models—from spinal cord injury and abdominal aortic aneurysm to hyperoxia-induced lung injury—is increasingly attributed to their capacity to modulate macrophage polarization [34] [10] [35]. This whitepaper focuses on the core molecular machinery embedded within MSC-exosomes that synergistically activates specific signaling pathways in macrophages, directing them toward the M2 phenotype. Understanding the integration of the CD73/adenosine axis with the EDA-FN/TLR and AKT/ERK pathways provides a rational foundation for predicting exosome potency and developing novel, cell-free immunomodulatory therapeutics.
Results: Uncovering the Synergistic Signaling Network
The Core Signaling Modules
Research has revealed that MSC-exosomes mediate M2-like macrophage polarization through at least two coordinated mechanisms. The first involves the exosomal surface protein Extra Domain A-Fibronectin (EDA-FN), which activates a MyD88-mediated Toll-like Receptor (TLR) signaling pathway in macrophages [34] [3] [33]. The second, more recently uncovered mechanism is driven by the ecto-5'-nucleotidase activity of exosomal CD73 [34] [3]. This enzyme catalyzes the conversion of adenosine monophosphate (AMP) to adenosine in the extracellular milieu. The generated adenosine then acts in a paracrine fashion by binding to adenosine receptors A2A and A2B on the macrophage surface [34] [3].
The critical finding is that these pathways are not independent; they converge to activate common downstream signaling nodes. Specifically, the engagement of both adenosine receptors (A2A and A2B) and TLR4 leads to the phosphorylation and activation of the kinases AKT and ERK [34] [3]. This synergistic activation is essential for the polarization process, as inhibition at any key point—CD73 activity, adenosine receptors, or AKT/ERK phosphorylation—abolishes the ability of MSC-exosomes to induce an M2-like phenotype [34]. This integrated signaling network ensures a robust and specific reprogramming of macrophage function.
Functional Outcomes of Pathway Integration
The synergistic signaling cascade culminates in a distinct shift in macrophage gene expression and function. Macrophages treated with MSC-exosomes demonstrate a marked decrease in pro-inflammatory M1 markers, such as iNOS, TNF-α, and IL-6, and a simultaneous increase in anti-inflammatory M2 markers, including CD206, ARG1, and IL-10 [3] [37] [35]. This polarization has direct functional consequences, such as enhanced tissue repair capabilities and suppression of damaging inflammation. In vivo, administration of MSC-exosomes has been shown to attenuate the development of abdominal aortic aneurysm, an effect that is dependent on their ability to promote M2 polarization within the inflammatory environment of the vessel wall [35].
Table 1: Summary of Key Quantitative Data from Mechanistic Studies
| Experimental Parameter | Measurement/Outcome | Significance/Implication |
|---|---|---|
| CD73/NT5E Activity in MSC-Exo Prep (Batch AC113) | 22.71 ± 0.54 mU/μg [3] | Serves as a critical potency attribute for immunomodulatory function. |
| MSC-Exo Particle Size | Modal diameter of 139.29 nm [3] | Confirms isolation of exosome-sized vesicles. |
| MSC-Exo Particle Concentration | 1.33 × 1011 particles/mg protein [3] | Provides a quantitative measure for dosing in experiments. |
| AAA Incidence (AngII-induced mouse model) | PBS: 70% (7/10); MSC-Exo: 50% (5/10) [35] | Demonstrates therapeutic efficacy of exosomes in vivo. |
| Maximal Abdominal Aorta Diameter | AngII+PBS: ~2.44 mm; AngII+Exo: ~1.49 mm [35] | Quantifies the structural protection conferred by exosome treatment. |
| Key Inhibitors Used | PSB12379 (CD73i) [3] | Validates the necessity of CD73 activity in the polarization mechanism. |
Experimental Protocols: Deconstructing the Key Methodologies
Preparation and Characterization of MSC Exosomes
Cell Source and Culture: The protocol often utilizes a clonal, immortalized human ESC-derived MSC line to ensure a scalable and consistent exosome supply [3] [33]. Cells are cultured in a chemically defined medium supplemented with FGF-2 and PDGF-AB for three days to condition the medium.
Exosome Isolation and Purification: The conditioned medium is processed using tangential flow filtration to remove larger particles and concentrated using a 100 kDa molecular weight cut-off membrane [3] [33]. Alternative methods for small-scale preparations include sequential ultracentrifugation, where the medium is centrifuged at lower speeds (e.g., 3,000 × g for 25 min) to remove cells and debris, followed by a high-speed centrifugation (e.g., 100,000 × g for 3 h) to pellet the exosomes [35].
Quality Control and Characterization: Rigorous characterization is essential and should include:
- Particle Analysis: Nanoparticle Tracking Analysis to determine particle size distribution and concentration [3] [35].
- Morphology: Transmission Electron Microscopy to confirm the classic cup-shaped, bilayer membrane structure [35].
- Protein Markers: Western blot analysis for positive exosomal markers (e.g., CD63, CD9, TSG101) [4] [35].
- Potency Attribute: Functional assay for CD73 activity using a phosphate detection system [3].
Macrophage Polarization and Inhibitor Assays
Macrophage Differentiation: Primary macrophages can be derived from rodent or human Peripheral Blood Mononuclear Cells. PBMCs are isolated via density gradient centrifugation and cultured for 7-9 days in medium containing Macrophage Colony-Stimulating Factor to drive differentiation into naïve macrophages [3].
Exosome Treatment and Polarization: Differentiated macrophages are treated with MSC-exosomes (e.g., at 10 μg/mL) for 24-48 hours to induce polarization [3]. A vehicle control should always be included.
Mechanistic Disruption using Inhibitors: To dissect the specific contribution of each pathway component, macrophages are co-treated with MSC-exosomes and specific inhibitors:
- CD73 Inhibition: PSB12379 (e.g., 10 nM) [3].
- Adenosine Receptor Antagonists: Selective inhibitors for A2A and A2B receptors.
- Kinase Inhibition: Inhibitors of AKT and ERK phosphorylation. Following treatment, polarization is assessed via:
- Immunofluorescence/Flow Cytometry: For M2 surface markers (CD206, CD163).
- Western Blot: For phosphorylation status of AKT and ERK, and expression of M2-related proteins.
- qPCR/ELISA: For M2-associated cytokines and gene expression profiles [3] [35].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Investigating MSC-Exosome Mediated Macrophage Polarization
| Reagent / Tool | Function / Target | Application in Research |
|---|---|---|
| PSB12379 | CD73 ecto-5'-nucleotidase inhibitor [3] | Validates the necessity of exosomal CD73 enzymatic activity in the polarization mechanism. |
| A2A & A2B Receptor Antagonists | Block adenosine signaling [3] | Determines the specific contribution of adenosine receptor subtypes downstream of CD73. |
| AKT/ERK Phosphorylation Inhibitors | Blocks downstream kinase signaling [3] | Confirms the convergence of CD73 and EDA-FN pathways on AKT/ERK activation. |
| Anti-CD206 (MMR) Antibody | Labels M2 macrophage surface receptor [37] [35] | Detects and quantifies M2 macrophage polarization via flow cytometry or immunofluorescence. |
| Anti-CD63 / CD9 / TSG101 Antibodies | Exosomal marker proteins [4] [35] | Characterizes and validates exosome preparations via Western blot. |
| Recombinant M-CSF | Macrophage Colony-Stimulating Factor | Differentiates primary monocytes into naïve macrophages for polarization assays [3]. |
Signaling Pathway and Experimental Workflow Visualization
Synergistic Signaling Pathway Diagram
Diagram Title: Synergistic Signaling for M2 Polarization
Experimental Workflow for Mechanistic Validation
Diagram Title: Experimental Workflow for Validation
The immunomodulatory power of MSC-exosomes is rooted in a sophisticated, multi-component signaling system. The integration of the CD73/adenosine axis with the EDA-FN/TLR pathway, converging on AKT/ERK activation, represents a robust synergistic mechanism for driving M2 macrophage polarization. This detailed mechanistic understanding is not merely academic; it provides a rational framework for standardizing MSC-exosome preparations based on CD73 activity and EDA-FN content as critical potency attributes [34] [3]. Furthermore, it opens new avenues for drug development, whether through engineering exosomes to enhance these specific pathways or by developing small molecule therapies that mimic this synergistic signaling to resolve inflammation and promote tissue repair across a wide spectrum of diseases.
From Bench to Bedside: Isolation Techniques and Therapeutic Applications in Disease Models
The field of extracellular vesicle (EV) research, particularly concerning exosomes derived from Mesenchymal Stem Cells (MSCs), has gained substantial momentum due to their profound therapeutic potential. These nanovesicles (30–150 nm) are pivotal mediators of intercellular communication, shuttling functional proteins, lipids, and nucleic acids between cells [38] [39]. Within the context of immunomodulation, a critical area of investigation is how MSC exosomes modulate macrophage polarization towards the anti-inflammatory M2 phenotype, a process with significant implications for regenerative medicine, cancer therapy, and the treatment of inflammatory diseases [40]. To ensure the reliability, reproducibility, and accurate interpretation of findings in this field, the implementation of standardized and rigorous methodologies for exosome isolation and characterization is paramount. This technical guide provides a detailed framework for the core techniques of ultracentrifugation, size-exclusion chromatography (SEC), Nanoparticle Tracking Analysis (NTA), and Western blotting, specifically framed within research on MSC exosome-mediated M2 macrophage polarization.
Standardized Isolation of MSC Exosomes
The isolation of high-purity exosomes is the critical first step for all downstream functional assays, including macrophage polarization studies. Co-isolation of contaminants can lead to misinterpretation of functional outcomes.
Ultracentrifugation-Based Isolation
Ultracentrifugation (UC) remains the most widely used method for exosome isolation due to its ability to handle large volumes of conditioned media [38] [39]. The following protocol is optimized for MSC-conditioned media.
Pre-processing of Cell Culture Supernatant:
- Cell Culture: Culture MSCs in media supplemented with EV-depleted fetal bovine serum (FBS) to avoid contamination with bovine exosomes. EV-depleted FBS is prepared by ultracentrifuging commercial FBS at 120,000 × g overnight (15 h) and collecting the upper 80% of the liquid [41].
- Collection: When MSCs reach 90% confluence, replace the medium with serum-free media and culture for 48 hours. Collect the conditioned supernatant [39].
- Clearing: Subject the supernatant to sequential centrifugation steps: 300–400 × g for 10 min to remove cells, and 2,000–10,000 × g for 30 min to remove cell debris and apoptotic bodies [41] [38] [39]. Filter the pre-processed supernatant through 0.45 µm and 0.22 µm polyvinylidene fluoride (PVDF) filters to remove large particles [41].
Differential Ultracentrifugation:
- Pelletting Exosomes: Ultracentrifuge the cleared supernatant at 100,000–120,000 × g for 70–90 minutes at 4°C using a swinging-bucket rotor (e.g., SW28 or SW32) [41] [38] [39].
- Washing: Carefully discard the supernatant and resuspend the often invisible exosome pellet in a large volume of sterile, particle-free phosphate-buffered saline (PBS). Repeat the ultracentrifugation step (100,000–120,000 × g, 70–90 min) to wash the exosomes [41].
- Resuspension: Finally, resuspend the purified exosome pellet in a small volume (e.g., 50–200 µL) of PBS and aliquot for storage at -80°C [41] [39].
Enhanced Protocol: Sucrose Cushion Ultracentrifugation To improve exosome yield and purity while preserving structural integrity, an improvised one-step sucrose cushion method is highly effective [39].
- After clearing the conditioned media, slowly layer it on top of a 4 mL cushion of 30% sucrose solution (in PBS) in an ultracentrifuge tube.
- Centrifuge at 100,000 × g at 4°C for 90 minutes. Exosomes will collect at the sample-sucrose interface due to their buoyant density (1.15-1.19 g/mL), while higher-density protein contaminants will pellet.
- Collect the sucrose layer, dilute it in a large volume of PBS, and ultracentrifuge again at 100,000 × g to pellet the exosomes. Resuspend the final pellet in PBS [39].
Size-Exclusion Chromatography (SEC)
SEC separates particles based on their hydrodynamic radius, allowing exosomes to elute in the void volume before soluble proteins and other small contaminants [41] [42]. It is excellent for obtaining high-purity exosomes suitable for functional studies.
Protocol:
- Column Selection: Use a commercially available SEC column, such as Superose 6, which has an optimal separation range for exosomes [41].
- Equilibration: Equilibrate the column with at least 2–3 column volumes of elution buffer (typically PBS, pH 7.4, filtered through a 0.22 µm filter) [41] [42].
- Sample Preparation and Loading: The pre-cleared conditioned media or a resuspended UC pellet can be used as input. Concentrate the sample if necessary and keep the loading volume small (e.g., 0.5–1 mL) for optimal resolution [42].
- Elution and Fraction Collection: Elute with PBS at a recommended flow rate of 0.5–1 mL/min. Collect sequential fractions (e.g., 0.5–1 mL each). Exosomes typically elute in the early fractions (fractions 5–7 for a 500 µL fraction size), while contaminating proteins elute later [41] [42].
- Concentration: Exosome fractions may be diluted and can be concentrated using centrifugal filters with a 100 kDa molecular weight cut-off [42].
Table 1: Comparison of Exosome Isolation Methods
| Parameter | Ultracentrifugation (UC) | Sucrose Cushion UC (SUC) | Size-Exclusion Chromatography (SEC) |
|---|---|---|---|
| Principle | Sequential centrifugal forces based on size/density | Buoyant density in a sucrose gradient | Size-based separation through a porous matrix |
| Relative Purity | Moderate, can co-pellet protein aggregates | High, reduces protein contamination | Very high, effectively removes soluble contaminants |
| Relative Yield | Moderate | Higher than UC [39] | Variable; high recovery but can be diluted |
| Time Consumption | High (often >4 hours) | High (similar to UC) | Moderate to Fast (~15 minutes post-setup) [42] |
| Equipment Cost | High (ultracentrifuge) | High (ultracentrifuge) | Moderate (columns) |
| Main Advantage | Handles large volumes; gold standard | Improved yield and integrity | High purity; maintains bioactivity |
| Main Disadvantage | Potential for particle deformation/aggregation | Additional sucrose removal step required | Sample dilution; limited sample loading volume |
Comprehensive Characterization of MSC Exosomes
A complete characterization strategy is mandatory to confirm the identity, purity, and integrity of isolated MSC exosomes before functional assays. The Minimal Experimental Requirements for EV studies (MISEV guidelines) recommend analyzing at least three different components.
Nanoparticle Tracking Analysis (NTA)
NTA measures the size distribution and concentration of particles in a liquid suspension based on their Brownian motion and light-scattering properties [43] [44].
Experimental Protocol:
- Sample Preparation: Dilute the exosome sample in sterile, particle-free PBS to achieve an ideal concentration of 20–100 particles per frame for accurate counting. Optimal measuring concentration is 1×10⁷–1×10⁹ particles/mL [43] [44].
- Instrument Calibration: Calibrate the NTA instrument (e.g., NanoSight) using latex beads of known size (e.g., 100 nm) [43].
- Data Acquisition: Inject the sample into the chamber. Record multiple videos (e.g., 3–5 videos of 30–60 seconds each) under consistent temperature, camera level, and detection threshold settings [43] [44].
- Data Analysis: The software tracks individual particles and uses the Stokes-Einstein equation to calculate the hydrodynamic diameter. Report the mode and mean size, concentration (particles/mL), and a size distribution profile [43] [44].
Fluorescence NTA (fl-NTA) for Specificity: For confirming the presence of exosomes and their cellular origin, fl-NTA is powerful. Exosomes can be labeled with antibodies against classic tetraspanin markers (CD63, CD81, CD9) conjugated to a fluorophore (e.g., Alexa Fluor 488). The fluorescent mode then allows for counting and sizing only the marker-positive vesicles, providing specificity beyond light-scattering alone [43] [44].
Table 2: Key Technical Specifications for NTA of MSC Exosomes [43] [44]
| Parameter | Specification | Application Note |
|---|---|---|
| Size Detection Range | ~30 nm to 1000 nm | MSC exosomes typically peak between 50-150 nm. |
| Concentration Range | 1×10⁷ to 1×10⁹ particles/mL | Requires precise dilution for accurate results. |
| Laser Wavelengths | 405, 488, 532, 640 nm | 488 nm is common for green fluorescent tags (e.g., CD63-Alexa488). |
| Sample Volume | ≥ 500 µL | Small volume requirement is advantageous. |
| Readouts | Size distribution (mode, D10, D50, D90), concentration (particles/mL), fluorescence-positive subpopulations. | Essential for dosing consistency in polarization experiments. |
Western Blotting
Western blotting is used to confirm the presence of exosome-enriched marker proteins and the absence of contaminants, validating the purity of the preparation.
Experimental Protocol:
- Protein Extraction: Lyse exosomes in RIPA buffer supplemented with protease inhibitors. Determine protein concentration using a BCA or other compatible assay [39].
- Gel Electrophoresis: Load equal amounts of protein (e.g., 10-20 µg) onto a 10-12.5% SDS-PAGE gel under reducing conditions for most proteins. For some markers like CD63, non-reducing conditions can be used to observe multimeric complexes [39].
- Membrane Transfer and Blocking: Transfer proteins to a PVDF membrane. Block the membrane with 5% non-fat milk or BSA in TBST for 1 hour.
- Antibody Incubation:
- Positive Markers: Probe for exosome-enriched tetraspanins (CD63, CD81, CD9) and proteins associated with the endosomal sorting complex required for transport (ESCRT) machinery, such as Alix and TSG101 [39].
- Negative Markers: Probe for contaminants like calnexin (endoplasmic reticulum marker) or GM130 (Golgi apparatus marker), which should be absent in pure exosome preparations.
- Detection: Incubate with appropriate HRP-conjugated secondary antibodies and develop using an enhanced chemiluminescence (ECL) substrate. Imaging should show strong signals for positive markers and no signal for negative markers.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for MSC Exosome Isolation and Characterization
| Item | Function / Application | Example / Note |
|---|---|---|
| EV-Depleted FBS | Cell culture supplement | Prevents contamination of sample with bovine EVs. Prepared by ultracentrifugation of standard FBS [41]. |
| Sucrose (for cushion) | Density gradient medium | Used in the SUC method to improve exosome purity and yield [39]. |
| Size-Exclusion Columns | High-purity exosome isolation | Columns like Superose 6 (GE Healthcare) effectively separate EVs from contaminants [41] [42]. |
| Anti-Tetraspanin Antibodies | Exosome characterization (WB, fl-NTA) | Antibodies against CD63, CD81, CD9 are standard for confirming exosome identity [43] [44] [39]. |
| NTA Instrument | Size and concentration analysis | Instruments like NanoSight (Malvern Panalytical) provide particle-by-particle data [43] [44]. |
| Fluorescent Conjugates | Specific subpopulation analysis (fl-NTA) | Antibody-fluorophore conjugates (e.g., CD63-Alexa488) enable detection of specific EV subpopulations [43] [44]. |
Application: Investigating MSC Exosome-Mediated M2 Macrophage Polarization
The methodologies described above are foundational for researching the role of MSC exosomes in promoting M2 macrophage polarization, a key anti-inflammatory and pro-regenerative mechanism.
Experimental Workflow
The following diagram illustrates the integrated workflow from exosome isolation to functional validation in macrophage polarization.
Key Signaling Pathways in M2 Polarization
MSC exosomes can induce M2 polarization through the transfer of various bioactive molecules (proteins, miRNAs) that modulate key signaling pathways in macrophages. Two documented pathways are shown below.
Pathway 1: SHP2-STAT3 Activation: MSC exosomes engineered to carry soluble fibrinogen-like protein 2 (sFgl2) can bind to the CD32b receptor on macrophages. This binding activates the tyrosine phosphatase SHP2, which in turn promotes the phosphorylation of the transcription factor STAT3. Phospho-STAT3 drives the expression of M2-associated genes (e.g., IL-10, Arg1), leading to the establishment of an M2 anti-inflammatory phenotype that can alleviate conditions like acute heart transplant rejection [45].
Pathway 2: FAK/ERK Suppression: In the context of prostate cancer, exosomes carrying a specific protein, PSM-E, have been shown to interact with the intracellular scaffold protein RACK1 in macrophages. This interaction recruits RACK1 and subsequently suppresses the phosphorylation and activity of FAK (Focal Adhesion Kinase) and ERK (Extracellular Signal-Regulated Kinase) signaling pathways. This suppression inhibits M2 polarization, which in this context leads to reduced tumor invasion and metastasis [46]. This highlights the context-dependent nature of exosome-mediated effects.
The rigorous and standardized application of ultracentrifugation (potentially enhanced with a sucrose cushion), size-exclusion chromatography, nanoparticle tracking analysis, and Western blotting is non-negotiable for producing high-quality, well-characterized MSC exosome preparations. By adhering to these detailed protocols, researchers can ensure the reliability and reproducibility of their data. This foundational work is critical for elucidating the precise mechanisms—such as the delivery of sFgl2 or PSM-E—by which MSC exosomes orchestrate macrophage polarization, thereby accelerating the translation of these promising nanovesicles into targeted therapeutic applications for a range of inflammatory and degenerative diseases.
The polarization of macrophages towards the M2 phenotype is a critical process in immune regulation, tissue repair, and resolution of inflammation. Within the broader thesis investigating how mesenchymal stem cell (MSC)-derived exosomes modulate macrophage polarization to the M2 phenotype, reliable and standardized in vitro polarization assays are fundamental research tools. These assays allow researchers to accurately identify and characterize M2 macrophages through specific surface markers, gene expression profiles, and functional cytokine secretion patterns. This technical guide provides a comprehensive framework for establishing robust in vitro polarization assays, with a specific focus on evaluating the canonical M2 markers CD206 and Arginase-1 (Arg-1), while detailing the cytokine profiles that define the M2 functional state. The protocols and analytical methods described herein serve as essential methodologies for elucidating the mechanisms by which MSC exosomes and other immunomodulatory agents promote M2 polarization, enabling drug development professionals to quantify therapeutic effects with precision and reproducibility.
Macrophage Polarization: Fundamental Concepts
Macrophages exhibit remarkable plasticity, allowing them to differentiate into distinct functional phenotypes in response to specific microenvironmental signals. The classical activation of naïve macrophages (M0) with interferon-gamma (IFN-γ) and lipopolysaccharide (LPS) drives polarization towards the pro-inflammatory M1 phenotype [47] [48]. In contrast, alternative activation with interleukin-4 (IL-4) and interleukin-13 (IL-13) promotes polarization towards the anti-inflammatory M2 phenotype, which plays key roles in immune regulation, tissue repair, and resolution of inflammation [47] [48] [4]. The M2 phenotype is not monolithic but encompasses several subtypes; however, this guide focuses on the IL-4/IL-13-induced M2a subtype, which is highly relevant for therapeutic applications. The core principle of in vitro polarization assays is to subject primary monocyte-derived macrophages or macrophage cell lines to a controlled cytokine milieu, then comprehensively assess the resulting phenotypic changes using multiple validation methods. Understanding these fundamental polarization pathways provides the context for evaluating how MSC-derived exosomes and other therapeutic agents can manipulate this process.
Experimental Protocols for M2 Polarization
Generation of Human Monocyte-Derived Macrophages (hMDMs)
The isolation and differentiation of human monocytes from peripheral blood represents a gold standard for generating physiologically relevant macrophages. The following protocol, adapted from current methodologies, ensures high purity and viability [47] [48]:
- Peripheral Blood Mononuclear Cell (PBMC) Isolation: Collect human venous blood in EDTA anticoagulant tubes. Isolate PBMCs using density gradient centrifugation with Ficoll-Paque Plus. Centrifuge at 400-500 × g for 30-40 minutes at room temperature without brake. Carefully collect the buffy coat layer containing PBMCs and wash cells 2-3 times with DPBS [47] [48].
- Monocyte Isolation: Resuspend PBMCs in pre-warmed monocyte attachment medium and transfer to T75 cell-culture flasks. Incubate at 37°C with 5% CO₂ for 90 minutes to allow monocyte adhesion. Discard supernatant and wash gently with pre-warmed complete medium to remove non-adherent cells (lymphocytes) [48]. For higher purity, CD14+ monocytes can be isolated using magnetic bead negative selection kits, achieving ~90% purity as verified by flow cytometry [47].
- Differentiation to M0 Macrophages: Culture adherent monocytes in RPMI-1640 or DMEM medium supplemented with 10% FBS, 1% penicillin/streptomycin, and 40-50 ng/mL Macrophage Colony-Stimulating Factor (M-CSF). Replace medium every 2-3 days. Mature, naïve (M0) macrophages are typically obtained after 6-7 days of differentiation [47] [48].
Table 1: Key Reagents for Monocyte-Derived Macrophage Generation
| Reagent | Function | Concentration/Duration |
|---|---|---|
| Ficoll-Paque Plus | Density gradient medium for PBMC isolation | - |
| M-CSF (Macrophage Colony-Stimulating Factor) | Drives monocyte-to-macrophage differentiation | 40-50 ng/mL for 6-7 days |
| RPMI-1640/DMEM Medium | Base culture medium | - |
| Fetal Bovine Serum (FBS) | Serum supplement for cell growth | 10% (v/v) |
| Penicillin/Streptomycin | Antibiotic to prevent bacterial contamination | 100 U/mL and 100 μg/mL |
Polarization to M2 Phenotype
Once M0 macrophages are obtained, polarization to the M2 phenotype is achieved through cytokine stimulation:
- Stimulus Preparation: Prepare complete medium containing 20 ng/mL of both IL-4 and IL-13. These cytokines are the primary drivers of M2a polarization [48] [4].
- Polarization Process: Remove the differentiation medium from M0 macrophages and add the polarization medium containing IL-4 and IL-13. Incubate cells for 24-48 hours at 37°C with 5% CO₂ [47] [48].
- Control Groups: Include appropriate controls in every experiment:
The following workflow diagram summarizes the complete process from monocyte isolation to polarized macrophage characterization:
Characterization of M2 Macrophages
Surface Marker Analysis by Flow Cytometry
CD206 (mannose receptor) is a highly specific surface marker for M2 macrophages and serves as a primary identifier for successfully polarized cells [49] [4]. The following protocol details its detection:
- Cell Harvest: After polarization, detach cells using non-enzymatic cell dissociation solution or gentle scraping. Wash cells twice with FACS buffer (PBS with 1% BSA).
- Antibody Staining: Resuspend cell pellets (approximately 1×10⁶ cells per sample) in FACS buffer. Add fluorochrome-conjugated anti-human CD206 antibody or isotype control. Follow manufacturer's recommended antibody dilution (typically 1:100-1:200). Incubate for 30 minutes at 4°C in the dark.
- Analysis: Wash cells twice with FACS buffer to remove unbound antibody. Resuspend in FACS buffer and analyze immediately using flow cytometry. Analyze a minimum of 10,000 events per sample. M2-polarized macrophages typically show significant increase in CD206 positivity compared to M0 and M1 controls [5].
Gene Expression Analysis by RT-qPCR
Gene expression analysis provides quantitative validation of polarization success. Key M2-associated genes include MRC1 (encodes CD206) and ARG1 (encodes Arginase-1) [47] [50].
- RNA Extraction: Lyse polarized macrophages directly in culture plates using TRIzol reagent. Follow standard RNA extraction protocols, including DNase treatment to remove genomic DNA contamination. Quantify RNA concentration using a spectrophotometer [49].
- cDNA Synthesis: Reverse transcribe 0.5-1 µg of total RNA to cDNA using a commercial reverse transcription kit with oligo(dT) and/or random primers.
- Quantitative PCR: Prepare reactions using SYBR Green or TaqMan Master Mix. Use the following cycling conditions: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Include melt curve analysis for SYBR Green assays. Normalize target gene expression to the housekeeping gene β-actin using the 2^–ΔΔCt method [49] [47].
Table 2: Key M2 Markers for Phenotypic Characterization
| Marker Type | Target | Detection Method | Significance in M2 Polarization |
|---|---|---|---|
| Surface Receptor | CD206 | Flow Cytometry, Immunofluorescence | Highly specific surface marker for M2 macrophages [49] [4] |
| Metabolic Enzyme | Arginase-1 (Arg-1) | RT-qPCR, Western Blot, ELISA | Competes with iNOS for arginine metabolism, promotes tissue repair [47] [50] |
| Transcription Factor | PPAR-γ | RT-qPCR, Western Blot | Regulates genes involved in alternative activation [50] |
Cytokine Secretion Profile Analysis
The functional output of M2 macrophages is characterized by their distinct cytokine secretion profile, which can be quantified using enzyme-linked immunosorbent assay (ELISA) or multiplex bead-based arrays:
- Sample Collection: Collect cell culture supernatant 24-48 hours after polarization. Centrifuge at 300 × g for 5 minutes to remove cellular debris. Aliquot and store supernatants at –80°C until analysis [48].
- ELISA Procedure: Following kit manufacturer's instructions, add standards and samples to pre-coated wells. Incubate with detection antibodies and enzyme conjugate. Develop with substrate solution and measure absorbance using a microplate reader. Calculate cytokine concentrations from the standard curve [48].
- Key Cytokines: M2 macrophages typically secrete elevated levels of IL-10, TGF-β, CCL17, and CCL22, while showing reduced secretion of pro-inflammatory cytokines like TNF-α and IL-12 [49] [4]. The table below summarizes the characteristic secretion profile of M2 macrophages.
Table 3: Characteristic Cytokine Secretion Profiles of M1 vs. M2 Macrophages
| Cytokine/Chemokine | M1 Macrophages | M2 Macrophages | Function |
|---|---|---|---|
| TNF-α | Significantly Elevated | Low | Pro-inflammatory mediator [47] [48] |
| IL-12 | Significantly Elevated | Low | Promotes Th1 responses [47] |
| IL-10 | Low | Significantly Elevated | Potent anti-inflammatory cytokine [49] [4] |
| TGF-β | Low | Elevated | Immunosuppression and tissue repair [4] |
| CCL17 | Low | Elevated | Recruitment of Th2 cells [49] |
| CCL22 | Low | Elevated | Recruitment of regulatory T cells [49] |
| CCL13 | Low | Elevated | Marker for alternative activation [48] |
MSC Exosomes in M2 Polarization: Mechanisms and Assessment
Mechanistic Insights
MSC-derived exosomes promote M2 macrophage polarization through several documented mechanisms, which can be investigated using the aforementioned polarization assays:
- CD73/Adenosine Pathway: MSC exosomes express CD73 (ecto-5'-nucleotidase), which catalyzes the conversion of AMP to adenosine. Adenosine then binds to A2A and A2B receptors on macrophages, activating AKT/ERK-dependent signaling pathways that drive M2 polarization [3]. This pathway can be inhibited using PSB12379 (CD73 inhibitor), which attenuates the polarizing effect.
- miRNA Transfer: MSC exosomes contain microRNAs that modulate macrophage polarization. For instance, miR-182 delivered by MSC exosomes targets TLR4, shifting macrophages toward the M2 phenotype [51]. Similarly, exosomal miR-let7 mediates M2 polarization in atherosclerosis models [4].
- Protein-Mediated Signaling: MSC exosomes carry immunomodulatory proteins like TGF-β, IL-10, and extra domain A-fibronectin (EDA-FN). EDA-FN activates the MyD88-dependent TLR signaling pathway in macrophages, promoting M2 polarization [3] [52].
The following diagram illustrates these primary mechanisms through which MSC exosomes modulate macrophage polarization:
Assessing Exosome-Mediated Polarization
To evaluate the effect of MSC exosomes on macrophage polarization:
- Exosome Treatment: Isolate exosomes from MSC culture supernatants using sequential ultracentrifugation or tangential flow filtration. Characterize exosomes by nanoparticle tracking analysis (size ~50-150 nm), transmission electron microscopy (cup-shaped morphology), and western blotting for markers (CD9, CD63, CD81, TSG101) [3] [51]. Treat M0 macrophages with 10-50 μg/mL MSC exosomes simultaneously with or without polarizing cytokines for 24-48 hours [3] [5].
- Functional Validation: Assess the functional consequences of exosome-induced M2 polarization using phagocytosis assays, metabolic profiling, and gene expression analysis. M2 macrophages typically demonstrate increased oxidative phosphorylation compared to glycolysis-dependent M1 macrophages [47]. Phagocytic capacity can be measured using pHrodo-labeled E. coli BioParticles or zymosan [47].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagent Solutions for M2 Polarization Assays
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Polarization Cytokines | Recombinant Human IL-4, IL-13 | Induce M2a macrophage polarization [48] |
| M1 Polarization Cytokines | Recombinant Human IFN-γ, LPS (E. coli) | Generate M1 controls for assay validation [48] |
| Differentiation Factor | Recombinant Human M-CSF | Drives monocyte to macrophage differentiation [48] |
| Surface Marker Antibodies | Anti-human CD206 (for flow/IF), Anti-human CD86 (M1 control) | Identification of M2 macrophages via surface staining [48] |
| ELISA Kits | IL-10, TGF-β, TNF-α, CCL13 | Quantify cytokine secretion profiles [48] |
| qPCR Reagents | Primers for MRC1, ARG1, ACTB (control) | Validate polarization at transcriptional level [49] [47] |
| MSC Exosome Isolation Kits | Total Exosome Isolation Kit, TEI Reagent | Isolate exosomes from MSC conditioned media [3] [5] |
Troubleshooting and Technical Considerations
- Low Polarization Efficiency: If CD206 and Arg-1 expression remains low after stimulation, verify cytokine activity and concentration using dose-response experiments. Ensure FBS batches are not inhibitory; consider using exosome-depleted FBS when studying exosome effects [3].
- High M1 Marker Expression in M2 Cultures: This indicates potential contamination with LPS or IFN-γ. Use low-endotoxin reagents and dedicated media for M2 polarization. Include M1 controls to confirm specific polarization.
- Exosome Variability: Different MSC sources (bone marrow, dental pulp, adipose) produce exosomes with varying potencies. Characterize exosome protein content and CD73 activity to standardize preparations [3] [5]. For MSC exosome studies, include controls with exosomes alone (without IL-4/IL-13) to determine their intrinsic polarizing capacity [3] [4].
- Metabolic Considerations: For comprehensive phenotyping, incorporate metabolic assessments. M2 macrophages preferentially use oxidative phosphorylation for ATP generation, while M1 macrophages rely on glycolysis [47].
The pursuit of novel therapeutic strategies for autoimmune diseases has led to significant interest in the immunomodulatory capabilities of mesenchymal stem cell-derived exosomes (MSC-Exos). These nano-sized extracellular vesicles, typically 30-150 nanometers in diameter, function as key mediators of intercellular communication by transferring bioactive molecules—including proteins, lipids, and genetic material—from parent MSCs to recipient immune cells [53] [54]. Within the context of autoimmune pathogenesis, MSC-Exos have demonstrated a remarkable ability to influence macrophage polarization, particularly toward the anti-inflammatory M2 phenotype, thereby shifting the immune environment from pro-inflammatory to regulatory and reparative [53] [45]. This whitepaper comprehensively reviews the preclinical evidence validating the efficacy of MSC-Exos in models of Systemic Lupus Erythematosus (SLE) and Rheumatoid Arthritis (RA), detailing the underlying molecular mechanisms, summarizing quantitative outcomes, and providing standardized experimental protocols for research and development professionals.
Molecular Mechanisms: How MSC Exosomes Drive M2 Polarization
Key Signaling Pathways
MSC-Exos promote M2 macrophage polarization through the delivery of a diverse cargo that modulates several critical signaling pathways. The table below summarizes the primary mechanisms identified in preclinical studies.
Table 1: Key Signaling Pathways in MSC-Exo-Mediated M2 Macrophage Polarization
| Pathway/Component | Mechanism of Action | Biological Effect | Experimental Model |
|---|---|---|---|
| SHP2-STAT3 Pathway | sFgl2 in engineered exosomes binds CD32b receptor, activating SHP2 and subsequent STAT3 phosphorylation [45]. | Promotes polarization to M2 phenotype; reduces inflammatory cell infiltration. | Mouse heart transplantation (Acute rejection model) [45]. |
| miRNA Cargo (e.g., miR-223, miR-146a) | Exosomes from M2 macrophages are enriched with these miRNAs, which dampen inflammatory responses and facilitate macrophage reprogramming [53]. | Immunomodulation; transition to anti-inflammatory state. | In vitro macrophage culture studies [53]. |
| Cytokine Delivery (e.g., IL-10, TGF-β) | Anti-inflammatory cytokines are shuttled via exosomes to recipient immune cells [53]. | Supports tissue repair and resolution of inflammation. | General autoimmune disease models [53]. |
| TGF-β/Smad Pathway | MSC-Exos can act as negative regulators, inducing PTEN expression or directly downregulating Thbs2 to inhibit TGF-β signaling [54]. | Inhibits fibroblast-to-myofibroblast differentiation and collagen synthesis. | Preclinical models of pulmonary fibrosis [54]. |
The following diagram illustrates the central SHP2-STAT3 pathway, identified as a crucial mechanism for engineered MSC exosomes.
The Functional Spectrum of Macrophage Polarization
The M1/M2 paradigm provides a foundational model for understanding macrophage plasticity. The polarization state of the parent macrophage is faithfully reflected in, and amplified by, the exosomes it releases [53].
Preclinical Efficacy in Autoimmune Disease Models
Systemic Lupus Erythematosus (SLE) Models
SLE is a heterogeneous systemic autoimmune disease characterized by loss of tolerance to nuclear antigens, production of autoantibodies, and immune complex deposition leading to organ damage [55] [56]. The therapeutic goal has shifted towards achieving sustained remission, as defined by frameworks like the Definition of Remission in SLE (DORIS) and the Lupus Low Disease Activity State (LLDAS) [55] [57]. While recent B-cell-targeted biologics and CAR-T therapies have shown promise, MSC exosomes offer a cell-free, potentially safer alternative with direct effects on innate immune players like macrophages [57] [58].
Table 2: Preclinical Efficacy of MSC Exosomes in SLE and Related Inflammatory Models
| Therapeutic Agent | Disease Model | Key Efficacy Findings | Proposed Mechanism Related to M2 |
|---|---|---|---|
| sFgl2-MSC-Exos [45] | Mouse heart transplantation (acute rejection) | - Prolonged graft survival- Reduced myocardial necrosis & inflammatory infiltration- ↑ M2 macrophages in grafts- ↑ Tregs, ↓ CD4+ T cells | Engineered exosomes promote M2 polarization via SHP2-STAT3 pathway, creating an anti-inflammatory milieu. |
| M2 Macrophage-Derived Exosomes [53] | General Autoimmunity (Lupus context) | - Dampened inflammatory signaling- Support for tissue repair and immune regulation | Innately carry anti-inflammatory miRNAs (miR-223, miR-146a) and cytokines (IL-10, TGF-β). |
| Allogeneic CD19 CAR NK-cell therapy [57] | Human SLE (Refractory) | - Durable drug-free remission- Deep B-cell depletion | Highlights the therapeutic success of immune cell reprogramming, a key goal of MSC-Exo therapy. |
Rheumatoid Arthritis (RA) and Related Arthritic Models
RA is a chronic inflammatory autoimmune disorder primarily affecting the joints, with a well-defined pre-RA phase characterized by autoantibodies like ACPA and RF, and subclinical inflammation on imaging [59]. Although direct studies on MSC exosomes in RA models in the provided search results are limited, the established role of macrophage polarization in RA pathogenesis and the effects of exosomes in other inflammatory models provide a strong rationale for their efficacy.
Evidence from related research underscores the importance of macrophage polarization. Exosomes from M1 macrophages have been shown to amplify inflammation in conditions like RA by enhancing dendritic cell maturation and recruiting other inflammatory cells [53]. Conversely, strategies that promote M2 polarization are considered a promising therapeutic avenue. Furthermore, recent clinical trials presented at EULAR 2025 for novel RA therapeutics like rosnilimab (a T-cell depleter) and CPL'116 (a dual JAK/ROCK inhibitor) highlight the clinical momentum toward targeted immunomodulation, a space where MSC exosomes could play a future role [58].
Detailed Experimental Protocols for Preclinical Validation
Protocol: Validating Efficacy in a Mouse Acute Rejection Model
This protocol is adapted from a study investigating sFgl2-MSC-Exos in mouse heart transplantation [45].
1. Exosome Preparation and Characterization:
- Source: Isolate MSCs from human umbilical cord (UC-MSCs) or bone marrow (BM-MSCs) and culture under standard conditions [60].
- Engineering: Transduce MSCs with lentivirus encoding sFgl2 to generate sFgl2-overexpressing MSCs.
- Isolation: Harvest cell culture supernatant. Isolate exosomes via ultracentrifugation (100,000 × g for 70 min) or commercial exosome isolation kits.
- Characterization:
- NTA (Nanoparticle Tracking Analysis): Determine particle size distribution and concentration.
- TEM (Transmission Electron Microscopy): Confirm cup-shaped morphology.
- Western Blot: Verify presence of exosomal markers (CD63, CD81, TSG101) and absence of negative markers (Calnexin).
2. In Vivo Model and Treatment:
- Model: Establish a mouse heterotopic heart transplantation model using microsurgical techniques.
- Randomization: Divide recipients into: (i) Untreated group, (ii) MSC-Exo treatment group, (iii) sFgl2-MSC-Exo treatment group.
- Dosing: Administer exosomes (e.g., 100 µg in 100 µL PBS) via intravenous injection post-transplantation.
3. Outcome Assessment:
- Graft Survival: Monitor daily for cessation of heartbeat.
- Histopathology: At day 7 post-op, harvest grafts for H&E staining to evaluate rejection scores and immunohistochemistry (IHC) for macrophage (e.g., F4/80, iNOS for M1, CD206 for M2) and T cell (CD4, Foxp3 for Tregs) infiltration.
- Flow Cytometry: Analyze macrophage subsets (M1: CD86+, M2: CD206+) and Tregs in peripheral blood and splenocytes.
- Mechanistic Profiling: Perform Western blot on graft tissue to assess phosphorylation levels of key proteins in the SHP2-STAT3 pathway.
Protocol: In Vitro Macrophage Polarization Assay
1. Macrophage Generation and Co-culture:
- BMDM Isolation: Isolate bone marrow-derived macrophages (BMDMs) from mouse femurs and tibias. Differentiate them into M0 macrophages using M-CSF (e.g., 20 ng/mL) for 7 days.
- Polarization and Treatment:
- Polarize M0 macrophages to M1 using LPS (100 ng/mL) and IFN-γ (20 ng/mL).
- Treat M1 macrophages with MSC-Exos or sFgl2-MSC-Exos (e.g., 50 µg/mL) for 24-48 hours.
- Include a control group with a SHP2 inhibitor to confirm pathway specificity.
2. Analysis of Polarization:
- Flow Cytometry: Quantify M1 (iNOS+, CD86+) and M2 (Arg1+, CD206+) surface/intracellular markers.
- ELISA: Measure cytokine levels in supernatant (TNF-α, IL-6 for M1; IL-10, TGF-β for M2).
- qPCR: Analyze expression of M1/M2-associated genes.
The following diagram outlines this core in vitro workflow.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating MSC Exosomes and Macrophage Polarization
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| MSC Sources | Human Umbilical Cord (UC-MSCs), Bone Marrow (BM-MSCs), Adipose Tissue (AD-MSCs) [60]. | Source of exosomes; different sources may have varying immunomodulatory potencies. |
| Macrophage Markers (Flow/IHC) | M1: iNOS, CD86, HLA-DR; M2: CD206, Arg1, CD163 [53] [45]. | Identification and quantification of macrophage polarization states. |
| Cytokine Kits | ELISA Kits for TNF-α, IL-6, IL-1β (M1); IL-10, TGF-β (M2) [45]. | Quantifying secretory profiles to confirm functional polarization. |
| Exosome Isolation Kits | Ultracentrifugation-based methods; Commercial kits (e.g., Total Exosome Isolation kit) [45] [54]. | Purification of exosomes from cell culture supernatant. |
| Exosome Characterization | Nanoparticle Tracking Analyzer (NTA); Transmission Electron Microscope (TEM); Antibodies for CD63, CD81, TSG101 [45] [54]. | Validating the size, morphology, and biomarker presence of isolated exosomes. |
| Signaling Pathway Reagents | Antibodies for p-STAT3, STAT3, p-SHP2, SHP2; SHP2 inhibitor (e.g, NSC-87877) [45]. | Elucidating mechanistic pathways involved in M2 polarization. |
Preclinical evidence robustly supports the efficacy of MSC-derived exosomes in mitigating pathology in autoimmune disease models, primarily through the reprogramming of macrophages from a pro-inflammatory M1 to an anti-inflammatory M2 phenotype. Key mechanisms involve the delivery of specific miRNAs, anti-inflammatory cytokines, and the activation of intracellular pathways such as SHP2-STAT3 [53] [45]. The successful application of engineered exosomes, like sFgl2-MSC-Exos, points toward a future of precision bioengineering where exosome cargo can be tailored for enhanced potency and target specificity [45] [54].
Future work should focus on standardizing isolation protocols for clinical-grade exosomes, conducting large-scale animal studies in canonical SLE and RA models to directly quantify efficacy and durability, and exploring combination therapies with existing biologic agents. As the field advances, MSC exosomes represent a promising cell-free therapeutic paradigm, potentially offering the immunomodulatory benefits of stem cell therapy while circumventing the risks and challenges associated with the administration of whole cells.
Mesenchymal stem cell-derived exosomes (MSC-Exos) have emerged as pivotal mediators of tissue repair, orchestrating complex processes in both hindlimb ischemia and bone regeneration through sophisticated immunomodulatory mechanisms. Central to their therapeutic effect is the capacity to modulate macrophage polarization toward the anti-inflammatory M2 phenotype, which creates a conducive microenvironment for angiogenesis and osteogenesis. This technical review synthesizes current mechanistic insights, experimental data, and methodological protocols, highlighting how MSC-Exos mediate crosstalk between immune and tissue-forming cells. By integrating quantitative findings from preclinical studies and detailing standardized experimental workflows, this guide provides a foundation for advancing exosome-based therapeutic development for ischemic and regenerative applications.
Exosomes are nanoscale extracellular vesicles (30-150 nm) that function as critical intermediaries of intercellular communication by transferring bioactive molecules—including proteins, lipids, and nucleic acids—to recipient cells [61]. In regenerative medicine, MSC-Exos have garnered significant attention as a cell-free therapeutic alternative, mitigating risks associated with whole-cell transplantation such as immunogenicity, tumor formation, and vascular occlusion [61] [60]. Their therapeutic profile is characterized by three principal mechanisms: promotion of angiogenesis, stimulation of tissue-specific regeneration (osteogenesis in bone), and immunomodulation via macrophage polarization [61] [45] [40].
The overarching premise of this review is that the modulation of macrophage polarization to the M2 phenotype is a fundamental mechanism through which MSC-Exos coordinate tissue repair in both hindlimb ischemia and bone regeneration. The M2 macrophage phenotype, induced by exosomal cues, secretes anti-inflammatory cytokines (e.g., IL-10), promotes vascularization, and supports a regenerative microenvironment, thereby bridging innate immune responses with structured tissue healing [45] [40].
Molecular Mechanisms of MSC Exosomes in Macrophage Polarization
Key Signaling Pathways in M2 Polarization
MSC-Exos promote M2 macrophage polarization through the delivery of specific molecular cargo, initiating defined signaling cascades that reprogram macrophage function.
- sFgl2/CD32b/SHP2/STAT3 Pathway: A primary mechanism involves the exosomal delivery of soluble fibrinogen-like protein 2 (sFgl2). This protein binds to the CD32b receptor on macrophages, triggering a phosphorylation cascade that activates SHP2 and subsequently STAT3. This signaling axis promotes the transcription of M2-associated genes while suppressing pro-inflammatory M1 markers [45]. Blocking the CD32b receptor or inhibiting SHP2 activation abolishes this polarizing effect, confirming the specificity of this pathway [45].
- Exosomal miRNA Cargo: MSC-Exos are enriched with microRNAs (miRNAs) that target transcripts sustaining the M1 phenotype. For instance, miR-21-5p, miR-146a, and let-7b are known to downregulate Toll-like receptor signaling and NF-κB pathway components, thereby reducing the production of pro-inflammatory cytokines like TNF-α and IL-6 and creating a permissive environment for M2 polarization [61] [62]. This post-transcriptional regulation fine-tunes the macrophage response to injury.
- Metabolic Reprogramming: Emerging evidence suggests that MSC-Exos can alter intracellular metabolism in macrophages, shifting them toward an oxidative phosphorylation state that is characteristic of the M2 phenotype. This metabolic shift is a prerequisite for the sustained anti-inflammatory and pro-reparative functions of M2 macrophages in tissue repair [40].
The following diagram illustrates the primary signaling pathway through which MSC-Exos drive M2 macrophage polarization:
Functional Outcomes of M2 Polarization in Tissue Repair
The shift in macrophage populations from a pro-inflammatory M1 to an anti-inflammatory M2 phenotype has direct and consequential effects on the tissue microenvironment:
- In Hindlimb Ischemia: M2 macrophages secrete factors like Vascular Endothelial Growth Factor (VEGF) and IL-10, which directly promote the sprouting and stabilization of new blood vessels, a process critical for restoring blood flow to ischemic tissues [45] [63]. This angiogenic response is further supported by the dampening of destructive inflammation, which can exacerbate tissue damage.
- In Bone Regeneration: Within bone defects, M2 macrophages contribute to a favorable osteoimmune environment. They secrete bone morphogenetic proteins (BMPs) and other factors that directly stimulate the osteogenic differentiation of MSCs while simultaneously resolving inflammation that can impede the healing process [61] [40]. This coordination is vital for the successful integration of bone grafts and prostheses [64].
Quantitative Data from Preclinical Models
The therapeutic efficacy of MSC-Exos has been quantitatively demonstrated in multiple animal models of hindlimb ischemia and bone defects. The tables below summarize key outcomes from pivotal studies.
Table 1: Therapeutic Outcomes of MSC-Exos in Hindlimb Ischemia Mouse Models
| Treatment Group | Oxygen Saturation (SpO2) Recovery | New Capillary Density (vessels/HPF) | Limb Necrosis Incidence | Functional Recovery Score |
|---|---|---|---|---|
| PBS Control | Slow, incomplete | Low | 60% (Grades I-IV) | Poor |
| MSC-Exos | Rapid, significant improvement | High (dense capillary networks) | 33.3% (Grades I-II only) | Significant improvement |
| ADSC-Exos | Enhanced perfusion | Significant increase vs. control | Reduced necrosis rate | Improved limb function |
Table 2: Efficacy of Engineered MSC-Exos in Bone Regeneration Models
| Exosome Type | Bone Volume Regenerated | Bone Mineral Density (BMD) | Angiogenic Markers (VEGF, etc.) | Osteogenic Markers (RUNX2, etc.) |
|---|---|---|---|---|
| Native MSC-Exos | Moderate increase | Moderate improvement | Upregulated | Upregulated |
| sFgl2-MSC-Exos | Significant increase | High improvement | Highly upregulated | Highly upregulated |
| BMP-2 loaded Exos | Maximum bone formation | Highest BMD | Sustained high levels | Sustained high levels |
Data derived from these models confirm that MSC-Exos treatment leads to superior outcomes compared to controls, with further enhancements achievable through exosome engineering [65] [63] [45].
Experimental Protocols for Key Investigations
Protocol: Evaluating Therapeutic Angiogenesis in Hindlimb Ischemia
This established protocol details the creation of a murine hindlimb ischemia model and the subsequent evaluation of exosome therapy [65] [63].
Hindlimb Ischemia Model Induction:
- Anesthesia: Induce and maintain anesthesia in C57BL/6 mice (8-10 weeks old) using inhaled isoflurane (2-3% in oxygen).
- Surgery: Make a skin incision along the medial thigh. Gently dissect to expose the femoral artery and its branches.
- Ligation: Double-ligate the proximal femoral artery just distal to the superficial caudal epigastric branch. A second ligation is performed proximal to the popliteal artery bifurcation.
- Excision: Excise the arterial segment between the two ligations to prevent spontaneous reperfusion.
- Closure: Suture the muscle layer and skin incision.
Exosome Treatment and Analysis:
- Treatment Administration: One day post-surgery, administer a local intramuscular injection of 100 µg of MSC-Exos (in 100 µL PBS) into the ischemic limb. Control groups receive an equal volume of PBS.
- Longitudinal Monitoring:
- Limb Perfusion: Use Laser Doppler Perfusion Imaging weekly for 4 weeks to quantify blood flow. Express results as a ratio (ischemic/contralateral limb).
- Functional Scoring: Employ a standardized limb function score (0=normal use, 5=severe impairment/auto-amputation).
- Necrosis Grading: Grade limb necrosis from I (toenail loss) to IV (limb loss).
- Endpoint Analysis:
- Histology: Harvest gastrocnemius muscle at day 28. Process for H&E staining to assess muscle fiber architecture and capillary density (CD31+ immunostaining).
- Oxygen Saturation: Measure tissue SpO2 at the endpoint using a veterinary pulse oximeter.
Protocol: Tracking Macrophage Polarization In Vivo and In Vitro
This protocol outlines methods to investigate the effect of MSC-Exos on macrophage polarization [45].
In Vivo Tracking and Analysis:
- Exosome Labeling: Label purified MSC-Exos with a near-infrared lipophilic dye (e.g., DIR).
- Administration: Inject DIR-labeled exosomes intravenously into mice with induced hindlimb ischemia or a bone defect model.
- In Vivo Imaging: Use an in vivo imaging system (IVIS) at 24, 48, and 72 hours post-injection to track exosome homing to the injury site.
- Tissue Analysis: At day 7, harvest tissues for:
- Flow Cytometry: Create a single-cell suspension from tissues. Stain cells with antibodies against F4/80 (macrophages), CD86 (M1 marker), and CD206 (M2 marker). Quantify the M1/M2 ratio.
- Immunohistochemistry: Stain tissue sections with anti-CD86 and anti-CD206 antibodies to visualize the spatial distribution of macrophage subsets.
In Vitro Polarization Assay:
- Cell Culture: Isolate and differentiate bone marrow-derived macrophages (BMDMs) from mice using M-CSF.
- Polarization and Treatment:
- Polarize BMDMs to an M1 state with LPS (100 ng/mL) and IFN-γ (20 ng/mL).
- Treat M1-polarized macrophages with MSC-Exos (50 µg/mL) for 48 hours.
- Analysis:
- Flow Cytometry: Analyze cells for CD86 and CD206 expression.
- ELISA: Collect culture supernatant and measure concentrations of TNF-α (M1) and IL-10 (M2) cytokines.
- Western Blot: Analyze cell lysates for phosphorylation levels of SHP2 and STAT3 to confirm pathway activation.
The workflow for the comprehensive analysis of macrophage polarization is depicted below:
The Scientist's Toolkit: Research Reagent Solutions
Successful research in this field relies on a suite of critical reagents and tools, as cataloged below.
Table 3: Essential Research Reagents for MSC Exosome and Macrophage Studies
| Reagent / Tool | Specific Example | Research Function |
|---|---|---|
| Exosome Isolation Kits | Total Exosome Isolation Kit | Precipitation-based isolation of exosomes from cell culture supernatant. |
| Antibodies for Characterization | Anti-CD63, CD81, CD9 | Confirm exosome identity via flow cytometry or Western blot [63]. |
| Macrophage Polarization Cytokines | Recombinant IL-4, IL-13, LPS, IFN-γ | Induce M2 (IL-4/IL-13) or M1 (LPS/IFN-γ) polarization in vitro [45] [40]. |
| Flow Cytometry Antibodies | Anti-F4/80, CD86, CD206 | Identify macrophages (F4/80) and distinguish M1 (CD86) from M2 (CD206) phenotypes [45]. |
| In Vivo Imaging Dyes | DIR, DiR Lipophilic Dyes | Label exosomes for tracking their biodistribution and homing in live animals [45]. |
| ELISA Kits | Mouse TNF-α, IL-10, IL-6 ELISA | Quantify secreted pro-inflammatory and anti-inflammatory cytokines in cell supernatant or serum. |
| Signal Pathway Inhibitors | SHP2 inhibitor (PHPS1), STAT3 inhibitor | Mechanistic validation of specific signaling pathways involved in polarization [45]. |
The body of evidence unequivocally establishes MSC exosomes as powerful orchestrators of tissue repair in hindlimb ischemia and bone regeneration, with their ability to polarize macrophages toward the M2 phenotype being a central mechanism. The standardized experimental protocols and quantitative data presented here provide a robust framework for researchers to validate and build upon these findings.
Future research trajectories should focus on the clinical translation of these preclinical insights. Key challenges include the standardization of exosome manufacturing, optimization of dosing regimens and delivery routes (e.g., using biomaterial scaffolds for sustained release in bone defects), and rigorous evaluation of long-term safety [61] [66]. Furthermore, the engineering of exosomes—through surface functionalization for enhanced targeting or pre-loading with specific therapeutic cargo like sFgl2 or BMP-2—represents the next frontier in amplifying their regenerative potential [45] [40]. By systematically addressing these translational hurdles, MSC exosome-based therapies hold exceptional promise for becoming a mainstream treatment paradigm for ischemic diseases and complex bone regeneration.
Mesenchymal stem/stromal cell (MSC)-derived exosomes have emerged as powerful therapeutic agents in respiratory and metabolic disease interventions, particularly for alleviating acute lung injury (ALI) and airway inflammation in asthma. These nano-sized extracellular vesicles (30-150 nm in diameter) mediate the paracrine effects of their parent cells by transferring bioactive molecules—including proteins, lipids, and nucleic acids—to recipient cells [67] [68]. Within the context of respiratory diseases, a central mechanism underlying their therapeutic efficacy is the strategic reprogramming of macrophage polarization from a pro-inflammatory M1 phenotype toward an anti-inflammatory, tissue-reparative M2 phenotype [3] [4]. This phenotypic switching represents a critical immunomodulatory axis in restoring pulmonary homeostasis, resolving inflammation, and facilitating tissue repair in damaged lungs [69] [70]. The focus of this technical guide is to elucidate the specific molecular mechanisms through which MSC exosomes orchestrate this macrophage polarization, thereby alleviating pathology in experimental models of ALI and asthma.
Molecular Mechanisms of M2 Macrophage Polarization
MSC exosomes promote M2-like macrophage polarization through multiple, often synergistic, molecular pathways. The key mechanisms identified in recent studies involve specific exosomal cargo and their subsequent signaling cascades.
CD73-Mediated Adenosine Signaling
A critical mechanism involves exosomal CD73 (ecto-5'-nucleotidase) activity. MSC exosomes express CD73, which catalyzes the conversion of adenosine monophosphate (AMP) to adenosine [3]. The generated adenosine then binds to and activates adenosine receptors A2A and A2B on macrophages. This receptor binding initiates an intracellular signaling cascade that involves the phosphorylation of AKT and ERK. Activation of this CD73/adenosine/A2A/A2B/AKT/ERK axis ultimately drives the genetic and functional reprogramming of macrophages toward the M2-like phenotype [3]. The essential role of this pathway is confirmed by inhibitor studies, where polarization is abolished upon exposure to CD73 inhibitors or antagonists of A2A and A2B receptors [3].
TRAF1-Dependent Regulation of NF-κB and PI3K/AKT
A second potent mechanism involves the tumor necrosis factor receptor-associated factor 1 (TRAF1) protein. Quantitative proteomic analyses revealed that MSC exosome treatment significantly modulates TRAF1 expression in macrophages [70]. Functional studies demonstrated that knockdown of TRAF1 enhances M1 polarization while impairing M2 polarization, whereas overexpression produces the opposite effect. Mechanistically, TRAF1 mediates its effects through the suppression of the NF-κB signaling pathway (a key pro-inflammatory pathway) and simultaneous activation of the PI3K/AKT pathway, thereby shifting the balance toward an anti-inflammatory state [70].
MyD88-Dependent Toll-like Receptor Signaling
An additional pathway is activated by extra domain A-fibronectin (EDA-FN) present on MSC exosomes. EDA-FN engages Toll-like receptors (TLRs) on macrophages, leading to the activation of the MyD88-mediated TLR signaling pathway [3]. This MyD88-dependent signaling represents another established mechanism contributing to the induction of the M2-like macrophage phenotype.
The following diagram synthesizes these primary mechanisms into a unified signaling network:
Diagram 1: MSC Exosome-Mediated M2 Macrophage Polarization Signaling Network
This integrated signaling network underscores the multi-faceted approach through which MSC exosomes reprogram macrophage function, converging on key regulatory nodes to promote an anti-inflammatory, tissue-reparative environment.
The efficacy of MSC exosomes in preclinical models is demonstrated by significant improvements across physiological, histological, and molecular endpoints.
Table 1: Summary of Quantitative Therapeutic Outcomes in Preclinical Models
| Disease Model | Intervention | Key Macrophage Polarization Outcomes | Physiological/Histological Outcomes | Citation |
|---|---|---|---|---|
| Severe Steroid-Resistant Asthma (SSRA) | hUCMSC-Exo (100 μg, intratracheal) | ↓ M1 markers (iNOS, TNF-α, IL-1β, IL-6); ↑ M2 markers (Arg-1, CD206, IL-10) | Reversed AHR, reduced inflammation, improved lung histopathology | [70] |
| Acute Lung Injury (ALI) | MSC-Exos (dose varied by study) | Induced M2 polarization; ↓ LPS-induced glycolysis in macrophages | Alleviated lung injury, improved vascular permeability, increased survival rate | [69] |
| Hyperoxia-Induced Lung Injury | MSC-Exos (dose varied by study) | ↑ Infiltration of M2-like macrophages; ↓ M1-like macrophages | Reduced pro-inflammatory cytokines (TNF-α) | [3] |
| ALI (Severe Burn Model) | hUCMSC-Exos | Modulated macrophage M2 polarization via miR-451/MIF/PI3K/AKT | Reduced inflammation and oxidative stress | [69] |
Table 2: Quantitative Changes in Inflammatory Mediators Post-MSC Exosome Treatment
| Analyte | Change Post-Treatment | Significance / Associated Pathway |
|---|---|---|
| CD73/NT5E Activity | 22.71 ± 0.54 mU/μg (in exosome prep) | Critical for adenosine production; AKT/ERK dependent [3] |
| Pro-inflammatory Cytokines (TNF-α, IL-1β, IL-6) | Significant decrease | Correlates with M1 suppression and clinical improvement [3] [70] |
| Anti-inflammatory Cytokines (IL-10) | Significant increase | Correlates with M2 activation and tissue repair [70] |
| TRAF1 Protein Level | Significantly modulated | Central regulator of NF-κB suppression and PI3K/AKT activation [70] |
Detailed Experimental Protocols
To ensure reproducibility and facilitate further research, this section provides detailed methodologies for key experiments cited in this field.
Protocol: In Vitro Macrophage Polarization Assay
This protocol is adapted from studies investigating MSC exosome-mediated macrophage polarization [3] [70].
Primary Macrophage Differentiation:
- Isolation: Isolate peripheral blood mononuclear cells (PBMCs) from human donors or animal models (e.g., Sprague Dawley rats) using Ficoll-Paque density gradient centrifugation.
- Culture: Seed PBMCs at a density of 0.5 × 10^6 cells/mL in RPMI medium supplemented with 10% heat-inactivated FBS and 1% Penicillin-Streptomycin.
- Differentiation: After overnight incubation, rinse off non-adherent cells. Culture the remaining monocytes for 7-9 days in RPMI medium containing 40 ng/mL Macrophage Colony-Stimulating Factor (M-CSF), with media changes every 2-3 days, to generate M0 macrophages.
Exosome Treatment & Polarization:
- Treatment: Treat mature M0 macrophages with 10 μg/mL of characterized MSC exosomes. Include a PBS vehicle control.
- Incubation: Incubate for 24-48 hours under standard culture conditions (37°C, 5% CO2).
Inhibition Studies (Mechanistic Validation):
- To probe specific pathways, co-treat macrophages with MSC exosomes and specific inhibitors.
- Common Inhibitors:
- CD73 inhibitor (e.g., PSB12379 at 10 nM)
- A2A/A2B adenosine receptor antagonists
- PI3K/AKT pathway inhibitors
Protocol: In Vivo Assessment in a Steroid-Resistant Asthma Model
This protocol is based on the established OVA/CFA-induced SSRA murine model [70].
Animal Model Generation:
- Sensitization: Sensitize female BALB/c mice (18-22 g) via intraperitoneal injection of a mixture of 20 μg Ovalbumin (OVA) in 75 μl Complete Freund's Adjuvant (CFA) and 25 μl PBS on days 0, 7, and 14.
- Challenge: On days 21, 22, and 23, challenge the sensitized mice with aerosolized 2% OVA for 20-30 minutes using an inhalation exposure system.
Therapeutic Intervention:
- Administration: On day 21, administer MSC exosomes (100 μg in 50 μl PBS per mouse) via intratracheal instillation.
- Control Groups: Include groups treated with dexamethasone (2 mg/kg, i.p.) and a PBS vehicle control.
Endpoint Analysis (Harvest on Day 24):
- Airway Hyperresponsiveness (AHR): Measure lung resistance and dynamic compliance in anesthetized, ventilated mice in response to increasing doses of methacholine (6.25-50 mg/mL).
- Bronchoalveolar Lavage Fluid (BALF): Collect BALF for total and differential immune cell counting (eosinophils, neutrophils, macrophages, lymphocytes).
- Cytokine Analysis: Analyze BALF and lung homogenates for cytokines (e.g., TNF-α, IL-1β, IL-6, IL-10) via ELISA or multiplex assays.
- Histopathology: Fix lung tissues in paraformaldehyde, embed in paraffin, section, and stain with Hematoxylin and Eosin (H&E) for general inflammation and Masson's Trichrome for collagen deposition.
- Macrophage Phenotyping: Analyze lung single-cell suspensions or tissue sections via flow cytometry (for CD86, iNOS, CD206, Arg-1) or immunofluorescence.
The workflow for establishing and analyzing the in vivo asthma model is summarized below:
Diagram 2: In Vivo SSRA Model Workflow and Analysis
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for MSC Exosome and Macrophage Research
| Reagent / Tool | Function / Application | Specific Examples / Notes |
|---|---|---|
| Exosome Isolation Kits | Isolation of exosomes from MSC-conditioned medium. | exoEasy Maxi Kit (Qiagen); Polymer-based precipitation kits (e.g., PEG-based) [70]. |
| Characterization Instruments | Physical and biomolecular analysis of purified exosomes. | NTA (ZetaView): Size/concentration; TEM: Morphology; Flow Cytometry: Surface markers (CD63, CD81) [70]. |
| CD73/NT5E Activity Assay | Functional potency assay for MSC exosome preparations. | PiColorLock Gold Phosphate Detection System [3]. |
| M-CSF | Differentiation of monocytes into M0 macrophages in culture. | Recombinant human or rodent M-CSF, used at 40 ng/mL [3]. |
| Polarization & Pathway Inhibitors | Mechanistic validation of signaling pathways in macrophage polarization. | PSB12379: CD73 inhibitor; A2A/A2B Antagonists; AKT/ERK inhibitors [3]. |
| Clodronate Liposomes | Systemic depletion of macrophages in vivo. | Critical for establishing the functional role of macrophages in therapeutic efficacy [70]. |
| Antibodies for Flow Cytometry | Identification and polarization analysis of macrophages. | M1 Markers: CD86, iNOS; M2 Markers: CD206, Arg-1 [70]. |
Engineering Strategies for Enhanced Therapeutic Efficacy
Native MSC exosomes have inherent therapeutic potential, but engineering strategies can significantly enhance their targeting, delivery, and potency.
Preconditioning: Culturing MSCs under simulated disease conditions (e.g., hypoxia, inflammation) can enrich exosomal cargo with beneficial miRNAs and proteins. For instance, hypoxic preconditioning enhances the loading of angiogenic and anti-inflammatory miRNAs [54] [71].
Surface Modification: Engineering the exosome membrane to display homing peptides or ligands (e.g., using lentiviral vectors to express fusion proteins like iRGD-Lamp2b) can confer tissue-specific or cell-specific targeting capabilities. This improves accumulation at the site of injury, such as the inflamed lung [71].
Cargo Loading: Direct loading of therapeutic molecules (e.g., anti-miRNA oligonucleotides, small molecule drugs) into isolated exosomes can be achieved using techniques like electroporation. This allows for the creation of targeted, nano-scale drug delivery vehicles with natural biocompatibility [71].
MSC exosomes represent a promising cell-free therapeutic paradigm for acute lung injury and steroid-resistant asthma, with their ability to drive M2 macrophage polarization being a central mechanism of action. The detailed molecular pathways—including CD73/adenosine signaling, TRAF1-mediated regulation, and TLR/MyD88 activation—provide a robust scientific foundation for their efficacy. The standardized experimental protocols and reagent toolkit outlined in this guide equip researchers with the necessary resources to further investigate and validate these mechanisms. Future work focused on optimizing exosome engineering and manufacturing will be crucial for translating these compelling preclinical findings into effective clinical therapies for debilitating respiratory diseases.
Enhancing Therapeutic Potency: Preconditioning Strategies and Engineering Approaches
The therapeutic potential of mesenchymal stem cell-derived exosomes (MSC-exosomes) is increasingly recognized in regenerative medicine and immunomodulation. These nano-sized extracellular vesicles mediate the paracrine effects of their parent cells by transferring functional proteins, lipids, and nucleic acids to recipient cells. Within the context of macrophage polarization research, MSC-exosomes demonstrate a remarkable capacity to modulate immune responses, particularly in promoting the anti-inflammatory M2 phenotype. However, this bioactivity is not uniform across all MSC sources. This technical review provides a comprehensive comparison of exosomes derived from adipose tissue (AD-MSC), bone marrow (BM-MSC), and gingival tissue (G-MSC), focusing on their source-dependent molecular signatures and differential effects on macrophage polarization. We present quantitative data on their characteristic biomarkers, nucleic acid cargo, and functional efficacy, supplemented with detailed experimental protocols and signaling pathway visualizations to guide research in therapeutic development.
Mesenchymal stem cells (MSCs) are multipotent stromal cells found in various adult tissues, including adipose tissue, bone marrow, and gingival tissue [72]. They are characterized by their plastic-adherent properties, specific surface marker expression (CD73, CD90, CD105), and lack of hematopoietic markers (CD34, CD45) [72]. Rather than through direct differentiation, MSCs primarily exert their therapeutic effects via paracrine signaling, with exosomes serving as key mediators of intercellular communication [73] [74].
Exosomes are nanoscale extracellular vesicles (30-150 nm in diameter) that transport functional cargo—including miRNA, mRNA, proteins, and lipids—from parent MSCs to recipient cells [73]. This transfer can reprogram target cell behavior, making exosomes promising cell-free therapeutic agents with improved safety profiles compared to whole-cell therapies [75] [76].
In the context of immunomodulation, MSC-exosomes have demonstrated a potent ability to modulate macrophage polarization—a critical process in immune homeostasis, tissue repair, and resolution of inflammation [77]. Macrophages can differentiate into pro-inflammatory M1 phenotypes or anti-inflammatory M2 phenotypes, with the latter promoting tissue repair and suppressing excessive inflammation. The specific mechanisms by which MSC-exosomes from different tissue sources drive this polarization toward the M2 phenotype remain an area of intense investigation, with significant implications for treating inflammatory and autoimmune conditions.
Source-Specific Characteristics of MSC-Exosomes
Molecular Profiles and Cargo Composition
Exosomes derived from different MSC sources exhibit distinct molecular signatures that influence their functional properties. The table below summarizes key characteristics of exosomes from adipose, bone marrow, and gingival MSCs.
Table 1: Comparative Characteristics of MSC-Exosomes from Different Tissues
| Characteristic | Adipose-derived (AD-MSC) | Bone Marrow-derived (BM-MSC) | Gingival-derived (G-MSC) |
|---|---|---|---|
| Key Protein Markers | CD73, CD90, CD105; Adipokine-related proteins | CD73, CD90, CD105; High levels of immunomodulatory proteins (e.g., IDO, PGE2) [72] | CD73, CD90, CD105; Unique tissue-specific markers under investigation |
| Characteristic miRNAs | miRNAs involved in adipogenesis (e.g., miR-148a, miR-26b) [73] | miRNAs with immunomodulatory functions (e.g., miR-21, miR-221-3p) [75] [78] | miRNAs related to oral mucosal repair and inflammation |
| Lipid Composition | Enriched in lipids common to all exosomes; potential variations in sphingomyelin and cholesterol [73] | Conserved lipid content with high proportions of cholesterol [73] | Similar conserved lipid profile; tissue-specific variations possible |
| Reported Immunomodulatory Strength | Potent; supported by clinical trials for Crohn's disease [79] | Very potent; gold standard source with extensive data on macrophage polarization [77] | Promising; evidence suggests strong regenerative and immunomodulatory capacity |
| Key Functional Evidence | Safe and clinically efficacious in multiple inflammatory conditions [79] | Regulates glioma-associated macrophages, inhibiting M2 polarization [77] | Associated with excellent wound healing and anti-inflammatory effects |
Technical Insights on Exosome Heterogeneity
Recent advances in separation technologies have revealed significant heterogeneity even within exosome populations from a single source. A 2025 study demonstrated that human umbilical cord MSC-exosomes can be separated into distinct subpopulations (S1-sEVs and S2-sEVs) using tangential flow filtration combined with size exclusion chromatography [80]. These subpopulations differ in size, membrane markers, cargo, and function:
- S1-sEVs: Generated via the ESCRT-dependent pathway, enriched in proteins like HGS. They demonstrate superior immunomodulatory capacity, effectively polarizing macrophages toward the M2 phenotype [80].
- S2-sEVs: Generated via ESCRT-independent pathways involving lipid rafts (marked by flotillin proteins). They excel in promoting cell proliferation and metabolic regulation [80].
This underscores that functional variability is influenced not only by tissue source but also by the specific exosome subpopulations isolated.
Experimental Protocols for Evaluating MSC-Exosomes in Macrophage Polarization
Protocol 1: Isolation and Characterization of MSC-Exosomes
Objective: To isolate and characterize exosomes from adipose, bone marrow, and gingival MSCs.
Materials:
- MSC Culture Medium: α-MEM complete medium supplemented with fetal bovine serum (exosome-depleted) [81]
- Isolation Reagents: Phosphate-buffered saline (PBS), protease inhibitors [73]
- Characterization Antibodies: Anti-CD63, Anti-CD81, Anti-CD9, Anti-Alix, Anti-TSG101 [81] [77]
Methodology:
- MSC Culture and Conditioned Media Collection: Culture MSCs from each source until 70-80% confluence. Replace with serum-free medium and culture for 48 hours. Collect conditioned media [81].
- Differential Ultracentrifugation:
- Centrifuge at 500 × g for 10 minutes to remove cells.
- Centrifuge at 2,000 × g for 20 minutes to remove dead cells.
- Centrifuge at 10,000 × g for 30 minutes to remove cell debris and apoptotic bodies.
- Ultracentrifuge at 100,000 × g for 70 minutes to pellet exosomes [73].
- Exosome Characterization:
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration [80].
- Transmission Electron Microscopy (TEM): Visualize exosome morphology and structure [81] [77].
- Western Blotting: Confirm presence of exosomal markers (CD63, CD81, Alix, TSG101) and absence of negative markers (calnexin) [81] [77].
Protocol 2: Assessing Macrophage Polarization In Vitro
Objective: To evaluate the effect of different MSC-exosomes on macrophage polarization to the M2 phenotype.
Materials:
- Cell Lines: Primary human monocytes or macrophage cell lines (e.g., THP-1)
- Polarization Reagents: Phorbol 12-myristate 13-acetate (PMA), Lipopolysaccharide (LPS), Interleukin-4 (IL-4)
- Detection Antibodies: Anti-CD68 (M1 marker), Anti-CD206 (M2 marker)
Methodology:
- Macrophage Differentiation and Polarization:
- Differentiate THP-1 monocytes into M0 macrophages using 100 ng/mL PMA for 24-48 hours.
- Polarize macrophages toward M1 phenotype using 100 ng/mL LPS or toward M2 phenotype using 20 ng/mL IL-4 [77].
- Exosome Treatment:
- Treat M0, M1, or M2 macrophages with MSC-exosomes (e.g., 50-100 μg/mL) from different sources for 24-48 hours.
- Use PKH67 fluorescent dye to label exosomes and track their uptake by macrophages [77].
- Analysis of Polarization Status:
- Quantitative PCR (qPCR): Measure expression of M1 markers (iNOS, CD68) and M2 markers (Arginase, CD206) [81] [77].
- ELISA: Quantify secretion of M1-related cytokines (IL-1β, TNF-α) and M2-related cytokines (CCL22, TGF-β) in cell culture supernatant [77].
- Flow Cytometry: Analyze surface expression of M1 (CD68) and M2 (CD206) markers [77].
- Immunofluorescence: Visualize macrophage morphology and marker expression.
Diagram 1: Macrophage polarization assay workflow
Signaling Pathways in MSC-Exosome Mediated M2 Polarization
MSC-exosomes from different sources utilize multiple signaling pathways to promote M2 macrophage polarization. The specific mechanisms vary based on their molecular cargo.
Key Pathway Components:
- miRNA-Mediated Regulation:
Metabolic Reprogramming:
Surface Protein Interactions:
- Exosomes express surface proteins (e.g., FASL, PD-L1) that directly interact with immune receptors on macrophages, influencing their polarization state [72].
Diagram 2: MSC-exosome signaling in M2 macrophage polarization
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagents for MSC-Exosome and Macrophage Polarization Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| MSC Culture Media | α-MEM complete medium; Serum-free media for conditioning | MSC expansion and exosome production [81] |
| Exosome Isolation Kits | Total Exosome Isolation Kits; TFF-SEC systems | Rapid isolation of exosomes from conditioned media [80] |
| Characterization Antibodies | Anti-CD63, CD81, CD9, Alix, TSG101; CD73, CD90, CD105 | Identification and validation of exosomes and MSCs [81] [72] |
| Macrophage Polarization Agents | PMA, LPS, IL-4 | Induce differentiation and polarization of macrophages [77] |
| Macrophage Marker Antibodies | Anti-CD68 (M1), Anti-CD206 (M2) | Detection and quantification of macrophage phenotypes [77] |
| Detection Assays | ELISA kits for IL-1β, TNF-α, CCL22, TGF-β | Quantify cytokine secretion profiles [77] |
| Cell Tracking Dyes | PKH67, PKH26 | Labeling and tracking exosome uptake by macrophages [77] |
MSC-derived exosomes from adipose tissue, bone marrow, and gingival tissue present distinct molecular and functional profiles that significantly impact their efficacy in driving macrophage polarization toward the anti-inflammatory M2 phenotype. BM-MSC-exosomes have the most substantial evidence base for immunomodulation, while AD-MSC-exosomes offer practical advantages for clinical translation. G-MSC-exosomes represent a promising but less characterized source. The emerging recognition of functional subpopulations within exosome preparations further complicates the source-dependent variability narrative. Future research should focus on standardized isolation protocols for specific exosome subpopulations and detailed mechanistic studies to fully elucidate how exosomes from different sources reprogram macrophages. This knowledge will be crucial for developing targeted exosome-based therapeutics for inflammatory and autoimmune diseases.
Mesenchymal stromal cell (MSC)-derived exosomes have emerged as potent mediators of immunomodulation, with their ability to promote anti-inflammatory M2 macrophage polarization representing a key mechanism in their therapeutic effects [3]. This polarization process is critically important in tissue repair and regeneration, as M2 macrophages facilitate the resolution of inflammation and promote healing through the release of anti-inflammatory mediators such as IL-10 and TGF-β [3]. The efficacy of MSC exosomes is not static but can be significantly enhanced through inflammatory preconditioning—a strategy that primes MSCs with specific cytokines to optimize the immunomodulatory cargo of their secreted vesicles [82] [83].
Synergistic preconditioning with TNF-α and IFN-α represents a particularly promising approach that uniquely boosts the expression of two critical molecules in exosomes: CD73 (an ecto-5'-nucleotidase) and CD5L (CD5 molecule-like) [83]. This technical guide examines the mechanisms through which this specific preconditioning strategy enhances the M2-polarizing capacity of MSC exosomes, providing researchers with detailed methodologies, signaling pathways, and experimental data to facilitate implementation in both basic research and therapeutic development contexts.
Molecular Mechanisms of Synergistic Preconditioning
CD73: Adenosine-Mediated Immunomodulation
CD73, also known as ecto-5'-nucleotidase (NT5E), is a surface enzyme that catalyzes the conversion of adenosine monophosphate (AMP) to adenosine, a potent immunosuppressive mediator [3] [84]. In the context of macrophage polarization, adenosine exerts its effects through binding to ubiquitously expressed adenosine receptors A2A and A2B, subsequently activating AKT/ERK-dependent signaling pathways that drive M2 polarization [3]. Research has demonstrated that MSC exosomes promote M2-like macrophage polarization through exosomal CD73 activity, as this polarization is abolished in the presence of inhibitors targeting CD73 activity, adenosine receptors A2A and A2B, or AKT/ERK phosphorylation [3].
CD5L: A Novel Player in Macrophage Polarization
CD5L (CD5 molecule-like), also known as apoptosis inhibitor of macrophage (AIM), is a soluble scavenger cysteine-rich protein that circulates in the blood at relatively high levels [85]. While initially identified as an apoptosis inhibitor protecting macrophage survival, recent evidence indicates it plays a broader role in inflammatory diseases [85]. In the context of TNF-α/IFN-α preconditioning, CD5L emerges as a critical component in exosome-mediated M2 polarization, with studies indicating that exosomal CD5L is a prerequisite for the synergistic effect of EVs in promoting M2 macrophage polarization [83]. This represents a distinct mechanism from the adenosine-mediated pathway of CD73.
Signaling Pathways Activated by TNF-α/IFN-α Preconditioning
The synergistic effect of TNF-α and IFN-α preconditioning on CD73 expression is mediated through specific intracellular signaling cascades. Research using gingival tissue-derived MSCs (GMSCs) has revealed that this combination treatment activates the mTOR-HIF-1α axis [83]. Specifically, dual cytokine stimulation synergistically enhances the expression of HIF-1α mRNA and protein, with significant nuclear accumulation of HIF-1α protein observed within 24 hours—a key indicator of HIF-1α activation [83].
For CD5L upregulation, TNF-α/IFN-α treatment significantly increases CD5L mRNA expression through the transcription factor DNA-binding protein inhibitor ID3 and liver X receptor [83]. This distinct pathway highlights how preconditioning simultaneously activates multiple mechanisms to enhance the immunomodulatory cargo of MSC exosomes.
Figure 1: Signaling Pathways in TNF-α/IFN-α Preconditioning. Dual cytokine stimulation activates distinct pathways for CD73 and CD5L upregulation in MSC exosomes, converging on enhanced M2 macrophage polarization.
Quantitative Effects of Preconditioning on Exosomal Cargo
The synergistic effect of TNF-α and IFN-α preconditioning on CD73 and CD5L expression has been quantitatively demonstrated through multiple experimental approaches. The table below summarizes key quantitative findings from preconditioning studies:
Table 1: Quantitative Effects of TNF-α/IFN-α Preconditioning on MSC Exosomes
| Parameter | Experimental Finding | Measurement Method | Citation |
|---|---|---|---|
| CD73 mRNA | Synergistic increase after 24h stimulation | qRT-PCR | [83] |
| CD73 Protein in EVs | Significantly increased with TNF-α and TNF-α/IFN-α | Western Blot | [83] |
| EV Particle Size | Mode: 167 ± 5.7 nm (vs 160 ± 2.1 nm control) | Nanoparticle Tracking Analysis | [83] |
| HIF-1α mRNA | Synergistic enhancement with dual stimulation | qRT-PCR | [83] |
| CD5L mRNA | Significantly increased via ID3 and LXR | qRT-PCR | [83] |
The efficacy of inflammatory preconditioning is not limited to TNF-α/IFN-α combinations. Other cytokine combinations have also been explored, though with potentially different mechanisms and outcomes:
Table 2: Comparative Analysis of MSC Preconditioning Strategies
| Preconditioning Method | Key Effects on MSC Exosomes/EVs | Functional Outcomes | Citation |
|---|---|---|---|
| TNF-α + IFN-α | Enhanced CD73 and CD5L expression | Synergistic promotion of M2 macrophage polarization | [83] |
| TNF-α + IFN-γ | Increased pro-angiogenic protein content | Stimulated HUVEC migration, proliferation, tube formation | [86] |
| TNF-α alone | Increased CD73 expression | Enhanced M2 macrophage polarization | [82] |
| Inflammatory Environment | Increased ICAM-1 expression | Improved immunoregulatory capacity through cell contact | [87] |
Experimental Protocols for Preconditioning and Evaluation
MSC Culture and Preconditioning Methodology
Cell Source and Culture:
- Gingival tissue-derived MSCs (GMSCs) are particularly suitable due to their unique immunoregulatory capacity and secretion of large amounts of EVs [83].
- Culture GMSCs in standard mesenchymal stem cell growth medium consisting of MEM Alpha Medium supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% Penicillin-Streptomycin [86].
- Maintain cultures at 37°C, 5% CO2, and 95% humidity with medium changes three times per week until reaching 80% confluence [86].
Preconditioning Protocol:
- At approximately 80% confluence, wash cells at least three times with sterile 1x PBS [86].
- Treat GMSCs with a combination of 100 ng/mL TNF-α and 50 ng/mL IFN-α for 48 hours [83].
- For comparative studies, include control groups without preconditioning and with individual cytokines alone.
- After preconditioning, collect conditioned medium for EV isolation.
EV Isolation and Characterization
Isolation Method:
- Isolate EVs from conditioned medium using tangential flow filtration and concentrate 50× using a membrane with a molecular weight cut-off of 100 kDa [3].
- Alternative methods include ultracentrifugation (100,000 × g for 70 minutes) or commercial EV isolation kits [83].
Characterization Parameters:
- Protein concentration: Measure using Coomassie Plus protein assay [3].
- Particle size distribution and concentration: Analyze by Nanoparticle Tracking Analysis (NTA) or ZetaView [3] [83].
- Modal size: Expected range of 157-167 nm [83].
- CD73/NT5E activity: Assess using PiColorLock Gold Phosphate Detection System [3].
- Morphology: Verify spherical morphology under transmission electron microscopy (TEM) [83].
Macrophage Polarization Assays
Macrophage Culture:
- Isolate peripheral blood mononuclear cells (PBMCs) from blood by Ficoll-Paque density gradient centrifugation [3].
- Culture PBMCs in RPMI medium supplemented with 1% penicillin-streptomycin and 10% heat-inactivated FBS [3].
- Differentiate macrophages using 40 ng/mL macrophage colony-stimulating factor (M-CSF) for 7-9 days, with media changes every 2-3 days [3].
Polarization Assessment:
- Treat macrophages with preconditioned EVs (10 μg/mL) or PBS vehicle for 24 and 48 hours [3].
- For inhibition studies, co-treat with 10 nM PSB12379 (CD73 inhibitor) to confirm mechanism [3].
- Evaluate M2 markers through:
Figure 2: Experimental Workflow for Preconditioning Studies. Comprehensive methodology from MSC culture and preconditioning through EV isolation and macrophage polarization assessment.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Preconditioning Studies
| Reagent/Category | Specific Examples | Function/Application | Citation |
|---|---|---|---|
| Cytokines | TNF-α (100 ng/mL), IFN-α (50 ng/mL) | MSC preconditioning to enhance CD73/CD5L | [83] |
| Cell Culture Media | MEM Alpha Medium, RPMI-1640 | MSC and macrophage culture | [3] [86] |
| EV Isolation Tools | Tangential flow filtration system, 100 kDa cut-off membranes | EV isolation and concentration | [3] |
| Characterization Instruments | Nanoparticle Tracking Analyzer, Transmission Electron Microscope | EV size, concentration, morphology | [83] |
| Macrophage Differentiation | M-CSF (40 ng/mL) | PBMC to macrophage differentiation | [3] |
| Inhibitors | PSB12379 (CD73 inhibitor, 10 nM) | Mechanistic studies of adenosine pathway | [3] |
| Antibodies for Characterization | CD90-FITC, CD105-FITC, CD73-PE, CD34-PE, CD45-PE | MSC phenotyping by flow cytometry | [86] |
| M2 Macrophage Markers | CD206, CD163, Arg1, IL-1RN | Assessment of M2 polarization | [3] |
Discussion and Research Implications
The strategic preconditioning of MSCs with TNF-α and IFN-α represents a significant advancement in the field of cell-free regenerative therapy. By synergistically enhancing the expression of both CD73 and CD5L in MSC-derived exosomes, this approach leverages multiple molecular pathways to achieve robust M2 macrophage polarization [83]. The dual mechanism of action—combining adenosine-mediated immunomodulation via CD73 with the distinct functions of CD5L—provides a compelling strategy for enhancing therapeutic efficacy.
From a translational perspective, the consistency and scalability of this approach are critical considerations. Research has demonstrated that immortalized MSC lines can provide an unlimited supply of MSCs for scalable production of MSC exosomes in a consistent and reproducible manner [3]. This addresses important challenges related to the heterogeneity of MSC sources and isolation protocols that can influence therapeutic efficacy [3].
Future research directions should focus on optimizing cytokine concentrations and timing for specific therapeutic applications, exploring the potential of other cytokine combinations, and investigating the role of additional molecules that may work synergistically with CD73 and CD5L. Furthermore, comprehensive safety profiles and manufacturing standardization will be essential for clinical translation of preconditioned MSC exosomes as therapeutic agents for inflammatory diseases, autoimmune conditions, and tissue repair applications.
The integration of preconditioning strategies with emerging technologies in EV engineering and targeted delivery holds particular promise for developing next-generation cell-free therapies with enhanced specificity and potency for clinical applications.
Hypoxic priming represents a critical adaptive response where Mesenchymal Stem Cells (MSCs) exposed to low oxygen tension undergo significant molecular reprogramming, leading to the secretion of exosomes with enhanced bioactivity. These hypoxia-conditioned exosomes exhibit enriched cargo of pro-angiogenic factors and immunomodulatory molecules that orchestrate tissue repair and regenerative processes. Within the context of macrophage polarization research, hypoxic MSC exosomes function as sophisticated communication vehicles that promote a shift toward the anti-inflammatory M2 phenotype through multiple synergistic mechanisms. This whitepaper comprehensively examines the molecular machinery driving hypoxic exosome biogenesis, details the specific cargo alterations, delineates the mechanistic pathways through which these vesicles modulate macrophage polarization, and provides standardized experimental frameworks for investigating these phenomena. The insights presented herein establish hypoxic priming as a fundamental regulatory process with substantial implications for therapeutic development in inflammatory diseases, cancer, and regenerative medicine.
Exosomes are nanoscale extracellular vesicles (30-150 nm in diameter) originating from the endosomal pathway through the formation of multivesicular bodies (MVBs) that subsequently fuse with the plasma membrane to release their intraluminal vesicles into the extracellular space [88] [89]. These vesicles serve as fundamental vehicles for intercellular communication, transporting functional proteins, lipids, mRNAs, and non-coding RNAs to recipient cells [90] [91]. Under normoxic conditions, exosome secretion maintains baseline physiological communication; however, when cells experience hypoxia, profound changes occur in both the quantity and quality of secreted exosomes.
Hypoxic priming refers to the process whereby MSCs preconditioned in low oxygen environments (typically 1-5% O₂) significantly alter their exosomal secretion profile. Research demonstrates that hypoxic stress induces a 2–6 fold increase in exosome production across various cell types, including MSCs [88] [89]. This enhanced biogenesis is coupled with selective cargo sorting that enriches specific pro-angiogenic and immunomodulatory molecules within exosomes. The resulting vesicles possess enhanced capacity to influence recipient cell behavior, particularly in directing macrophage polarization toward the M2 phenotype, which plays a central role in resolving inflammation and promoting tissue repair [3] [4]. The hypoxic tumor microenvironment similarly exploits these mechanisms, where tumor-derived exosomes educate macrophages toward tumor-associated macrophages (TAMs) that support immunosuppression and angiogenesis [90] [92].
Molecular Mechanisms of Hypoxic Priming
HIF-Dependent Signaling Pathways
The primary molecular responder to hypoxia is the hypoxia-inducible factor (HIF) family of transcription factors, which serves as the master regulator of cellular adaptation to low oxygen tension [90] [93] [89]. HIF functions as a heterodimer consisting of an oxygen-sensitive α-subunit (HIF-1α, HIF-2α, or HIF-3α) and a constitutively expressed β-subunit (HIF-1β) [93]. Under normoxic conditions, HIF-α subunits undergo prolyl hydroxylation by prolyl hydroxylases (PHDs), leading to their recognition by the von Hippel-Lindau (pVHL) E3 ubiquitin ligase complex and subsequent proteasomal degradation [93] [89]. Under hypoxic conditions, PHD activity is inhibited, resulting in HIF-α stabilization, nuclear translocation, dimerization with HIF-1β, and binding to hypoxia-response elements (HREs) in target gene promoters [90] [93].
Table 1: HIF-Dependent Mechanisms in Exosome Biogenesis and Cargo Sorting
| HIF-Target Gene | Function in Exosome Biology | Biological Outcome |
|---|---|---|
| Rab GTPases (Rab22A) | Regulates MVB formation and trafficking | Increased exosome secretion [89] |
| Tetraspanins (CD63) | Facilitates endosomal membrane invagination | Altered exosome cargo composition [91] [89] |
| Pyruvate Kinase M2 (PKM2) | Metabolic reprogramming of donor cells | Enhanced pro-angiogenic cargo loading [88] |
| VEGF-A | Direct packaging into exosomes | Enhanced angiogenic signaling [90] |
| microRNAs (miR-210) | Selective sorting into exosomes | Metabolic reprogramming of recipient cells [93] [89] |
HIF activation directly influences exosome biogenesis through transcriptional upregulation of Rab GTPases, particularly Rab22A, which promotes MVB formation and trafficking [89]. Additionally, HIF stimulates expression of tetraspanin proteins (CD9, CD63, CD81) that facilitate endosomal membrane invagination and cargo selection [91]. Beyond biogenesis, HIF activation directs the selective packaging of specific cargo molecules into exosomes, including pro-angiogenic factors like VEGF-A and matrix metalloproteinases, and immunomodulatory miRNAs such as miR-210 and let-7 family members [90] [88] [93].
HIF-Independent Regulatory Mechanisms
While HIF signaling represents the cornerstone of hypoxic response, several HIF-independent pathways contribute to hypoxic priming of exosomes. These include:
- NF-κB Signaling: Hypoxia activates NF-κB through reactive oxygen species (ROS) and calcium-mediated signaling, promoting inflammation and enhancing exosome secretion [88] [89].
- Metabolic Reprogramming: Hypoxic conditions shift cellular metabolism toward glycolysis, increasing lactate production and extracellular acidification that influence exosome release and uptake [88].
- Calcium Signaling: Elevated intracellular calcium under hypoxia activates calcium-dependent enzymes that modulate MVB fusion with the plasma membrane [91].
- Ceramide Pathway: Hypoxia upregulates neutral sphingomyelinase 2 (nSMase2), which catalyzes ceramide production, facilitating inward budding of endosomal membranes and exosome formation [91] [88].
These coordinated molecular events create a comprehensive adaptive response that significantly enhances both the production and functional capacity of MSC-derived exosomes under hypoxic conditions.
Alterations in Exosome Cargo Under Hypoxia
Protein Cargo Modifications
Hypoxic priming substantially reshapes the proteomic profile of MSC-derived exosomes. Quantitative analyses reveal significant enrichment of specific protein classes that enhance angiogenic and immunomodulatory functions:
Table 2: Key Protein Cargo Enriched in Hypoxic MSC Exosomes
| Protein Category | Specific Examples | Functional Significance |
|---|---|---|
| Angiogenic Factors | VEGF-A, FGF2, PDGF-AB, Angiopoietin-1 | Promote endothelial cell proliferation, migration, and tube formation [90] [93] |
| Immunomodulators | CD73, TGF-β1, IL-10, EDA-Fibronectin | Polarize macrophages to M2 phenotype; suppress pro-inflammatory responses [3] [4] |
| Matrix Remodelers | MMP-2, MMP-9, Lysyl Oxidase (LOX), TIMP-1 | Degrade extracellular matrix; facilitate vascular sprouting and cell migration [90] [89] |
| Adhesion Molecules | Integrins, ICAM-1 | Mediate exosome targeting to specific recipient cells [91] [89] |
The ecto-5'-nucleotidase CD73 represents a particularly critical protein cargo that catalyzes the conversion of AMP to adenosine, which then activates adenosine receptors (A2A and A2B) on macrophages to promote M2 polarization through AKT/ERK-dependent pathways [3]. Similarly, EDA-Fibronectin in hypoxic exosomes activates TLR-mediated signaling in macrophages, further reinforcing M2 polarization [3].
Nucleic Acid Cargo Modifications
The nucleic acid composition of exosomes undergoes dramatic restructuring under hypoxia, with selective enrichment of non-coding RNAs that regulate recipient cell gene expression:
microRNAs: Hypoxic exosomes show increased levels of miR-210 (regulates mitochondrial metabolism), miR-126 (promotes angiogenesis), let-7 family (modulates immune responses), and miR-21-5p (activates IL-6/STAT3 signaling) [88] [92] [4]. These miRNAs suppress specific target genes in recipient macrophages to direct polarization toward the M2 phenotype.
Long Non-coding RNAs: Hypoxia upregulates specific lncRNAs such as lnc01060 in glioma stem cells, which promotes tumor progression when transferred via exosomes [90]. Similar mechanisms likely operate in MSC systems.
mRNAs: Full-length mRNAs for transcription factors (e.g., STAT3) and angiogenic factors (e.g., VEGFA) are packaged into hypoxic exosomes and can be translated in recipient cells, directly modifying their functional state [93] [89].
The selective sorting of nucleic acids into exosomes under hypoxia involves RNA-binding proteins (e.g., hnRNPs) that recognize specific motifs in target RNAs, though the precise molecular machinery remains an active research area [88].
Experimental Models and Methodologies
Standardized Hypoxic Priming Protocol
Establishing consistent hypoxic conditions is essential for reproducible research on primed exosomes. The following protocol represents current best practices:
Cell Culture: Culture MSCs in standard growth medium (DMEM with 10% FBS) until 70-80% confluence. Use low passage numbers (P3-P7) to maintain stemness [3].
Hypoxic Exposure: Place cells in a specialized hypoxic chamber or multi-gas incubator with precise environmental control:
Exosome Isolation: Collect conditioned medium and perform sequential centrifugation:
Exosome Characterization:
- Nanoparticle Tracking Analysis: Determine particle size distribution (expected mode: 130-140 nm) and concentration (typically 1-5 × 10¹¹ particles/mg protein) [3]
- Western Blotting: Confirm presence of exosomal markers (CD9, CD63, CD81, TSG101) and absence of contaminants (calnexin, GM130) [3]
- Electron Microscopy: Visualize vesicle morphology [3]
- Functional Assays: CD73/NT5E activity (expected: 20-25 mU/μg) [3]
Macrophage Polarization Assays
To evaluate the functional impact of hypoxic exosomes on macrophage polarization:
Macrophage Differentiation: Isolate peripheral blood mononuclear cells (PBMCs) from human blood or rodent specimens by Ficoll-Paque density gradient centrifugation. Differentiate monocytes into macrophages using M-CSF (40 ng/mL) for 7-9 days [3].
Exosome Treatment: Treat differentiated macrophages (M0) with:
- Experimental: Hypoxic MSC exosomes (10 μg/mL)
- Control: Normoxic MSC exosomes (10 μg/mL) or PBS vehicle
- Duration: 24-48 hours [3]
Inhibition Studies: To elucidate mechanisms, co-treat with specific inhibitors:
- CD73 inhibitor: PSB12379 (10 nM)
- A2A/A2B adenosine receptor antagonists
- AKT/ERK pathway inhibitors [3]
Phenotype Assessment:
- Flow Cytometry: Quantify M2 markers (CD206, CD163) vs. M1 markers (CD86, CD80)
- qPCR: Measure M2 gene expression (Arg1, IL-10, TGF-β) vs. M1 genes (iNOS, TNF-α, IL-1β)
- Cytokine Array: Analyze secreted factors (IL-10, TGF-β vs. TNF-α, IL-6, IL-12) [3] [4]
- Functional Assays: Phagocytosis capacity, metabolic profiling [3]
Diagram 1: Molecular Mechanisms of Hypoxic Exosome-Mediated M2 Macrophage Polarization
Research Reagent Solutions
Table 3: Essential Research Reagents for Investigating Hypoxic Exosomes
| Reagent Category | Specific Examples | Research Application | Key Findings Enabled |
|---|---|---|---|
| CD73 Inhibitors | PSB12379 (10 nM) | Elucidating adenosine-dependent polarization | CD73/adenosine axis essential for M2 polarization via A2A/A2B receptors [3] |
| Adenosine Receptor Antagonists | A2A antagonist (SCH58261), A2B antagonist (PSB1115) | Determining receptor specificity | Both A2A and A2B receptors required for optimal M2 polarization [3] |
| Pathway Inhibitors | AKT inhibitor (MK-2206), ERK inhibitor (SCH772984) | Mapping signaling pathways | AKT/ERK activation downstream of adenosine receptors [3] |
| HIF Stabilizers | Dimethyloxalylglycine (DMOG), CoCl₂ | Mimicking hypoxic conditions | Confirmed HIF role in exosome biogenesis and cargo sorting [88] [89] |
| Exosome Isolation Kits | Total Exosome Isolation Kit, TFF systems | Vesicle purification | High-purity exosome preparation for functional studies [3] |
| Characterization Antibodies | Anti-CD63/CD81/CD9, Anti-CD206/ CD163 | Phenotype validation | Confirmed exosome identity and M2 macrophage polarization [3] [4] |
Hypoxic priming represents a fundamental adaptive mechanism whereby MSCs significantly enhance the production and functional capacity of their exosomal secretions. Through coordinated HIF-dependent and independent pathways, hypoxia reprograms exosome biogenesis, leading to selective enrichment of pro-angiogenic and immunomodulatory cargo that orchestrates macrophage polarization toward the M2 phenotype. The CD73/adenosine axis emerges as a particularly crucial mechanism, working in concert with EDA-Fibronectin/TLR signaling and miRNA-mediated gene regulation to establish a robust anti-inflammatory program in recipient macrophages.
From a translational perspective, hypoxic priming offers promising strategies for enhancing the therapeutic efficacy of MSC-derived exosomes in regenerative medicine, inflammatory diseases, and cancer immunotherapy. However, several challenges remain, including standardization of hypoxic preconditioning protocols, comprehensive characterization of cargo alterations, and development of manufacturing processes that maintain consistent product quality. Future research should focus on elucidating the precise molecular mechanisms governing cargo selection under hypoxia, developing engineered exosomes with enhanced targeting specificity, and validating the therapeutic potential of hypoxic exosomes in advanced disease models. The integration of hypoxic priming methodologies with emerging exosome engineering approaches holds significant promise for developing next-generation cell-free therapies capable of precise immunomodulation and tissue repair.
Within the broader scope of research on how mesenchymal stem cell (MSC) exosomes modulate macrophage polarization to an M2 phenotype, the targeted engineering of specific microRNAs (miRNAs) presents a promising therapeutic strategy. This technical guide delves into the mechanistic roles of miR-21 and miR-16, two critical regulators carried by MSC-derived exosomes, in promoting M2-like macrophage polarization. We provide an in-depth analysis of their target pathways, quantitative data summaries, detailed experimental protocols for manipulating these miRNAs, and essential research tools required for advancing this field. The content is structured to serve researchers, scientists, and drug development professionals working at the intersection of immunomodulation and regenerative medicine.
Mesenchymal stem cell (MSC)-derived exosomes have emerged as potent mediators of immunomodulation, with their ability to promote anti-inflammatory M2 macrophage polarization being a key mechanism of action [94] [95]. These nano-sized extracellular vesicles (30-150 nm) facilitate intercellular communication by transferring bioactive cargo, including proteins, lipids, and nucleic acids, from donor MSCs to recipient immune cells such as macrophages [96] [3]. The therapeutic efficacy of MSC exosomes has been demonstrated across various disease models, including myocardial infarction, systemic lupus erythematosus, and sepsis, where their promotion of M2 polarization contributes to inflammation resolution and tissue repair [94] [95] [97].
The polarization process involves shifting macrophages from a pro-inflammatory M1 state to an anti-inflammatory M2 state characterized by specific surface markers and cytokine profiles [3]. M2 macrophages express signature markers including CD206, CD163, Arg-1 (arginase-1), and IL-10, which distinguish them from M1 macrophages that typically express iNOS, CD86, and pro-inflammatory cytokines like TNF-α and IL-6 [95] [3]. MSC exosomes modulate this polarization through multiple mechanisms, including the delivery of regulatory miRNAs such as miR-21-5p and miR-16, which post-transcriptionally regulate gene expression in target macrophages [94] [95].
Mechanistic Insights: How miR-21 and miR-16 Drive M2 Polarization
miR-21-5p Signaling Pathways
miR-21-5p has been identified as a principal mediator of MSC exosome-induced M2 polarization. The mechanism involves direct targeting of negative regulators in the polarization pathway:
- PDCD4/NF-κB Axis: miR-21 directly binds to the 3' untranslated region (UTR) of PDCD4 (programmed cell death 4) mRNA, leading to its translational repression. This downregulation relieves PDCD4-mediated inhibition of the NF-κB pathway, facilitating M2 polarization [97]. In sepsis models, this mechanism has been shown to reduce inflammation and ameliorate disease severity [97].
- PTEN/AKT Pathway: Simultaneously, miR-21 targets PTEN (phosphatase and tensin homolog), a negative regulator of the AKT signaling pathway. PTEN suppression activates AKT signaling, which promotes M2 polarization and enhances tissue repair capabilities [97].
The following diagram illustrates the coordinated signaling mechanism by which exosomal miR-21 promotes M2 macrophage polarization:
miR-16 Signaling Pathways
miR-16 works synergistically with miR-21 to reinforce M2 polarization through distinct molecular targets:
- PDCD4 Regulation: Similar to miR-21, miR-16 also targets PDCD4, creating a synergistic effect that amplifies M2 polarization signals [95]. This dual regulation ensures robust suppression of this key negative regulator.
- Coordinated Action: Studies using MSC exosomes depleted of both miR-16 and miR-21 demonstrated attenuated anti-inflammatory polarization effects, confirming their cooperative role in modulating macrophage phenotype [95].
The coordinated action of these miRNAs enables precise control over macrophage polarization, making them ideal targets for therapeutic engineering.
The effects of miR-21 and miR-16 on macrophage polarization markers and functional outcomes have been quantified across multiple experimental systems. The tables below summarize key findings from peer-reviewed studies.
Table 1: Effects of miRNA Manipulation on Macrophage Polarization Markers
| miRNA | Experimental System | M1 Marker Change | M2 Marker Change | Significance | Citation |
|---|---|---|---|---|---|
| miR-21-5p | Mouse sepsis model; IL-1β-primed MSC exosomes | iNOS ↓, IL-1β ↓, IL-6 ↓ | Arg-1 ↑, IL-10 ↑ | P<0.05 | [94] [97] |
| miR-21-5p | RAW264.7 cells + LPS | CD86 ↓ | CD206 ↑, B7H4 ↑ | P<0.05 | [95] |
| miR-16 | MRL/lpr mouse kidney macrophages | - | CD206 ↑, B7H4 ↑, CD138 ↑, Arg-1 ↑ | P<0.05 | [95] |
| miR-21 + miR-16 | MSC exosomes (miR-16/21 depleted) | - | CD206 ↓, B7H4 ↓, Arg-1 ↓ | Attenuated polarization | [95] |
Table 2: Functional Outcomes of miRNA-Induced M2 Polarization
| Functional Assay | System | Key Findings | Impact | Citation |
|---|---|---|---|---|
| Cytokine Secretion | Mouse myocardial tissue | IL-6 ↓, IL-10 ↑ | Reduced inflammation | [94] |
| Efferocytosis | MRL/lpr mouse macrophages | Phagocytic capacity ↑ | Enhanced apoptotic debris clearance | [95] |
| ROS Production | MRL/lpr mouse macrophages | ROS levels ↓ | Reduced oxidative stress | [95] |
| Treg Recruitment | Pristane-induced lupus model | IL-17+ Treg cells ↑ | Enhanced immunoregulation | [95] |
| Cardiac Function | Mouse myocardial I/R injury | EF ↑, FS ↑ | Improved heart repair | [94] |
Detailed Experimental Protocols
Generating miRNA-Modified MSC Exosomes
This protocol outlines the process for creating MSC exosomes with engineered miRNA content for macrophage polarization studies:
MSC Culture and Activation:
- Culture human MSCs (e.g., E1-MYC 16.3 line) in DMEM supplemented with 10% exosome-depleted FBS, 1% non-essential amino acids, 1% glutamine, and 1% insulin-transferrin-selenium-X [3].
- For enhanced miRNA expression, prime MSCs with 10 ng/mL IL-1β for 12 hours to upregulate immunomodulatory factors (COX-2, IDO-1, TSG-6) [97].
- Validate MSC surface markers (CD29, CD44, CD73, CD90, CD105 positive; CD11b, CD14, CD19, CD34 negative) using flow cytometry [97].
miRNA Transfection:
- Transfect MSCs at 60-70% confluency using Lipofectamine 2000 with:
- Incubate for 48 hours, then replace with fresh, serum-free, chemically-defined medium for exosome production [3].
Exosome Isolation and Purification:
- Collect conditioned medium after 72 hours and centrifuge at 2,000 × g for 30 minutes to remove cells and debris.
- Concentrate medium using tangential flow filtration with a 100 kDa molecular weight cut-off membrane [3].
- Isolate exosomes using size-exclusion chromatography (e.g., qEV columns) or precipitation kits (e.g., ExoQuick) [95] [3].
- Validate exosome isolation by nanoparticle tracking analysis (size: 50-150 nm; concentration: >10^10 particles/mL) and Western blot for markers (CD9, CD63, TSG101, Alix) [98] [3].
The following workflow diagram summarizes the complete process from MSC culture to exosome characterization:
Macrophage Polarization Assays
Comprehensive assessment of miRNA-induced M2 polarization requires multiple validation methods:
Macrophage Culture and Treatment:
- Differentiate human monocyte THP-1 cells with 100 ng/mL PMA for 48 hours or isolate primary macrophages from mouse peritoneal cavity or bone marrow using M-CSF (40 ng/mL) for 7-9 days [94] [3].
- Treat macrophages with isolated exosomes (10 μg/mL) for 24-48 hours in the presence of LPS (100 ng/mL) to simulate inflammatory conditions [3].
Polarization Validation:
- Flow Cytometry: Analyze surface markers using anti-CD86 (M1) and anti-CD206 (M2) antibodies. Calculate polarization ratio as CD206+ vs. CD86+ populations [95].
- qPCR Analysis: Extract total RNA and measure expression of:
- Western Blot: Detect intracellular proteins iNOS (M1) and Arg-1 (M2) using specific antibodies [95].
- ELISA: Quantify secreted cytokines in supernatant: IL-6, IL-12 (M1) vs. IL-10, TGF-β (M2) [94] [95].
Functional Assays:
- Efferocytosis: Incubate macrophages with pHrodo Red-labeled apoptotic cells for 2.5 hours and quantify phagocytosis by flow cytometry [95].
- ROS Production: Use Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit to measure reactive oxygen species in culture supernatant [95].
- Treg Recruitment: Co-culture polarized macrophages with T cells and analyze Foxp3+ IL-17+ regulatory T cell population by flow cytometry [95].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for MSC Exosome and Macrophage Polarization Studies
| Reagent/Cell Line | Specifications | Application | Supplier Examples |
|---|---|---|---|
| MSC Lines | E1-MYC 16.3 (human ESC-derived), C57BL/6 mouse BM-MSC | Consistent exosome production | Cyagen, ATCC |
| Macrophage Lines | RAW264.7 (mouse), THP-1 (human), primary rat/mouse macrophages | Polarization assays | Ningbo Mingzhou, ATCC |
| miRNA Modulators | miR-21-5p mimics/inhibitors, miR-16 mimics/inhibitors, scrambled controls | miRNA engineering | Dharmacon, Sigma-Aldrich |
| Exosome Isolation Kits | qEV columns, ExoQuick-TC, Total Exosome Isolation | Exosome purification | iZON, System Biosciences |
| Characterization Antibodies | Anti-CD9, CD63, TSG101, Alix (exosome markers); CD86, CD206, F4/80 (macrophage markers) | Validation assays | BioLegend, Abcam |
| Cell Culture Media | DMEM, RPMI-1640 with exosome-depleted FBS | Cell maintenance | Thermo Fisher, Cytiva |
| Cytokines & Factors | IL-1β, M-CSF, LPS, IL-4, IL-13 | Cell priming/polarization | PeproTech, R&D Systems |
| Assay Kits | Amplex Red ROS, PiColorLock Gold Phosphate Detection (CD73 activity) | Functional assays | Thermo Fisher, Innova Biosciences |
The targeted engineering of miR-21 and miR-16 in MSC-derived exosomes represents a sophisticated approach for controlling macrophage polarization with significant therapeutic potential. The mechanistic insights, experimental protocols, and research tools outlined in this technical guide provide a foundation for advancing this promising field. As research progresses, optimizing delivery systems for engineered exosomes, exploring synergistic miRNA combinations, and establishing standardized potency metrics will be crucial for translational applications. The ability to precisely manipulate macrophage phenotype through engineered exosomes holds promise for treating a wide range of inflammatory and autoimmune diseases where M2 macrophage polarization can promote resolution of inflammation and tissue repair.
The therapeutic efficacy of mesenchymal stem cell-derived exosomes (MSC-Exos) in modulating macrophage polarization to the M2 phenotype is increasingly recognized as a key mechanism in promoting tissue repair and resolving inflammation. This whitepaper establishes a comprehensive quality control framework identifying two critical quality attributes (CQAs) essential for predicting and ensuring MSC-Exos potency: CD73 ecto-5'-nucleotidase activity and specific microRNA (miRNA) cargo profiles. We present validated experimental protocols for quantifying these CQAs, including detailed methodologies for CD73 enzymatic activity assessment via high-pressure liquid chromatography (HPLC) and miRNA content analysis through quantitative PCR (qPCR) and sequencing. Furthermore, we integrate these assays into a holistic potency testing strategy that correlates CQA measurements with functional outcomes in macrophage polarization assays. This technical guide provides researchers and drug development professionals with standardized approaches for quality control, ultimately accelerating the clinical translation of MSC-Exos as advanced therapy medicinal products.
Mesenchymal stem cell-derived exosomes have emerged as promising cell-free therapeutics for a range of inflammatory and autoimmune conditions, with their ability to modulate macrophage polarization representing a primary mechanism of action [99]. These nanoscale extracellular vesicles (30-150 nm in diameter) transfer bioactive molecules—including proteins, lipids, and nucleic acids—from MSCs to recipient immune cells, thereby reprogramming their function [100]. Specifically, MSC-Exos have demonstrated a remarkable capacity to shift macrophage polarization from the pro-inflammatory M1 phenotype toward the anti-inflammatory, pro-reparative M2 phenotype, making them particularly valuable for treating conditions characterized by excessive inflammation [10].
The transition from MSC-Exos as research tools to standardized therapeutic products requires rigorous quality control measures. While MSC-Exos are defined by surface tetraspanins (CD9, CD63, CD81) and parental cell markers (CD73, CD90, CD44) [99], these characteristics alone are insufficient potency indicators. This whitepaper establishes CD73 activity and specific miRNA content as CQAs directly linked to the mechanism of action in macrophage polarization, providing a scientific framework for their assessment and implementation in quality control systems.
Biological Basis: How MSC Exosomes Drive M2 Polarization
Dual Pathway Model of Macrophage Reprogramming
MSC-Exos modulate macrophage polarization through two primary, interconnected mechanisms: CD73-mediated adenosine signaling and miRNA-directed gene regulation. The CD73 pathway operates extracellularly, while miRNA cargo functions intracellularly within recipient macrophages, creating a coordinated mechanism that effectively promotes the M2 phenotype.
Table 1: Key Mechanisms of MSC-Exos in M2 Macrophage Polarization
| Mechanism | Key Components | Biological Effect | Functional Outcome |
|---|---|---|---|
| CD73/Adenosine Pathway | CD39/CD73 enzymes, AMP substrate, Adenosine, A2a receptor | Immunosuppressive adenosine signaling, cAMP elevation in macrophages | Inhibition of pro-inflammatory pathways, Promotion of M2 genetic programs [101] [102] |
| miRNA Transfer | miR-146a, miR-125a-3p, miR-223, miR-21-5p | Post-transcriptional regulation of inflammatory genes, Modulation of signaling pathways (NF-κB, STAT) | Suppression of inflammatory activation, Enhanced M2-associated functions [99] [100] [103] |
| Surface Protein Interaction | Tetraspanins, Adhesion molecules, Co-stimulatory molecules | Direct receptor engagement, Activation of anti-inflammatory signaling cascades | Synergistic enhancement of polarization signals [100] |
CD73-Mediated Adenosine Immunomodulation
CD73 (ecto-5'-nucleotidase) is a glycosylphosphatidylinositol (GPI)-anchored membrane protein highly expressed on MSC-Exos that catalyzes the conversion of AMP to adenosine [102]. This enzymatic activity initiates a potent immunosuppressive cascade: extracellular adenosine engages A2A and A2B receptors on macrophages, triggering intracellular signaling events that suppress pro-inflammatory mediator production while promoting M2-associated gene expression [101] [102]. The CD39/CD73 axis effectively metabolizes pro-inflammatory extracellular ATP into immunosuppressive adenosine, representing a crucial metabolic switch that shapes the immune microenvironment [104] [102].
miRNA-Directed Phenotypic Switching
MSC-Exos carry a specific repertoire of miRNAs that directly target molecular networks governing macrophage polarization. For instance, miR-146a dampens NF-κB signaling, a primary driver of M1 polarization, while miR-125a-3p modulates inflammatory gene expression and supports M2 characteristics [99] [103]. These miRNAs are selectively packaged into exosomes and transferred to macrophages, where they fine-tune the transcriptome to establish and maintain the M2 phenotype, facilitating tissue repair and inflammation resolution [10] [100].
Diagram 1: MSC exosome M2 polarization mechanisms
Establishing Critical Quality Attributes (CQAs)
CD73 Enzymatic Activity as a CQA
CD73 activity serves as a direct functional CQA because it represents the exosomes' capacity to generate immunosuppressive adenosine in the tissue microenvironment. Quantitative assessment of CD73 activity provides a measurable indicator of this specific mechanism of action [104]. Research demonstrates that CD73-positive MSC-Exos effectively metabolize AMP to adenosine, while CD73-negative vesicles lack this capacity [104] [101]. Furthermore, CD73 activity can be directly inhibited using selective antagonists like PSB12379 or specific neutralizing antibodies, enabling demonstration of assay specificity and mechanism validation [104].
miRNA Content as a CQA
The miRNA cargo of MSC-Exos constitutes a second essential CQA, representing the exosomes' capacity to directly reprogram macrophage gene expression networks. Specific miRNA signatures correlate with functional efficacy in promoting M2 polarization, with particular miRNAs demonstrating direct roles in this process [99] [10]. For instance, miR-146a and miR-125a-3p have been identified as key mediators of MSC-Exos effects on macrophage polarization, making them candidate markers for quality control [99]. The stability of exosomal miRNAs—protected from degradation by the lipid bilayer—makes them particularly suitable as measurable quality attributes [103].
Table 2: miRNA CQA Candidates for M2 Macrophage Polarization
| miRNA | Expression in MSC-Exos | Target Pathways in Macrophages | Documented Effect on Polarization |
|---|---|---|---|
| miR-146a | Enriched | NF-κB signaling, IRAK1, TRAF6 | Suppresses M1 polarization, promotes M2 phenotype [99] [100] |
| miR-125a-3p | Present | Inflammatory gene networks | Suppresses T cell activity, modulates Th1/Th2 balance [99] |
| miR-21-5p | Detected | PDCD4, IL-12 expression | Inhibits dendritic cell maturation, anti-inflammatory effects [99] |
| miR-223 | Transferable | NLRP3 inflammasome | Dampens inflammatory responses, promotes M2 characteristics [53] |
| miR-155-5p | Variable | SHIP1, SOCS1 | Context-dependent roles in inflammation regulation [99] |
Experimental Protocols for CQA Assessment
CD73 Activity Assay Protocol
The following protocol adapts the methodology successfully applied to breast cancer-derived small extracellular vesicles for assessment of CD73 activity in MSC-Exos [104].
Sample Preparation
- Exosome Isolation: Isolate MSC-Exos from cell culture supernatant using differential ultracentrifugation or size-exclusion chromatography. Validate exosome identity through nanoparticle tracking analysis (NTA), transmission electron microscopy (TEM), and western blotting for surface markers (CD9, CD63, CD81, CD73) [101].
- Protein Quantification: Determine exosomal protein concentration using a BCA protein assay kit to standardize input across experimental conditions [101].
- Inhibitor Controls: Prepare separate aliquots with CD73 inhibitor PSB12379 (10µM) or anti-CD73 neutralizing antibodies to demonstrate assay specificity.
HPLC-Based CD73 Activity Measurement
- Reaction Setup: Incubate MSC-Exos (50µg protein) with the fluorescent substrate N6-etheno-AMP (100µM) in reaction buffer (50mM Tris-HCl, pH 7.4, 5mM MgCl₂) for 60 minutes at 37°C [104].
- Reaction Termination: Stop enzymatic activity by heating to 95°C for 5 minutes.
- Product Separation: Analyze samples using reverse-phase HPLC with fluorescence detection (excitation λ=230nm, emission λ=410nm) to separate and quantify the substrate (N6-etheno-AMP) from product (N6-etheno-adenosine) [104].
- Quantification: Calculate CD73 activity as the rate of N6-etheno-adenosine formation normalized to exosomal protein content and reaction time (pmol/µg/min).
Diagram 2: CD73 activity assay workflow
Data Interpretation and Quality Standards
Establish acceptance criteria for CD73 activity based on correlation with functional potency in macrophage polarization assays. CD73 activity should be significantly reduced (≥80% inhibition) in samples treated with CD73-specific inhibitors, confirming assay specificity [104].
miRNA Cargo Analysis Protocol
Comprehensive miRNA profiling ensures consistent cargo in MSC-Exos therapeutic products. This protocol integrates established methodologies from multiple studies [99] [10] [103].
RNA Isolation and Quality Control
- RNA Extraction: Isolate total RNA from MSC-Exos (100-200μg exosomal protein) using phenol-chloroform extraction (TRIzol/LS) combined with silica membrane purification columns. Incorporate synthetic spike-in miRNAs (e.g., cel-miR-39) during lysis to monitor extraction efficiency.
- Quality Assessment: Evaluate RNA quantity and purity using spectrophotometry (NanoDrop) or fluorometry (Qubit). Verify the absence of cellular RNA contamination by assessing the 18S/28S ribosomal RNA ratio—exosomal preparations should show minimal ribosomal peaks.
miRNA Quantification and Profiling
- Reverse Transcription: Convert RNA to cDNA using stem-loop reverse transcription primers specifically designed for miRNAs of interest (e.g., miR-146a, miR-125a-3p, miR-223).
- qPCR Analysis: Perform quantitative PCR using miRNA-specific TaqMan assays. Normalize cycle threshold (Ct) values using spiked-in controls and stable reference miRNAs (e.g., miR-16-5p, miR-26a-5p) that demonstrate consistent expression across MSC-Exos batches.
- Next-Generation Sequencing: For comprehensive profiling, prepare miRNA sequencing libraries using adaptor ligation methods. Sequence on appropriate platforms (Illumina) to obtain unbiased miRNA expression profiles.
Data Analysis and Acceptance Criteria
Calculate relative expression of target miRNAs using the 2^(-ΔΔCt) method for qPCR data. Process sequencing data through standardized pipelines (alignment, quantification, differential expression). Establish acceptable ranges for key polarization-associated miRNAs based on correlation with functional potency in macrophage polarization assays.
Functional Correlation with Macrophage Polarization
To validate CD73 activity and miRNA content as meaningful CQAs, establish correlation with functional outcomes using robust macrophage polarization assays.
Macrophage Polarization Potency Assay
- Macrophage Culture: Differentiate human monocyte-derived macrophages (from primary monocytes or THP-1 cell line) with GM-CSF (for M1 bias) or M-CSF (for M2 bias).
- Exosome Treatment: Treat M1-biased macrophages with standardized MSC-Exos doses (e.g., 50-100μg exosomal protein/mL) for 24-48 hours.
- Phenotype Assessment:
- Surface Markers: Analyze M2 markers (CD206, CD163) and M1 markers (CD86, CD80) using flow cytometry.
- Cytokine Secretion: Quantify M2-associated cytokines (IL-10, TGF-β) and M1-associated cytokines (TNF-α, IL-12, IL-6) using ELISA or multiplex immunoassays.
- Gene Expression: Measure M2-associated genes (ARG1, MRC1, FIZZ1) and M1-associated genes (NOS2, IL1B, CXCL10) using qPCR.
Establishing Correlation with CQAs
Correlate CD73 activity measurements and specific miRNA levels with quantitative measures of M2 polarization (e.g., CD206 mean fluorescence intensity, IL-10 secretion). Establish minimum CD73 activity thresholds and miRNA expression ranges that predict functional potency.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for CQA Assessment
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| CD73 Activity Assay | N6-etheno-AMP substrate, PSB12379 inhibitor, Anti-CD73 neutralizing antibodies | CD73 enzymatic activity quantification, Assay specificity controls | HPLC system with fluorescence detection required [104] |
| Exosome Isolation | Differential ultracentrifugation protocol, Size-exclusion chromatography columns, Commercial isolation kits | MSC-Exos purification from conditioned media | Maintain cold temperature throughout isolation [101] |
| Characterization | CD9/CD63/CD81 antibodies, CD73 antibodies, Nanoparticle Tracking Analyzer, Transmission Electron Microscope | Exosome identification, quantification, and morphology | Multi-method characterization recommended [99] [101] |
| miRNA Analysis | TRIzol LS reagent, miRNA sequencing kits, Stem-loop RT primers, TaqMan miRNA assays | Comprehensive miRNA profiling, Targeted miRNA quantification | Include spike-in controls for normalization [103] |
| Cell Culture | Primary human monocytes, THP-1 cell line, M-CSF, GM-CSF, Polarizing cytokines (IFN-γ, IL-4) | Functional macrophage polarization assays | Use consistent cell sources between experiments [100] |
Quality Control Implementation Strategy
Implementing CD73 activity and miRNA content as CQAs requires integration into a comprehensive quality control system throughout the therapeutic development process.
Release Testing Specifications
Establish specification ranges for CQAs based on extensive batch data correlation with functional potency. For example:
- CD73 Activity: ≥ 50 pmol/μg/min (or batch-specific relative percentage)
- miRNA Signature: Expression levels of key miRNAs (e.g., miR-146a, miR-125a-3p) within 2-fold of reference therapeutic batch
- Potency Correlation: Demonstration of significant M2 polarization capacity in macrophage assays
Stability Monitoring
Include CQA assessment in stability studies to ensure maintenance of therapeutic potency throughout product shelf life. Monitor CD73 activity and miRNA integrity at predetermined time points under recommended storage conditions.
The establishment of CD73 enzymatic activity and specific miRNA content as critical quality attributes represents a significant advancement in the quality control of MSC-Exos therapeutics targeting macrophage polarization. The experimental protocols outlined in this whitepaper provide standardized methodologies for quantifying these CQAs, enabling researchers and drug development professionals to ensure consistent product quality and predictable therapeutic outcomes. Implementation of this comprehensive quality framework will facilitate the clinical translation of MSC-Exos as robust, well-characterized therapeutic products for inflammatory diseases and tissue repair applications.
Evaluating Efficacy and Safety: Analytical Methods and Comparative Profiles
Within the broader research on how mesenchymal stem cell (MSC) exosomes modulate macrophage polarization to the M2 phenotype, functional validation is a critical step. This guide details the core experimental methodologies used to confirm the biological activity of MSC exosomes, focusing on three pivotal processes: their enhancement of macrophage efferocytosis (the clearance of apoptotic cells), their role in recruiting regulatory T (Treg) cells, and the techniques for in vivo tracking of exosome fate. Mastering these assays is essential for researchers and drug development professionals aiming to provide mechanistic proof for the therapeutic immunomodulation claims of exosome-based therapies.
Efferocytosis Assays: Quantifying Apoptotic Cell Clearance
A key functional outcome of MSC-exosome-induced M2 macrophage polarization is the enhanced ability to clear apoptotic cells, a process known as efferocytosis. This is typically measured using flow cytometry-based phagocytosis assays.
Experimental Protocol for Efferocytosis Assay
The following protocol, adapted from a study on systemic lupus erythematosus (SLE) nephritis, outlines the key steps for quantifying macrophage efferocytosis [105].
- Induction of Apoptosis: Mouse kidney epithelial cells (TCMK-1 cell line) are incubated at 45°C for 10 minutes, followed by 4 hours of culture under standard conditions to induce apoptosis. The percentage of apoptotic cells should be confirmed by flow cytometry (e.g., using Annexin V/PI staining) [105].
- Fluorescent Labeling of Apoptotic Cells: The induced apoptotic cells are washed with phosphate-buffered saline (PBS) twice and then labeled with pHrodo Red (or a similar pH-sensitive fluorescent dye). pHrodo Red is ideal because its fluorescence increases dramatically in the acidic environment of phagolysosomes, minimizing background signal from non-internalized cells [105].
- Co-culture and Phagocytosis: Labeled apoptotic cells (1x10^6) are added to a culture of macrophages (1x10^6) that have been pre-treated with MSC exosomes or a control. The co-culture is incubated for 2.5 hours at 37°C to allow for phagocytosis [105].
- Cell Harvest and Staining: After incubation, macrophages are washed thoroughly with ice-cold D-Hanks solution to remove any non-phagocytosed apoptotic cells. The macrophages are then stained with a FITC-conjugated anti-mouse CD11b antibody (or another macrophage-specific surface marker like F4/80) for 45 minutes at 4°C [105].
- Flow Cytometry Analysis: The cells are analyzed by flow cytometry. Macrophages are gated as CD11b-positive (FITC+) cells, and efferocytic activity is quantified by measuring the percentage of CD11b+ macrophages that are also pHrodo Red-positive [105].
Key Signaling Pathways in Exosome-Enhanced Efferocytosis
Mechanistic studies go beyond quantification to elucidate the signaling pathways by which MSC exosomes enhance efferocytosis. Proteomic profiling of MSC-exosomes has identified Growth Arrest-Specific 6 (GAS6) as an enriched protein [106]. GAS6 is a bridging molecule that facilitates efferocytosis by linking phosphatidylserine on apoptotic cells to the MerTK (Myeloid-epithelial-reproductive tyrosine kinase) receptor on macrophages.
The proposed signaling cascade, as identified in a model of hepatic ischemia-reperfusion injury, is as follows [106]:
- MSC-exosome-derived GAS6 activates MerTK on macrophages.
- This activation triggers phosphorylation of the ERK (Extracellular signal-Regulated Kinase) pathway.
- Activated ERK upregulates Cyclooxygenase-2 (COX2).
- The resultant signaling enhances the efficiency of efferocytosis.
This pathway can be validated using inhibitors, such as UNC2025 (a MerTK small molecule inhibitor), which has been shown to attenuate the protective effects of MSC exosomes on macrophage efferocytosis and subsequent liver injury [106].
Quantitative Data from Efferocytosis Assays
The functional outcome of MSC exosome treatment on macrophage efferocytosis can be quantified as shown in the table below, which summarizes typical experimental data.
Table 1: Summary of Quantitative Data from Efferocytosis Assays
| Experimental Model | Exosome Treatment | Key Measured Parameters | Experimental Outcome | Citation |
|---|---|---|---|---|
| In vitro assay with macrophages from MRL/lpr mouse kidney | BM-MSC exosomes | Percentage of macrophages performing efferocytosis (pHrodo Red+ CD11b+ cells) | Significant upregulation of efferocytosis activity | [105] |
| In vivo hepatic ischemia-reperfusion injury (HIRI) mouse model | BM-MSC exosomes (systemic administration) | TUNEL+ apoptotic cells in liver tissue; Efferocytosis efficiency via imaging | Significant reduction of apoptotic cells; enhanced clearance | [106] |
Treg Cell Recruitment and Induction Assays
The immunomodulatory function of MSC-exosome-educated macrophages extends to the adaptive immune system, particularly in promoting regulatory T cells (Tregs). This can be assessed through co-culture systems and flow cytometry.
Experimental Protocol for Treg Cell Analysis
The following protocol is used to evaluate the role of exosome-primed macrophages in Treg recruitment and induction, both in vitro and in vivo [105] [107].
In Vitro T Cell Proliferation and Treg Induction:
- Macrophage Education: Macrophages are treated with MSC exosomes or a control for 24-48 hours to induce an M2-like, anti-inflammatory phenotype [105] [107].
- Co-culture with T Cells: Educated macrophages are co-cultured with CD4+ T cells isolated from spleen or peripheral blood. To assess T cell proliferation, the T cells can be pre-labeled with Carboxyfluorescein succinimidyl ester (CFSE) or their proliferation measured using a Bromodeoxyuridine (BrdU) assay [107].
- Flow Cytometry for Tregs: After co-culture, cells are harvested, blocked, and stained for Treg markers. A standard staining panel includes:
- Analysis is performed by flow cytometry, gating on CD3+CD4+ T cells and then determining the percentage that are Foxp3+.
In Vivo Treg Analysis:
- Disease Model Treatment: An animal disease model (e.g., pristane-induced lupus nephritis or autoimmune dacryoadenitis) is treated with MSC exosomes or a control via injection (e.g., intravenous or subconjunctival) [105] [107].
- Cell Isolation and Staining: After a predetermined period, spleens or target organs (e.g., lacrimal glands, kidneys) are harvested and processed into single-cell suspensions.
- The cell suspension is then stained using the same antibody panel (CD3, CD4, Foxp3) and analyzed by flow cytometry to determine the proportion of Tregs in the target tissue or systemically [105] [107].
Key Molecular Mediators in Treg Induction
The mechanism by which MSC exosomes ultimately lead to Treg induction often involves the transfer of specific microRNAs (miRNAs) to macrophages. For example:
- miR-100-5p: Highly enriched in human umbilical cord MSC exosomes, it has been shown to promote M2 macrophage polarization. Knockdown of miR-100-5p in exosomes blunted their ability to induce M2 polarization and subsequently attenuated Treg expansion [107].
- miR-16 and miR-21: Found in bone marrow MSC exosomes, these miRNAs target PDCD4 and PTEN in macrophages, respectively, to drive anti-inflammatory polarization. Depletion of these miRNAs attenuated exosome-induced Treg recruitment in a lupus nephritis model [105].
Quantitative Data on Treg Recruitment
The impact of MSC exosome treatment on Treg populations is summarized in the table below.
Table 2: Summary of Quantitative Data on Treg Cell Induction and Recruitment
| Experimental Model | Exosome Treatment | Key Measured Parameters | Experimental Outcome | Citation |
|---|---|---|---|---|
| In vitro macrophage-CD4+ T cell co-culture | hUC-MSC-sEVs | Percentage of Foxp3+ Tregs among CD4+ T cells; T cell proliferation (BrdU assay) | Significant increase in Treg proportion; inhibition of CD4+ T cell proliferation | [107] |
| In vivo pristane-induced murine lupus nephritis | BM-MSC exosomes (injection) | Proportion of IL-17+ Treg cells in the disease model | Significant increase in Treg production | [105] |
| In vivo rabbit autoimmune dacryoadenitis | hUC-MSC-sEVs (subconjunctival injection) | Proportion of Foxp3+ Tregs in lacrimal glands | Significant elevation of local Tregs, correlated with improved clinical scores | [107] |
In Vivo Tracking of Exosome Fate
Understanding the biodistribution and pharmacokinetics of administered MSC exosomes is crucial for validating their therapeutic potential and optimizing delivery routes. Multiple imaging modalities are employed for this purpose, each with advantages and limitations.
Experimental Workflow for In Vivo Exosome Tracking
The general process for tracking exosomes in live animals involves labeling, administration, and imaging.
Labeling Strategies and Imaging Modalities
A critical choice in tracking studies is the labeling method, which dictates the imaging modality used.
Table 3: Key Modalities for In Vivo Exosome Imaging
| Imaging Modality | Labeling Strategy | Representative Probes | Key Advantages | Key Limitations | Citation |
|---|---|---|---|---|---|
| Fluorescence Imaging | Lipophilic dyes; Genetic reporters | DiR, Cy5/Cy7, CD63-GFP | High sensitivity; real-time imaging; relatively simple | Shallow tissue penetration; low resolution; potential dye aggregation | [108] [109] [110] |
| Bioluminescence Imaging (BLI) | Genetic fusion of luciferase to exosome proteins | CD63-NanoLuc, RLuc | Very high sensitivity; low background; quantitative | Signal loss with tissue depth; surface-weighted signals | [108] [109] |
| Positron Emission Tomography (PET) | Labeling with positron-emitting radionuclides | ^89^Zr-DFO, ^64^Cu-NOTA | Extremely high sensitivity; excellent for quantification; deep tissue penetration | Short half-life of tracers; requires cyclotron | [108] |
| Magnetic Resonance Imaging (MRI) | Loading with contrast agents | SPIONs (Superparamagnetic iron oxide nanoparticles) | High spatial resolution; deep penetration; no radiation | Low sensitivity; high cost; long scan times | [108] [110] |
Application Example: In a hepatic ischemia-reperfusion injury (HIRI) mouse model, MSC exosomes labeled with Cy5.5 (a NIR dye) were administered via tail vein. In vivo imaging system (IVIS) analysis at 6 hours post-injection showed that the fluorescence signal predominantly accumulated in the injured liver. Subsequent immunofluorescence staining of liver tissue confirmed co-localization of the Cy5.5 signal with F4/80+ macrophages, demonstrating targeted delivery to hepatic macrophages [106].
The Scientist's Toolkit: Essential Research Reagents
The following table catalogs key reagents and their functions essential for conducting the functional validation assays described in this guide.
Table 4: Essential Research Reagents for Functional Validation of MSC Exosomes
| Reagent / Assay | Function / Application | Specific Example |
|---|---|---|
| pHrodo Red | pH-sensitive fluorescent dye for labeling apoptotic cells in efferocytosis assays; fluorescence increases upon phagolysosomal internalization. | Used in flow cytometry-based phagocytosis assays [105]. |
| Anti-CD11b / F4/80 Antibodies | Macrophage surface markers for identifying and gating on macrophage populations in flow cytometry. | Used to identify macrophages after co-culture with apoptotic cells [105]. |
| Anti-CD3 / CD4 / Foxp3 Antibodies | Antibody panel for identifying regulatory T cells (Tregs) via flow cytometry (requires intracellular staining for Foxp3). | Used to quantify Treg induction in co-culture systems and in vivo [105] [107]. |
| BrdU Assay / CFSE | Methods for measuring cell proliferation. BrdU is incorporated into DNA and detected by antibody, while CFSE dilution is measured by flow cytometry. | Used to assess suppression of CD4+ T cell proliferation by exosome-educated macrophages [107]. |
| Lipophilic Dyes (e.g., DiR, DiD) | Fluorescent dyes that incorporate into exosome lipid bilayers for in vivo and in vitro tracking. | DiR-labeled exosomes used to track biodistribution in osteosarcoma models [108] [109]. |
| NanoLuc Luciferase | Genetic reporter fused to exosome membrane proteins (e.g., CD63) for highly sensitive bioluminescence imaging (BLI). | CD63-NanoLuc reporter used for spatiotemporal tracking of endogenous exosome release in mice [108] [109]. |
| UNC2025 | A small molecule inhibitor of MerTK. Used to block the GAS6-MerTK signaling axis to validate its role in efferocytosis. | Partially abolished the protective effects of MSC exosomes on liver injury [106]. |
| miRNA Inhibitors | Oligonucleotides used to knock down specific microRNAs in parent MSCs. Validates the functional role of exosomal miRNAs. | Knockdown of miR-100-5p or miR-16/21 attenuated exosome-induced M2 polarization and Treg induction [105] [107]. |
Mesenchymal stem cell (MSC) therapy has emerged as a highly promising strategy in regenerative medicine due to the cells' self-renewal, pluripotency, and potent immunomodulatory properties [60]. Traditionally, the therapeutic potential of MSCs was attributed to their ability to engraft at injury sites and differentiate into specific cell types to replace damaged tissues. However, a paradigm shift has occurred with growing recognition that MSCs exert most of their therapeutic effects through paracrine signaling rather than direct cell replacement [60]. This revelation has spurred intense interest in the secreted bioactive molecules of MSCs, particularly extracellular vesicles (EVs) like exosomes, as potential cell-free therapeutic agents [111] [112].
Exosomes are nanoscale extracellular vesicles (typically 30-150 nm) secreted by nearly all cell types, including MSCs, and play critical roles in intercellular communication by transferring proteins, lipids, and nucleic acids to recipient cells [113] [29]. Within the context of immunomodulation, particularly macrophage polarization, both MSC whole cell therapies and MSC-derived exosomes have demonstrated significant potential, yet through distinct mechanisms and with different therapeutic profiles. This review provides a comprehensive technical comparison of these two approaches, with specific focus on their efficacy, mechanisms, and practical applications in polarizing macrophages toward the immunoregulatory M2 phenotype, a key process in resolving inflammation and promoting tissue repair [114] [45] [111].
Fundamental Comparison: Whole MSC Therapy vs. MSC-Derived Exosomes
Core Characteristics and Definitions
Whole MSC Therapies utilize living, metabolically active mesenchymal stem cells administered to patients. These cells are defined by three key criteria established by the International Society for Cellular Therapy (ISCT): (1) adherence to plastic under standard culture conditions; (2) expression of specific surface markers (CD73, CD90, CD105) while lacking hematopoietic markers; and (3) capacity for in vitro trilineage differentiation into osteoblasts, chondrocytes, and adipocytes [60]. These cells can be isolated from various tissues including bone marrow (BM-MSCs), adipose tissue (AD-MSCs), umbilical cord (UC-MSCs), and dental pulp [60]. Their therapeutic mechanism is multifaceted, involving direct cell-cell interactions, secretion of soluble factors, and release of extracellular vesicles including exosomes [113].
MSC-Derived Exosomes are a subset of extracellular vesicles of endocytic origin, typically ranging from 30-150 nm in size [113] [115]. They are generated within multivesicular bodies and released upon fusion with the plasma membrane. As therapeutic agents, they represent a cell-free approach that harnesses the paracrine activity of MSCs without administering viable cells. Their cargo includes proteins, lipids, mRNAs, and microRNAs that can modulate recipient cell behavior [111] [29]. Notably, their composition and therapeutic potential are significantly influenced by their cellular origin and production methods [115] [29].
Table 1: Fundamental Comparison of Whole MSC Therapy vs. MSC-Derived Exosomes
| Parameter | Whole MSC Therapy | MSC-Derived Exosomes |
|---|---|---|
| Nature of Agent | Living, metabolically active cells | Non-living, acellular nanoparticles |
| Size Range | 15-30 μm (cell diameter) | 30-150 nm (vesicle diameter) [115] |
| Key Mechanism | Direct differentiation, cell-cell contact, paracrine signaling (including exosome secretion) [113] | Cargo delivery (proteins, miRNAs, lipids) to recipient cells [111] |
| Primary Sources | Bone marrow, adipose tissue, umbilical cord, dental pulp [60] | Conditioned medium from MSC cultures [115] |
| Therapeutic Duration | Days to months (cells may persist) [113] | Hours to days (typically require sustained delivery systems) [114] |
| Manufacturing Complexity | High (requires strict viability control, sterility) | Moderate (challenges in scaling and purity) [113] |
Advantages and Clinical Considerations
Whole MSC therapies offer the advantage of a sustained therapeutic presence, as the living cells can potentially remain in the patient for extended periods (days to months), continuously secreting bioactive factors and exosomes in response to the local microenvironment [113]. However, they present challenges including potential immune reactions (though MSCs are generally considered immunoprivileged), risk of ectopic tissue formation, and complex logistical requirements for storage and administration [60].
MSC-derived exosomes present a cell-free alternative with several distinct advantages: reduced immunogenicity (lower risk of immune rejection), minimal risk of tumorigenicity (unlike whole cells, they cannot divide), ability to cross biological barriers more easily, and enhanced safety profile [111] [112]. Their primary challenges include rapid clearance from the body, limited retention at target sites, and difficulties in large-scale production of consistent, clinical-grade products [113] [114]. Storage stability also presents challenges, as exosomes are susceptible to degradation due to temperature fluctuations, which can cause them to break down, change size, or lose contents [113].
Molecular Mechanisms in Macrophage Polarization to M2 Phenotype
Key Signaling Pathways
Both whole MSCs and MSC-derived exosomes promote the polarization of macrophages from the pro-inflammatory M1 phenotype toward the immunoregulatory M2 phenotype, but they employ distinct molecular pathways to achieve this effect. Current research has elucidated several key mechanisms:
p38 MAPK Pathway Inhibition: Small extracellular vesicles derived from MSCs (MSC-sEVs) have been demonstrated to effectively inhibit the p38 MAPK signaling pathway in macrophages. Research on diabetic wound healing models showed that MSC-sEVs significantly decrease phosphorylation of both p38 and MAPKAPK2, with RNA sequencing identifying Mapk14 and Nfkbia as key regulators within this pathway [114]. This inhibition promotes a shift from M1 to M2 polarization, characterized by downregulation of iNOS and TNF-α while upregulating CD206 and IL-10 expression [114].
SHP2-STAT3 Pathway Activation: In models of heart transplantation, MSC-derived exosomes carrying soluble fibrinogen-like protein 2 (sFgl2) were found to activate the SHP2-STAT3 signaling pathway through engagement of the CD32b receptor on macrophages [45]. This pathway activation promotes M2 polarization while simultaneously inhibiting M1 polarization, resulting in reduced acute rejection of transplanted hearts. Blocking the CD32b receptor or inhibiting SHP2 activation effectively reversed this protective effect [45].
miRNA-Mediated Regulation: Exosomes carry various microRNAs that play crucial roles in regulating macrophage polarization. For instance, exosomes derived from hypoxia-preconditioned M2 macrophages were found to be enriched with miR-124-3p, which inhibits STAT3 expression in recipient cells and enhances therapeutic outcomes in osteoarthritis models [116]. Similarly, MSC-derived apoptotic vesicles (apoVs) carrying miR-191-5p promote M2 polarization by downregulating CDK6 protein expression and stabilizing mitochondrial membrane potential in macrophages [117].
Diagram 1: MSC Exosome Mechanisms in M2 Macrophage Polarization. This diagram illustrates the three primary molecular pathways through which MSC-derived exosomes promote macrophage polarization toward the immunoregulatory M2 phenotype.
Comparative Efficacy in Disease Models
Both whole MSC and MSC-exosome therapies have demonstrated efficacy in promoting M2 macrophage polarization across various disease models, though with different practical considerations:
Diabetic Wound Healing: In diabetic rat models, MSC-derived small extracellular vesicles (MSC-sEVs) delivered via GelMA microspheres enhanced M2 macrophage polarization and reduced inflammation, promoting wound closure within 28 days [114]. The sustained-release system addressed the challenge of rapid exosome clearance, highlighting a key consideration for exosome therapeutics.
Heart Transplantation: In mouse heart transplantation models, both MSC-derived exosomes and especially sFgl2-engineered exosomes significantly prolonged cardiac graft survival and reduced myocardial necrosis by promoting M2 macrophage infiltration while reducing CD4+ T cell presence [45]. The engineered exosomes demonstrated enhanced efficacy over natural exosomes.
Osteoarthritis: Exosomes derived from hypoxia-preconditioned M2 macrophages (which share similarities with MSC exosomes in their immunomodulatory functions) alleviated degeneration in knee osteoarthritis through the miR-124-3p/STAT3 axis, demonstrating enhanced effects compared to exosomes from normoxic conditions [116].
Early-Onset Preeclampsia (EOPE): MSC-derived apoptotic vesicles (apoVs) carrying miR-191-5p promoted M2 polarization in placental macrophages, enhancing trophoblast invasion and improving pregnancy outcomes in mouse models, including reduced blood pressure and decreased proteinuria [117].
Table 2: Comparative Efficacy in Preclinical Models of Macrophage-Mediated Diseases
| Disease Model | Whole MSC Therapy Effects | MSC-Exosome Therapy Effects | Key Mechanisms |
|---|---|---|---|
| Diabetic Foot Ulcers | Not specifically reported in search results | Enhanced wound closure, improved epidermal regeneration, reduced inflammation [114] | M2 polarization via p38 MAPK inhibition [114] |
| Heart Transplantation | Reduced acute rejection, modulated immune cell infiltration [60] | Prolonged graft survival, reduced myocardial necrosis, enhanced M2 infiltration [45] | SHP2-STAT3 activation via CD32b receptor [45] |
| Osteoarthritis | Chondroprotection, reduced inflammation [60] | Alleviated degeneration, enhanced cartilage repair [116] | miR-124-3p delivery and STAT3 inhibition [116] |
| Early-Onset Preeclampsia | Not specifically reported in search results | Improved pregnancy outcomes, enhanced trophoblast invasion [117] | miR-191-5p delivery, CDK6 downregulation [117] |
Experimental Protocols and Methodologies
Standard Protocol for MSC-sEV Isolation and Characterization
The isolation and characterization of MSC-derived small extracellular vesicles follows standardized methodologies critical for ensuring research reproducibility and therapeutic quality:
Cell Culture and Conditioning: Bone marrow-derived MSCs (BM-MSCs) are cultured in complete Dulbecco's Modified Eagle Medium (DMEM)/F12 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin [114]. For exosome production, cells at approximately 80% confluency are rinsed with PBS and cultured in pure DMEM/F12 for 48 hours to condition the medium [114]. Studies comparing culture media have found that Alpha Minimum Essential Medium (α-MEM) may yield higher particle concentrations compared to DMEM, though not statistically significant [115].
Exosome Isolation: Two primary methods are commonly employed:
- Ultracentrifugation (UC): The classical method involving sequential centrifugation steps: initial low-speed centrifugation (300 × g) to remove cells, followed by higher speeds (2000 × g) to remove dead cells and debris, then 10,000 × g to remove larger vesicles, and finally ultracentrifugation at 100,000 × g to pellet exosomes [114] [115].
- Tangential Flow Filtration (TFF): A more scalable method that demonstrates statistically higher particle yields compared to ultracentrifugation, making it more suitable for potential large-scale clinical production [115].
Exosome Characterization: Isolated vesicles must undergo comprehensive characterization:
- Nanoparticle Tracking Analysis (NTA): To determine particle size distribution and concentration using the ZetaView system [114] [115].
- Transmission Electron Microscopy (TEM): To visualize cup-shaped morphology characteristic of exosomes [114] [115].
- Western Blotting: To confirm presence of exosomal markers (CD9, CD63, TSG101) and absence of negative markers (calnexin) [115].
- Functional Uptake Assays: Using Dil-labeled MSC-sEVs co-cultured with bone marrow-derived macrophages (BMDMs) to confirm internalization, visualized via confocal microscopy [114].
Macrophage Polarization Assay Protocol
Macrophage Differentiation and Polarization:
- BMDMs Isolation: Bone marrow cells are isolated from rat leg bones, erythrocytes lysed, filtered through a 70 μm filter, and maintained in complete medium with 30 ng/mL M-CSF for 7 days to differentiate into BMDMs [114].
- Macrophage Identification: Confirmation via flow cytometry using antibodies against CD11b and CD68 [114].
- Polarization Assay: BMDMs are treated with 100 ng/mL LPS for 24 hours to induce M1 polarization, followed by treatment with 30 μg/mL MSC-sEVs for 48 hours to promote M2 polarization [114].
- Phenotype Analysis:
- Flow Cytometry: Using antibodies against M1 markers (CD86) and M2 markers (CD206) [45] [117].
- Gene Expression: RT-qPCR analysis of M1 markers (iNOS, TNF-α) and M2 markers (CD206, IL-10) [114].
- Cytokine Profiling: ELISA or multiplex assays to measure secretion of pro-inflammatory and anti-inflammatory cytokines [45].
Diagram 2: Experimental Workflow for MSC-sEV Isolation and Macrophage Polarization Assay. This diagram outlines the key methodological steps from MSC culture and exosome isolation through functional assessment of macrophage polarization.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful investigation of MSC exosomes and their effects on macrophage polarization requires specific reagents and methodological approaches. The following toolkit outlines essential materials and their applications in this research domain:
Table 3: Essential Research Reagents and Materials for MSC Exosome-Macrophage Studies
| Reagent/Material | Specific Examples | Application Purpose | Technical Notes |
|---|---|---|---|
| MSC Culture Media | α-MEM, DMEM/F12 [115] | Expansion of mesenchymal stem cells | α-MEM may yield higher particle concentrations [115] |
| Supplement | 10% FBS, 10% human platelet lysate (hPL) [115] | Cell growth and maintenance | For exosome production, switch to serum-free medium [114] |
| sEV Isolation Kits | Ultracentrifugation, Tangential Flow Filtration [115] | Isolation of small extracellular vesicles | TFF provides higher yield than UC [115] |
| Characterization Tools | NTA (ZetaView), TEM, Western Blot [114] [115] | Verification of sEV size, morphology, markers | Essential for confirming exosome identity (CD9, CD63, TSG101) [115] |
| Macrophage Differentiation | M-CSF (30 ng/mL) [114] | Generation of BMDMs from bone marrow | 7-day differentiation protocol [114] |
| Polarization Inducers | LPS (100 ng/mL) [114] | M1 macrophage induction | 24-hour treatment standard [114] |
| Flow Cytometry Antibodies | Anti-CD11b, CD68, CD86 (M1), CD206 (M2) [114] [45] | Macrophage identification and phenotyping | Critical for quantifying M1/M2 ratios [45] |
| sEV Labeling | Dil dye (20 μg/mL) [114] | Tracking sEV uptake by macrophages | Visualized via confocal microscopy [114] |
| Delivery Systems | GelMA microspheres [114] | Sustained release of sEVs in vivo | Enhances retention from 7 days [114] |
The comparative analysis of MSC exosomes versus whole cell therapies for immunomodulation reveals a complex landscape with distinct advantages for each approach. Whole MSC therapies offer the benefit of living, responsive cells that can dynamically adjust their secretory profile to the local microenvironment and potentially provide longer-term effects through engraftment [113] [60]. In contrast, MSC-derived exosomes present a cell-free alternative with enhanced safety profile, reduced immunogenicity, and more precise targeting capabilities [111] [112].
For macrophage polarization to the M2 phenotype specifically, MSC exosomes demonstrate potent efficacy through multiple well-defined molecular mechanisms, including p38 MAPK pathway inhibition, SHP2-STAT3 activation, and miRNA-mediated regulation [114] [45] [116]. Their nanoscale size facilitates tissue penetration and crossing of biological barriers, while their cargo of proteins, lipids, and nucleic acids allows for multi-faceted modulation of macrophage behavior.
Future research directions should focus on optimizing exosome production and delivery systems to overcome current limitations in scalability, stability, and target site retention [113] [29]. Engineering approaches, including preconditioning strategies (hypoxia, cytokine exposure), genetic modification of parent MSCs, and direct exosome loading, offer promising avenues for enhancing therapeutic efficacy [116] [29]. Additionally, advanced delivery systems such as hydrogels and microspheres show potential for prolonging exosome retention at target sites [114].
As the field progresses, standardized protocols for exosome isolation, characterization, and potency assessment will be crucial for translating these promising findings into clinically viable therapies. Both whole MSC and MSC-exosome therapies hold significant potential for modulating macrophage polarization in inflammatory and degenerative diseases, offering complementary approaches in the rapidly evolving landscape of immunomodulatory therapeutics.
Within the broader investigation of how mesenchymal stem cell (MSC) exosomes modulate macrophage polarization to the M2 phenotype, comparing the efficacy of exosomes from different cellular sources is a critical research question. Adipose-derived stem cell exosomes (ADSC-Exos) and bone marrow mesenchymal stem cell exosomes (BM-MSC-Exos) represent two of the most widely studied candidates for cell-free immunomodulatory therapy. Both carry a complex cargo of proteins, lipids, and nucleic acids that can reprogram macrophage function, but their relative potencies and mechanistic emphases differ. This whitepaper provides a technical comparison of ADSC-Exos and BM-MSC-Exos, synthesizing current research to guide scientists in selecting the appropriate exosome source for specific therapeutic applications targeting macrophage repolarization. The ability of these exosomes to shift macrophages from a pro-inflammatory M1 state to an anti-inflammatory, pro-repair M2 state is foundational to their therapeutic potential in inflammatory and degenerative diseases [4].
Comparative Efficacy: ADSC-Exos vs. BM-MSC-Exos
A direct comparative study of exosomes from ADSCs, BMSCs, and umbilical cord MSCs (UMSCs) provides the most clear-cut data on their relative efficacy. The findings indicate that while all MSC-derived exosomes are effective, they are not equivalent.
Table 1: Head-to-Head Comparison of ADSC-Exos and BM-MSC-Exos
| Parameter | ADSC-Exos | BM-MSC-Exos | Experimental Context |
|---|---|---|---|
| Anti-Inflammatory Efficacy | Moderate reduction of pro-inflammatory markers [118] | Superior suppression of pro-inflammatory markers (e.g., pp65) [118] | In vitro model of inflammation (IL-1β stimulation) [118] |
| Chondroprotective Effects | Enhanced expression of chondroprotective genes [118] | Superior enhancement of chondroprotective genes [118] | In vitro and ex vivo models of osteoarthritis [118] |
| Inhibition of Apoptosis | Inhibits chondrocyte apoptosis [118] | Displays superior efficacy in inhibiting chondrocyte apoptosis [118] | In vitro model of osteoarthritis [118] |
| Cell Migration | Markedly enhances chondrocyte motility [118] | Markedly enhances chondrocyte motility [118] | Chondrocyte migration assay [118] |
| Key Mechanistic Insight | Promotes tissue repair via activation of multiple pathways (e.g., Wnt/β-catenin, PI3K/Akt) [119] | Suppresses NF-κB and MAPK (p38, JNK, ERK) signaling pathways [118] | Western blot analysis of signaling pathways [118] [119] |
| Macrophage Polarization | Promotes M2-like polarization; exosomes from certain ADSCs can alleviate obesity via white adipose tissue beiging [4] | Promotes M2-like polarization; reduces M1 markers (IL-1β, TNF-α) [3] [120] | In vitro macrophage cultures and in vivo disease models [3] [120] [4] |
| Therapeutic Angiogenesis | Promotes angiogenesis, critical for wound healing [121] [119] | Promotes angiogenic tube formation in HUVECs [120] | HUVEC tube formation assay; in vivo wound healing and rotator cuff repair models [121] [120] |
Molecular Mechanisms of M2 Macrophage Polarization
The immunomodulatory effects of MSC-derived exosomes are mediated through the delivery of specific molecular cargoes to recipient macrophages, engaging multiple signaling pathways to promote an M2 phenotypic state.
Key Signaling Pathways and Exosomal Cargo
Table 2: Molecular Mechanisms of M2 Polarization by MSC Exosomes
| Mechanism Category | Specific Molecule/Pathway | Function in M2 Polarization | Exosome Source |
|---|---|---|---|
| Surface Proteins | CD73 (Ecto-5'-nucleotidase) [3] | Catalyzes production of adenosine, which binds A2A/A2B receptors to activate AKT/ERK pathways [3] | MSC Exosomes (Immortalized line) |
| Extra domain A-fibronectin (EDA-FN) [3] | Activates MyD88-dependent Toll-like receptor (TLR) signaling pathway [3] | MSC Exosomes | |
| MicroRNAs (miRNAs) | miR-223 [24] | Targets LACC1 to inhibit NLRP3 inflammasome activation and Caspase-1-dependent pyroptosis [24] | BM-MSC-Exos |
| miR-182 [4] | Regulates macrophage polarization in myocardial ischaemia-reperfusion injury models [4] | MSC-Exos | |
| let-7 miRNA family [4] | Mediates infiltration and polarization of M2 macrophages in atherosclerosis models [4] | MSC-Exos | |
| Signaling Pathways | PI3K/AKT/mTOR Pathway [4] | Promotes M2 polarization; activated by exosomal cargo [4] | ADSC-Exos, BM-MSC-Exos |
| NF-κB Pathway [118] [4] | Inhibition suppresses M1 polarization; BMSC-Exos show superior suppression of phosphorylated p65 [118] | Primarily BM-MSC-Exos | |
| JAK/STAT Pathway [4] | Involved in M2 gene expression; modulated by exosomes [4] | ADSC-Exos, BM-MSC-Exos | |
| Wnt/β-catenin Pathway [119] [4] | Activated by ADSC-Exos to promote tissue regeneration [119] | Primarily ADSC-Exos |
This diagram illustrates the distinct and shared molecular mechanisms by which ADSC-Exos and BM-MSC-Exos promote M2 macrophage polarization. BM-MSC-Exos are particularly noted for their superior inhibition of the NF-κB and MAPK pathways, while ADSC-Exos strongly activate the PI3K/AKT pathway. Both leverage surface proteins like CD73 and EDA-FN, and deliver specific microRNAs to reprogram macrophage function.
Detailed Experimental Protocols for Key assays
To ensure reproducibility and provide a technical resource, this section outlines core methodologies used to generate the data discussed in this whitepaper.
Protocol 1: In Vitro Macrophage Polarization Assay
This protocol is used to directly evaluate the effect of exosomes on macrophage phenotype.
- Cell Source: Primary human or rat Peripheral Blood Mononuclear Cells (PBMCs) isolated via Ficoll-Paque density gradient centrifugation [3] or human monocyte cell lines (e.g., THP-1) [122].
- Macrophage Differentiation: Differentiate monocytes into naïve M0 macrophages by culturing in RPMI-1640 medium supplemented with 40 ng/mL Macrophage Colony-Stimulating Factor (M-CSF) for 7-9 days [3]. For THP-1 cells, use 100 ng/mL Phorbol 12-myristate 13-acetate (PMA) for 48-72 hours [122].
- Exosome Treatment: Treat M0 macrophages with a standardized concentration (e.g., 10 μg/mL) of isolated ADSC-Exos or BM-MSC-Exos for 24-48 hours [3]. To probe specific mechanisms, co-treat with pathway inhibitors (e.g., CD73 inhibitor PSB12379 [3]).
- Phenotype Analysis:
Protocol 2: Characterization of Isolated Exosomes
This protocol is critical for verifying the quality and identity of exosome preparations prior to functional assays.
- Isolation: Use differential ultracentrifugation, density gradient centrifugation, or tangential flow filtration to isolate exosomes from MSC-conditioned medium [3] [120]. Culture MSCs in exosome-depleted FBS for 48 hours prior to collection [120].
- Characterization:
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration. Confirms vesicles are in the 30-200 nm range [118] [3].
- Transmission Electron Microscopy (TEM): Visualize morphology. Confirms classic cup-shaped, bilayer membrane structure [118].
- Western Blot: Confirm presence of exosomal marker proteins (CD63, CD81, CD9, TSG101, Alix) and absence of negative markers (e.g., Calnexin) [118] [121].
- Protein Quantification: Use bicinchoninic acid (BCA) assay to standardize exosome concentrations for experiments [120].
Protocol 3: In Vivo Tendon-Bone Healing Model
This protocol exemplifies how exosome efficacy is validated in a complex in vivo environment involving inflammation and repair.
- Animal Model: 54 Sprague-Dawley rats undergoing rotator cuff reconstruction surgery [120].
- Experimental Groups: Rats are divided into two groups: a PBS control group and a BMSC-Exos treatment group (each n=27) [120].
- Intervention: BMSC-Exos are administered via tail vein injection post-surgery [120].
- Outcome Analysis:
- Angiogenesis Analysis: At the tendon-bone interface, performed via immunofluorescence staining for CD31 and endomucin on tissue sections [120].
- Biomechanical Testing: Measure the breaking load and stiffness of the healed rotator cuff to assess functional recovery [120].
- Histology & Immunohistochemistry: Evaluate tissue structure, inflammation, and cell proliferation at the healing interface [120].
This workflow outlines the key stages of a comprehensive research pipeline, from exosome isolation and validation to functional testing in increasingly complex models, culminating in data analysis that confirms phenotypic and functional changes in macrophages.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for MSC Exosome and Macrophage Polarization Research
| Reagent/Cell Line | Function and Application | Example Source/Product |
|---|---|---|
| Cell Lines | ||
| RAW264.7 Cells | Mouse monocyte/macrophage cell line; widely used for in vitro polarization studies [24] | ATCC |
| THP-1 Cells | Human monocytic cell line; can be differentiated into macrophages with PMA for polarization assays [122] | ATCC |
| Cytokines & Inducers | ||
| Macrophage Colony-Stimulating Factor (M-CSF) | Differentiates primary monocytes into naïve M0 macrophages [3] | PeproTech |
| Phorbol 12-myristate 13-acetate (PMA) | Differentiates THP-1 monocytes into macrophages [122] | Sigma-Aldrich |
| Interleukin-4 (IL-4) & IL-13 | Classic inducers for M2 macrophage polarization [40] | PeproTech |
| Lipopolysaccharide (LPS) & Interferon-γ (IFN-γ) | Classic inducers for M1 macrophage polarization [120] [24] | Sigma-Aldrich |
| Inhibitors & Agonists | ||
| PSB12379 | Selective CD73 inhibitor; used to block exosome-mediated adenosine production [3] | Tocris Bioscience |
| Assay Kits | ||
| CCK-8 Assay Kit | Measures cell viability and proliferation after exosome treatment [118] [24] | Dojindo |
| ELISA Kits | Quantifies secreted cytokine levels (e.g., TNF-α, IL-1β, IL-6, IL-10) in culture supernatant [24] [122] | Various Vendors |
| Antibodies | ||
| Anti-CD63 / CD81 / CD9 | Surface markers for exosome characterization via Western Blot or Flow Cytometry [118] [121] | Abcam, System Biosciences |
| Anti-CD80 (M1 marker) | Used in flow cytometry to identify pro-inflammatory macrophages [122] | BioLegend |
| Anti-CD163 / CD206 (M2 markers) | Used in flow cytometry to identify anti-inflammatory macrophages [122] | BioLegend |
The choice between ADSC-Exos and BM-MSC-Exos for macrophage repolarization is context-dependent. BM-MSC-Exos demonstrate superior potency in suppressing key pro-inflammatory pathways (NF-κB/MAPK) and mitigating inflammation in models like osteoarthritis, making them a strong candidate for treating highly inflammatory conditions [118]. Their well-documented role in promoting M2 polarization through various mechanisms, including specific miRNA delivery like miR-223, further solidifies their therapeutic profile [24] [4]. ADSC-Exos, sourced from abundant and accessible tissue, exhibit robust pro-regenerative capabilities, particularly in promoting angiogenesis and modulating the immune response via pathways like PI3K/AKT [121] [119]. They are exceptionally promising for applications where tissue repair and vascularization are paramount, such as in wound healing. Future research should focus on standardizing exosome isolation, engineering exosomes to enhance targeting and cargo delivery, and conducting rigorous, direct comparative studies in standardized disease models to fully unlock the clinical potential of these powerful biological nanoparticles.
Multi-omics integration represents a transformative framework for characterizing the molecular cargo of mesenchymal stromal cell (MSC) exosomes and elucidating their mechanisms in macrophage polarization. This technical guide explores the combined application of proteomics and miRNA sequencing to comprehensively profile exosomal content, providing insights into how MSC exosomes mediate the transition of macrophages to the immunomodulatory M2 phenotype. By moving beyond single-omics approaches, researchers can uncover the complex synergistic relationships between proteins and miRNAs that underlie this therapeutic effect. This whitepaper details experimental methodologies, computational integration tools, and pathway analysis techniques to advance research in regenerative medicine and immunomodulation.
The analysis of MSC exosomes requires sophisticated analytical approaches due to their complex cargo of proteins, miRNAs, lipids, and other biomolecules. Single-omics approaches, such as transcriptomics or proteomics alone, provide limited insights into the integrated molecular networks responsible for exosome-mediated effects including macrophage polarization to the M2 phenotype [123]. Multi-omics strategies simultaneously quantify multiple molecular layers, enabling the identification of functional biomolecules and their interactions that would remain hidden in isolated analyses [124] [125].
MSC exosomes have demonstrated the ability to promote M2-like macrophage polarization through multiple mechanisms, including CD73-mediated adenosine production and EDA-FN activation of TLR signaling [3]. Comprehensive cargo profiling via proteomics and miRNA sequencing is essential to fully understand these mechanisms and develop standardized exosome-based therapeutics. This approach allows researchers to connect exosomal cargo to functional outcomes through integrated molecular signatures, providing a more complete picture of exosome biology than any single molecular analysis could achieve [124] [125].
Technical Frameworks for Multi-Omics Integration
Data Integration Methodologies
Multiple computational frameworks have been developed to integrate proteomic and miRNA sequencing data, each with distinct strengths for specific research applications:
Table 1: Multi-omics Integration Methods for Exosome Research
| Method Type | Examples | Key Principles | Advantages for Exosome Research |
|---|---|---|---|
| Matrix Factorization | intNMF, jNMF | Decomposes omics matrices into shared and individual factors | Identifies co-varying features across proteomics and miRNA data; useful for subtype discovery |
| Probabilistic Methods | iCluster, MOFA+ | Models shared latent factors across omics datasets with uncertainty estimation | Handles noise in exosome datasets; identifies robust molecular patterns |
| Correlation-Based | sGCCA, DIABLO | Maximizes correlation/covariance between omics datasets | Finds coordinated protein-miRNA changes linked to M2 polarization outcomes |
| Network-Based | Similarity networks | Constructs molecular interaction networks | Maps functional relationships between exosomal proteins and miRNAs |
| Deep Learning | VAEs, GAUDI | Learns non-linear relationships in lower-dimensional space | Captures complex, non-linear protein-miRNA interactions; powerful for large datasets |
These integration methods enable researchers to move beyond simple correlation analyses toward systems-level understanding of how exosomal cargo components interact to produce phenotypic effects [124] [126]. For MSC exosome research, these approaches can identify which combinations of proteins and miRNAs contribute most significantly to macrophage polarization.
Specialized Tools for miRNA-Proteome Integration
The MoPC (Multi-omics Partial Correlation) computational tool specifically addresses the challenge of integrating miRNA and proteomic data. This method uses partial correlation between mRNA and protein levels conditioned on miRNA expression to identify significant miRNA-target interactions that might be missed when analyzing either data type alone [127].
MoPC implementation involves:
- Input Requirements: Three matched expression matrices (mRNA, protein, and miRNA) from the same samples
- Core Algorithm: Calculation of partial correlation (r~yx.z~) between mRNA (x) and protein (y) expression while accounting for the effect of miRNA (z)
- Statistical Framework: Selection of miRNA-target interactions where partial correlation shows significant improvement over bivariate correlation
- Validation Integration: Automatic checking against miRDB, TargetScan, and miRTarBase databases [127]
Application of MoPC to cancer datasets has successfully identified biologically relevant miRNA-protein regulatory pairs, demonstrating its utility for discovering functional networks in MSC exosome research [127].
Experimental Design and Methodologies
MSC Exosome Preparation and Characterization
Standardized protocols for MSC exosome isolation and characterization are fundamental to generating reliable multi-omics data:
Cell Culture Conditions:
- Use immortalized E1-MYC 16.3 human ESC-derived MSCs or primary MSC sources
- Culture in chemically defined medium (DMEM with 1% non-essential amino acids, 1% glutamine, 1% insulin-transferrin-selenium-X, 1 mM sodium pyruvate, 0.05 mM β-mercaptoethanol, 5 ng/mL FGF-2, and 5 ng/mL PDGF-AB) [3]
- Collect conditioned medium after 3 days of culture
Exosome Isolation:
- Size fractionate conditioned medium using tangential flow filtration
- Concentrate 50× using a membrane with 100 kDa molecular weight cut-off [3]
- Alternative methods: Ultracentrifugation, size exclusion chromatography, or polymer-based precipitation
Quality Control Metrics:
- Protein concentration quantification (e.g., Coomassie Plus assay)
- Particle concentration and size distribution (e.g., ZetaView analysis)
- Modal diameter target: ~130-140 nm [3]
- CD73/NT5E activity assessment (e.g., PiColorLock Gold Phosphate Detection System)
- Western blot for exosomal markers (CD63, CD81, TSG101)
Table 2: Characterization Metrics for MSC Exosome Preparations
| Parameter | Target Specification | Assessment Method |
|---|---|---|
| Protein Concentration | ≥1 mg/mL | Colorimetric protein assay |
| Particle Concentration | ~1.33 × 10^11 particles/mg | Nanoparticle tracking analysis |
| Size Distribution | Modal diameter: 130-140 nm | Dynamic light scattering |
| CD73 Activity | ~22 mU/μg | Phosphate detection system |
| Exosomal Markers | CD63+, CD81+, TSG101+ | Western blot |
| Morphology | Cup-shaped spheres | Electron microscopy |
Proteomic Profiling Workflow
Mass spectrometry-based proteomics provides comprehensive characterization of exosomal protein cargo:
Sample Preparation:
- Protein extraction and denaturation
- Digestion with trypsin/Lys-C
- Peptide desalting and concentration
Liquid Chromatography:
- Utilize micro-flow LC systems for enhanced sensitivity
- Column options: PepMap C18 (optimal for plasma proteomics), HALO ES-C18, or Acquity UPLC Peptide BEH C18 [128]
- Gradient separation: 60-120 minutes depending on complexity
Mass Spectrometry Analysis:
- Electrospray ionization (ESI) sources
- Data-dependent acquisition (DDA) or data-independent acquisition (DIA) modes
- High-resolution mass analyzers (Orbitrap, TimeTOF)
Data Processing:
- Database search against human proteome databases
- Label-free or multiplexed (TMT, SILAC) quantification
- Statistical analysis for differential abundance
This workflow typically identifies thousands of proteins in MSC exosomes, including the immunomodulatory factor CD73 which has been mechanistically linked to M2 macrophage polarization [3] [125].
miRNA Sequencing Protocol
Comprehensive miRNA profiling captures the regulatory RNA component of exosomal cargo:
RNA Isolation:
- Use phenol-chloroform or column-based methods optimized for small RNAs
- Assess RNA quality (RIN >7) and quantity
- Input requirement: 1-10 ng small RNA
Library Preparation:
- 3' and 5' adapter ligation
- Reverse transcription and cDNA amplification
- Size selection for small RNAs (15-30 nt)
Sequencing:
- Platform: Illumina NextSeq, HiSeq, or NovaSeq
- Read length: 50-75 bp single-end
- Depth: 10-20 million reads per sample
Bioinformatic Analysis:
- Adapter trimming and quality control
- Alignment to reference genome
- miRNA quantification using miRBase annotations
- Differential expression analysis
- Target prediction and pathway enrichment
Integration of miRNA sequencing with proteomic data enables the identification of functional miRNA-mRNA-protein regulatory axes relevant to macrophage polarization [127].
Analytical Approaches for Macrophage Polarization Studies
Macrophage Polarization Assays
Primary Macrophage Culture:
- Isolate PBMCs from human or rat blood by Ficoll-Paque density gradient centrifugation
- Seed at 0.5 × 10^6 cells/mL in RPMI with 1% PS and 10% FBS
- Differentiate with 40 ng/mL M-CSF for 7-9 days [3]
- Confirm differentiation by flow cytometry (CD14, CD68 expression)
Exosome Treatment:
- Treat macrophages with 10 μg/mL MSC exosomes or PBS vehicle
- Incubate for 24-48 hours
- For inhibition studies, co-treat with:
- 10 nM PSB12379 (CD73 inhibitor)
- A2A and A2B adenosine receptor antagonists
- AKT/ERK phosphorylation inhibitors [3]
Phenotype Assessment:
- Flow cytometry for surface markers (CD206 for M2, CD80 for M1)
- Cytokine profiling by ELISA or multiplex assays
- Quantitative PCR for polarization genes
- Functional assays (phagocytosis, metabolic profiling)
Multi-Omics Pathway Analysis
The mitch R package enables multi-contrast pathway enrichment analysis for integrated omics data:
Workflow Implementation:
- Import differential expression results from proteomic and miRNA analyses
- Calculate directional significance scores combining p-value and fold change
- Perform multi-contrast enrichment using rank-MANOVA approach
- Identify pathways jointly enriched across multiple omics layers
- Visualize results through heatmaps and enrichment plots [129]
This approach identifies pathways that show coordinated regulation at both protein and miRNA levels, highlighting key mechanisms in MSC exosome-mediated macrophage polarization [129].
Signaling Pathways in MSC Exosome-Mediated Macrophage Polarization
Multi-omics approaches have elucidated two primary mechanisms through which MSC exosomes promote M2 macrophage polarization. The following diagram integrates these pathways based on current research:
Pathway 1: CD73/Adenosine Signaling
- Exosomal CD73 catalyzes conversion of AMP to adenosine
- Adenosine binds to A2A and A2B receptors on macrophages
- Downstream activation of AKT/ERK phosphorylation cascades
- Induction of M2-associated genes (CD206, Arg-1, IL-10, TGF-β) [3]
Pathway 2: EDA-FN/TLR Signaling
- Exosomal EDA-FN activates Toll-like receptors (TLRs)
- Recruitment of MyD88 adaptor protein
- Activation of M2-polarization gene programs [3]
Multi-omics approaches can quantify key components of these pathways (CD73 protein, regulatory miRNAs, downstream effectors) to build comprehensive models of their relative contributions to macrophage polarization.
Research Reagent Solutions
Table 3: Essential Research Reagents for MSC Exosome - Macrophage Studies
| Reagent/Category | Specific Examples | Research Function | Application Notes |
|---|---|---|---|
| MSC Sources | E1-MYC 16.3 immortalized MSCs, Primary bone marrow MSCs, SHED-MSCs | Consistent exosome production | Immortalized lines reduce heterogeneity; primary cells maintain physiological relevance [3] [23] |
| Exosome Isolation | Tangential flow filtration systems, Ultracentrifugation, Size exclusion columns | Particle concentration and purification | TFF offers scalability; SEC provides high purity but lower yield |
| Characterization | CD63/CD81/TSG101 antibodies, CD73 activity assays, Nanoparticle tracking | Quality control and standardization | Essential for batch-to-batch consistency and reproducibility [3] |
| Pathway Inhibitors | PSB12379 (CD73 inhibitor), A2A/A2B receptor antagonists, AKT/ERK inhibitors | Mechanistic validation | Confirm specific pathway involvement in macrophage polarization [3] |
| Macrophage Markers | CD14, CD68, CD206, CD80, CD86 antibodies | Phenotype quantification | Flow cytometry panels for M1/M2 differentiation status [3] [23] |
| Cytokine Assays | IL-10, TGF-β, IL-1β, TNF-α ELISA/multiplex kits | Functional immune profiling | Verify anti-inflammatory polarization at protein level [23] |
| Omics Technologies | LC-MS/MS systems, Next-generation sequencers, Multiplex immunoassays | Comprehensive cargo profiling | Enable multi-omics integration approaches [125] [123] |
The integration of proteomic and miRNA sequencing data provides an unprecedented opportunity to comprehensively profile MSC exosome cargo and elucidate the complex mechanisms underlying macrophage polarization to the M2 phenotype. The multi-omics approaches detailed in this technical guide enable researchers to move beyond descriptive cargo catalogs toward functional understanding of how specific biomolecules work in concert to produce immunomodulatory effects. As these methodologies continue to evolve with improvements in analytical sensitivity, computational integration, and single-cell resolution, they will accelerate the development of exosome-based therapeutics for inflammatory diseases, tissue repair, and immune modulation.
Within the broader investigation of how mesenchymal stem cell (MSC)-derived exosomes modulate macrophage polarization towards the M2 phenotype, a critical and parallel line of inquiry must address the safety and immunogenicity profile of these therapeutic agents. MSC exosomes, nano-sized extracellular vesicles (30-150 nm) carrying bioactive cargo like proteins, lipids, and nucleic acids, have emerged as promising cell-free therapeutic agents due to their immunomodulatory and regenerative properties [130] [131] [132]. Their therapeutic potential, particularly in directing macrophage polarization, is significant for applications in regenerative medicine, cancer therapy, and treatment of autoimmune diseases [133] [132] [134]. However, their translation to clinical use necessitates a rigorous assessment of two primary safety concerns in preclinical models: tumorigenic risk (the potential to initiate or promote tumors) and immune rejection (the activation of host immune responses against the therapeutic agent) [135] [136]. This review synthesizes current preclinical data and methodologies for evaluating these risks, providing a technical guide for researchers and drug development professionals.
Mechanisms of MSC Exosome-Mediated M2 Macrophage Polarization
Understanding the safety profile of MSC exosomes is intrinsically linked to their biological function, particularly their role in modulating immune cells like macrophages. Macrophages exist on a spectrum of activation states, commonly simplified to the pro-inflammatory M1 phenotype and the anti-inflammatory, pro-regenerative M2 phenotype [133]. MSC exosomes have been demonstrated to promote a shift towards the M2 phenotype, which is crucial for tissue repair, inflammation resolution, and immune regulation, but also a potential concern in the context of cancer [131] [133] [132].
The mechanisms underlying this polarization are multifaceted and involve the transfer of exosomal cargo to target cells. Key signaling pathways and components include:
- TLR/TRAF Signaling Pathway: Exosomal circular RNAs, such as circ-CBLB derived from synovial cells, have been shown to inhibit the TLR3/TRAF3 signaling axis, leading to a reduction in pro-inflammatory M1 polarization and a relative increase in M2-like phenotypes [137].
- miRNA Transfer: MSC exosomes are enriched with microRNAs (miRNAs) that directly target signaling pathways in macrophages. For instance, miR-148a and miR-20a-5p in adipose-derived stem cell exosomes (ADSC-Exos) can induce M2 polarization via STAT3/KLF6 and TGF-β/Smad signaling, respectively [130]. Similarly, miR-146a and miR-16-5p target TLR4/IRAK1/TRAF6 to inhibit NF-κB signaling, creating an anti-inflammatory environment conducive to M2 polarization [130].
- Cytokine and Factor Delivery: MSC exosomes carry anti-inflammatory cytokines and factors such as IL-10, IL-1ra, and PGE2, which directly drive macrophages toward an anti-inflammatory M2 phenotype [130] [131].
The following diagram illustrates the core signaling pathways through which MSC exosomes modulate macrophage polarization, highlighting key molecular players identified in preclinical studies.
Diagram 1: Core Signaling Pathways in MSC Exosome-Mediated Macrophage Polarization. MSC exosomes promote M2 polarization by delivering inhibitory circRNAs (e.g., circ-CBLB) and miRNAs (e.g., miR-148a), while also providing anti-inflammatory cytokines. Concurrently, they inhibit M1 polarization by suppressing key signaling pathways like TLR3/TRAF3 and PI3K/Akt.
Assessing Tumorigenic Risk in Preclinical Models
The potential for MSC exosomes to promote tumorigenesis is a paramount safety concern, particularly given their ability to modulate the tumor microenvironment (TME), enhance angiogenesis, and influence macrophage polarization towards a pro-tumorigenic M2 state [138] [133] [135]. Preclinical models are essential for evaluating this risk.
Key Mechanisms of Tumor Promotion
Preclinical studies have identified several mechanisms by which MSC exosomes could theoretically contribute to tumorigenesis:
- Modulation of the Tumor Microenvironment (TME): MSC exosomes can transfer oncogenic molecules (e.g., proteins, miRNAs) to cancer cells, altering their behavior and promoting progression [138]. They can also facilitate the formation of a pre-metastatic niche, preparing distant organs for tumor cell colonization [138].
- Angiogenesis: Exosomes from certain MSC sources carry pro-angiogenic factors like VEGF and FGF2, which can support tumor vascularization and growth [130] [135].
- Immunosuppression: The induction of M2-like tumor-associated macrophages (TAMs) is a critical mechanism. M2-like TAMs are known to promote tumor growth, metastasis, and drug resistance by secreting anti-inflammatory factors (e.g., IL-10, TGF-β), promoting angiogenesis, and suppressing anti-tumor immune responses [133]. MSC exosome-driven polarization towards this phenotype could therefore inadvertently support tumor immune evasion.
Preclinical Models and Methodologies for Tumorigenicity Assessment
A combination of in vitro and in vivo models is employed to assess these risks.
Table 1: Preclinical Models and Assays for Tumorigenicity Assessment
| Model Type | Specific Assay/Model | Key Readouts and Parameters Measured | Relevance to Tumorigenic Risk |
|---|---|---|---|
| In Vitro | Co-culture of MSC exosomes with cancer cell lines (e.g., osteosarcoma, prostate cancer) | Cell proliferation, migration, invasion, colony formation, epithelial-to-mesenchymal transition (EMT) markers [138] [139] | Measures direct effect on cancer cell aggressiveness and metastatic potential. |
| In Vitro | Tube formation assay (HUVECs) | Number of tubules, branches, and total tube length [130] [135] | Quantifies pro-angiogenic potential, a key factor in tumor growth. |
| In Vivo | Xenograft models (immunodeficient mice) | Tumor volume/growth kinetics, incidence, metastasis (via bioluminescence, histology) [139] | Assesses ability to promote tumor growth and spread in a living organism. |
| In Vivo | Syngeneic tumor models (immunocompetent mice) | Tumor growth, immune cell profiling (flow cytometry of TILs), cytokine levels [133] | Evaluates impact on tumor growth within a functional immune system, including TAM polarization. |
| Ex Vivo | Histopathological analysis of harvested tissues | Staining for proliferation markers (Ki-67), microvessel density (CD31), M2 macrophage markers (CD163, CD206) [133] [134] | Confirms pro-tumorigenic effects and underlying mechanisms in tissue. |
Evidence from Preclinical Studies
The current body of preclinical data presents a dualistic picture, underscoring the need for thorough, context-dependent safety testing.
- Tumor-Promoting Evidence: Some studies indicate that MSC exosomes can enhance tumor progression. For example, tumor-derived exosomes (TDEs) can modulate mesenchymal stem cells in the TME to promote cancer progression [138]. Furthermore, MSC exosomes have been shown to promote angiogenesis, a process vital for tumor survival [135].
- Tumor-Suppressing Evidence: Conversely, numerous studies highlight the anti-tumor potential of engineered MSC exosomes. For instance, MSC exosomes overexpressing miR-187 have been shown to inhibit malignant characteristics in prostate cancer cells by downregulating CD276 and suppressing the JAK2-STAT3 pathway [139]. Similarly, exosomal delivery of miR-29a-3p can inhibit migration and vasculogenic mimicry in glioma cells [139].
Evaluating Immune Rejection in Preclinical Models
A significant theoretical advantage of MSC exosomes over whole-cell therapies is their lower immunogenicity. However, their potential to elicit immune responses remains a critical parameter for safety assessment [136] [132].
Mechanisms of Immunogenicity and Immune Evasion
The immune profile of MSC exosomes is complex:
- Low Immunogenicity: MSC exosomes lack major histocompatibility complex (MHC) class II molecules and co-stimulatory signals on their surface, which contributes to their low immunogenic profile and makes them promising for allogeneic use [130] [132].
- Immunomodulatory Properties: Their primary mechanism of action is actively suppressing immune responses. They can inhibit the proliferation and maturation of B cells and T cells, induce regulatory T cells (Tregs), and, as detailed earlier, promote anti-inflammatory M2 macrophage polarization [131] [132].
Preclinical Models for Immunogenicity Testing
Testing for immune rejection involves assessing both innate and adaptive immune responses.
Table 2: Preclinical Models and Assays for Immunogenicity Assessment
| Immune Response | Preclinical Model / Assay | Key Readouts and Parameters Measured | Interpretation and Significance |
|---|---|---|---|
| Innate Immunity | Phagocytosis assay (e.g., with macrophages) | Exosome uptake rate, macrophage cytokine secretion profile (IL-6, TNF-α vs. IL-10) [138] [131] | Measures initial inflammatory response and clearance by innate immune cells. |
| Adaptive Immunity | Mixed lymphocyte reaction (MLR) | T-cell proliferation (CFSE dilution), activation markers (CD69, CD25) [131] [132] | Directly tests the ability of exosomes to trigger T-cell responses. |
| Adaptive Immunity | Allogeneic/ xenogeneic in vivo models | Serum levels of anti-exosome antibodies (IgG, IgM), T-cell infiltration at injection site [136] | Assesses potential for humoral and cellular immune rejection in a physiological context. |
| Systemic Immunomodulation | Disease models (e.g., collagen-induced arthritis, DTH) | Cytokine levels (IFN-γ, IL-17, IL-10), frequencies of Tregs and Bregs, disease severity scores [137] [131] [132] | Evaluates overall immunomodulatory potency and therapeutic safety in an inflammatory context. |
Evidence from Preclinical and Clinical Data
The consensus from preclinical and early clinical data is that MSC exosomes are generally well-tolerated. A review of 66 registered clinical trials noted that MSC-EVs and exosomes exhibited advantages such as "low immunogenicity, stability, comparable efficacy, and no risk of tumorigenesis or thrombosis" [136]. Their immunomodulatory cargo, including IL-10, TGF-β, and specific miRNAs, is actively exploited to treat autoimmune diseases like rheumatoid arthritis and systemic lupus erythematosus, demonstrating a suppressive effect on unwanted immune activation [132].
Standardized Experimental Protocols for Safety Assessment
To ensure reproducible and reliable safety data, standardized experimental protocols are crucial. Below are detailed methodologies for key experiments cited in this field.
Protocol for Assessing M2 Polarization In Vitro
This protocol is fundamental for linking MSC exosome treatment to its potential immunomodulatory and pro-tumorigenic effects via macrophage polarization.
- Macrophage Differentiation: Differentiate human THP-1 monocytes into M0 macrophages by treating with 100 ng/mL Phorbol 12-myristate 13-acetate (PMA) for 72 hours [137].
- Exosome Treatment: Co-culture M0 macrophages with isolated MSC exosomes (e.g., 50-100 µg/mL) for 48 hours. A transwell co-culture system can be used to separate exosomes from macrophages while allowing paracrine communication [137].
- Flow Cytometry Analysis: Harvest macrophages and stain with fluorescently conjugated antibodies against M2 surface markers (e.g., CD206, CD163) and M1 markers (e.g., CD86). Use an appropriate isotype control [137] [133].
- qRT-PCR Analysis: Extract RNA from macrophages and perform qRT-PCR to measure the expression of M2-associated genes (e.g., ARG1, MRC1, IL10) and M1-associated genes (e.g., NOS2, TNF, IL12) [137]. Calculate fold changes using the 2-ΔΔCt method with a housekeeping gene like β-actin for normalization.
- Cytokine Profiling: Collect cell culture supernatant and analyze the levels of M2-related cytokines (e.g., IL-10, TGF-β) and M1-related cytokines (e.g., TNF-α, IL-12) using ELISA or a multiplex cytokine array [131] [133].
Protocol for Tumorigenicity Testing In Vivo
This protocol assesses the potential of MSC exosomes to promote tumor growth in a pre-clinical setting.
- Animal Model: Use immunodeficient mice (e.g., NOD/SCID) for xenograft studies or immunocompetent syngeneic mice for a more comprehensive assessment of the immune role [133] [139].
- Tumor Cell Inoculation: Subcutaneously inject tumor cells (e.g., prostate cancer, glioma cells) into the flank of the mice.
- Exosome Administration: Once palpable tumors form, randomly group the mice and administer MSC exosomes via intravenous or intratumoral injection. Include a control group receiving vehicle (e.g., PBS). Doses used in preclinical studies can vary; a common range is 10-100 µg exosomal protein per injection, multiple times per week [136] [139].
- Monitoring: Measure tumor dimensions 2-3 times per week with calipers. Calculate tumor volume using the formula: Volume = (Length × Width²) / 2.
- Endpoint Analysis: At the study endpoint, euthanize the animals and harvest tumors. Weigh the tumors and process them for histology (H&E staining, immunohistochemistry for Ki-67, CD31, CD206) to assess proliferation, angiogenesis, and immune cell infiltration [133] [139].
The following diagram outlines the key decision points and assessments in a comprehensive preclinical safety evaluation workflow.
Diagram 2: Preclinical Safety and Immunogenicity Assessment Workflow. A comprehensive safety profile is built through a multi-phase process, beginning with in vitro screening of core biological functions, followed by in vivo validation in disease-relevant models, culminating in an integrated risk assessment.
The Scientist's Toolkit: Key Research Reagent Solutions
Successful and reproducible safety assessment relies on a standardized set of reagents and tools. The following table details essential materials for the featured experiments.
Table 3: Essential Research Reagents for Safety and Immunogenicity Studies
| Reagent / Tool | Specific Example | Function and Application in Safety/Immunogenicity Studies |
|---|---|---|
| Cell Lines | THP-1 (human monocyte), RAW 264.7 (mouse macrophage), HUVEC (human umbilical vein endothelial cell), Various cancer cell lines (e.g., PC-3, U87) | THP-1/RAW 264.7 for macrophage polarization studies; HUVEC for angiogenesis assays; cancer lines for tumorigenicity tests [137] [138] [139]. |
| Antibodies for Flow Cytometry | Anti-human CD206, CD163, CD86; Anti-mouse F4/80, CD206, CD80; Isotype controls | Surface marker staining to identify and quantify M1 vs. M2 macrophage populations in vitro and in vivo [137] [133]. |
| Cytokine Detection Kits | ELISA or Luminex kits for TNF-α, IL-6, IL-12, IL-10, TGF-β | Quantifying secreted cytokines from immune or cancer cells to profile inflammatory status and exosome immunomodulatory effects [131] [133]. |
| Exosome Isolation Kits | Total Exosome Isolation reagent, PEG-based precipitation kits | Rapid isolation of exosomes from cell culture supernatant for functional testing, though may co-precipitate contaminants [136]. |
| qRT-PCR Reagents | Primers for ARG1, NOS2, IL10, TNF; SYBR Green master mix; RNA extraction kit (e.g., TRIzol) | Gene expression analysis to validate macrophage polarization and inflammatory pathways at the transcriptional level [137]. |
| Animal Models | Immunodeficient mice (NOD/SCID), Immunocompetent syngeneic mice (e.g., C57BL/6) | In vivo assessment of tumorigenic risk (xenograft) and immunogenicity/immunomodulation (syngeneic) [133] [139]. |
The assessment of tumorigenic risk and immune rejection in preclinical models is a critical step in the development of MSC exosome-based therapies, especially within the context of their ability to polarize macrophages towards the M2 phenotype. Current evidence suggests that while MSC exosomes are generally low in immunogenicity and possess a favorable safety profile in early clinical trials, their tumorigenic potential is context-dependent and necessitates rigorous, standardized evaluation [135] [136]. The dual role of M2 macrophages—promoting tissue repair in degenerative diseases while potentially fostering tumor progression—highlights the need for disease-specific safety assessments. Future efforts must focus on standardizing isolation protocols, dosing metrics, and potency assays to better define the safety and efficacy boundaries of these promising therapeutic agents, ensuring their successful translation from the laboratory to the clinic [136] [139].
Conclusion
MSC exosomes represent a sophisticated, cell-free therapeutic platform that promotes M2 macrophage polarization through multiple interconnected mechanisms, primarily mediated by CD73/adenosine signaling and specific exosomal cargo such as miRNAs and proteins. The therapeutic efficacy of these exosomes has been validated across diverse inflammatory and autoimmune disease models, demonstrating significant potential for clinical translation. Future research should focus on standardizing isolation protocols, optimizing preconditioning strategies to enhance potency, conducting rigorous comparative studies between exosomes from different tissue sources, and advancing engineered exosomes for targeted delivery. As our understanding of their mechanistic actions deepens, MSC exosomes hold immense promise for developing novel immunomodulatory biologics that can be precisely controlled and consistently manufactured for clinical applications in regenerative medicine and immunotherapy.
References
- 1. Advances in the regulation of macrophage polarization by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12328869/]
- 2. A mechanistic integrative computational model of macrophage ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6860420/]
- 3. Mesenchymal Stromal Cell Exosomes Mediate M2-like ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10220822/]
- 4. Anti-inflammatory and M2 macrophage polarization- ... [https://www.sciencedirect.com/science/article/abs/pii/S1567576921004598]
- 5. The role of natural exosomes from SHED-MSC in ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11842380/]
- 6. Reprogram to heal: Macrophage phenotypes as living ... [https://www.sciencedirect.com/science/article/abs/pii/S0024320525002358]
- 7. Mesenchymal stem cell-derived exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC7477653/]
- 8. Mesenchymal stem cell-derived extracellular vesicles for ... [https://www.nature.com/articles/s41419-022-05034-x]
- 9. Mesenchymal stem cell-derived exosomes: A paradigm ... [https://www.sciencedirect.com/science/article/abs/pii/S0014482725002125]
- 10. Mesenchymal Stem Cell-Derived Exosomal MiRNAs Promote ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9319398/]
- 11. Mesenchymal stem cell-derived extracellular vesicles alter ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-020-01937-8]
- 12. Mechanism of Action of Mesenchymal Stem Cell-Derived ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.833840/full]
- 13. Extracellular Vesicles from Different Mesenchymal Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11989379/]
- 14. Mesenchymal Cell-Derived Exosomes as Novel Useful ... [https://brieflands.com/journals/ans/articles/98722]
- 15. Mesenchymal stromal/stem cell (MSC)-derived exosomes in ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-023-03287-7]
- 16. The progress and prospects of targeting the adenosine ... [https://biomarkerres.biomedcentral.com/articles/10.1186/s40364-025-00784-0]
- 17. The CD39-CD73-adenosine axis: Master regulator of ... [https://pubmed.ncbi.nlm.nih.gov/40930263/]
- 18. CD39 and CD73 - Adenosine Signaling [https://bpsbioscience.com/cd39-cd73-adenosine-signaling-immunotherapy]
- 19. Research progress of CD73-adenosine signaling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12105175/]
- 20. CD73 on cancer-associated fibroblasts enhanced by the ... [https://www.nature.com/articles/s41467-019-14060-x]
- 21. Signaling pathways involving adenosine A2A and A2B ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4854836/]
- 22. Potential of Adora2b as an immunotherapeutic target for ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1687675/full]
- 23. The role of natural exosomes from SHED-MSC in ... [https://pubmed.ncbi.nlm.nih.gov/39991155/]
- 24. Bone marrow mesenchymal stem cell exosome-derived ... [https://www.nature.com/articles/s41598-025-22511-3]
- 25. The role of macrophage subtypes and exosomes in ... [https://cmbl.biomedcentral.com/articles/10.1186/s11658-022-00384-y]
- 26. The immune regulatory role of exosomal miRNAs and ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11647038/]
- 27. Shining the light on mesenchymal stem cell-derived ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-023-03245-3]
- 28. The role of macrophage subtypes and exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9528143/]
- 29. Exosome Source Matters: A Comprehensive Review from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11858990/]
- 30. MFGE8 in exosomes derived from mesenchymal stem cells ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10986023/]
- 31. Extracellular vesicles containing MFGE8 from colorectal ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-024-01669-9]
- 32. Exosome-derived circ-001422 promotes tumor-associated ... [https://www.nature.com/articles/s42003-024-07134-0]
- 33. Mesenchymal Stromal Cell Exosomes Mediate M2-like ... [https://www.mdpi.com/1999-4923/15/5/1489]
- 34. Mesenchymal Stromal Cell Exosomes Mediate M2-like ... [https://pubmed.ncbi.nlm.nih.gov/37242732/]
- 35. Mesenchymal stromal cell-derived exosomes protect against ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-03808-y]
- 36. Mesenchymal Stem/Stromal Cell-Derived Exosomes for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7290908/]
- 37. Recent advances in adipose-derived mesenchymal stem ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1525466/full]
- 38. Isolation of Extracellular Vesicles Derived from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8032486/]
- 39. An improvised one-step sucrose cushion ultracentrifugation ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-018-0923-0]
- 40. Engineered M2 macrophage-derived exosomes ... [https://www.sciencedirect.com/science/article/abs/pii/S1742706125005379]
- 41. Characterization of Extracellular Vesicles by Size- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6802587/]
- 42. Size exclusion chromatography for exosome isolation [https://immunostep.com/2022/09/29/size-exclusion-chromatography-for-exosome-isolation/]
- 43. Nanoparticle Tracking Analysis: An Effective Tool to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11477862/]
- 44. NTA-Based Exosome Identification [https://www.creative-proteomics.com/subcell/nta-based-exosome-identification.htm]
- 45. MSC-derived exosomes carrying sFgl2 alleviate acute ... [https://pubmed.ncbi.nlm.nih.gov/41101233/]
- 46. Exosomal PSM-E inhibits macrophage M2 polarization to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11562865/]
- 47. Expanded characterization of in M0, M1, and... vitro polarized [https://pmc.ncbi.nlm.nih.gov/articles/PMC9980743/]
- 48. and Characterization of M1 and M2 Human... Polarization [https://www.jove.com/t/67180/polarization-characterization-m1-m2-human-monocyte-derived]
- 49. CD206 modulates the role of M2 macrophages in the origin of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10861823/]
- 50. Predictive screening of M1 and M2 macrophages reveals ... [https://www.nature.com/articles/srep40144]
- 51. Mesenchymal stromal cell-derived exosomes attenuate ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6529919/]
- 52. Mesenchymal Stem Cells Derived Extracellular Vesicles ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2021.811164/full]
- 53. Polarized Macrophages and Their Exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561894/]
- 54. Engineerable mesenchymal stem cell-derived extracellular ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04490-4]
- 55. How will we treat systemic lupus erythematosus in the next ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12495987/]
- 56. Systemic lupus erythematosus: updated insights on the ... [https://www.nature.com/articles/s41392-025-02168-0]
- 57. Efficacy and safety of allogeneic CD19 CAR NK-cell ... [https://www.sciencedirect.com/science/article/pii/S014067362501671X]
- 58. Post-EULAR 2025 Highlights: Innovations in Autoimmune ... [https://inderocro.com/post-eular-2025-highlights-innovations-in-autoimmune-disease-management/]
- 59. Rheumatoid arthritis: prediction of future clinically-apparent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10959682/]
- 60. Mesenchymal stem cells in treating human diseases [https://www.nature.com/articles/s41392-025-02313-9]
- 61. Exosome-based therapeutics in bone regeneration [https://pmc.ncbi.nlm.nih.gov/articles/PMC12512519/]
- 62. Frontiers | Mesenchymal stem cells and exosomes in ischemic brain... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2025.1639756/full]
- 63. from adipose-derived stem cells promote Exosomes ... angiogenesis [https://ijbms.mums.ac.ir/article_21837.html]
- 64. Therapeutic potential of stem cell-derived exosomes for ... [https://pubmed.ncbi.nlm.nih.gov/40291635/]
- 65. Molecular Imaging of Therapeutic Effect of Mesenchymal Stem... [https://pubmed.ncbi.nlm.nih.gov/30941719/]
- 66. Functionalized mesenchymal stem cells for enhanced bone ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04699-3]
- 67. The role of mesenchymal stem cell‑derived exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12012432/]
- 68. The role of mesenchymal stem cell‑derived exosomes ... [https://www.spandidos-publications.com/10.3892/mmr.2025.13531]
- 69. Mesenchymal stem cells for lung diseases - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12411635/]
- 70. Exosomes from human umbilical cord mesenchymal stem ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-021-02244-6]
- 71. Advancements in engineered mesenchymal stem cell ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-023-04729-9]
- 72. - Wikipedia Mesenchymal stem cell [https://en.wikipedia.org/wiki/Mesenchymal_stem_cell]
- 73. derived- Mesenchymal : a modern approach in... stem cell exosomes [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-020-02622-3]
- 74. : The Future Mesenchymal -Based Therapy? Stem Cell Exosomes MSC [https://link.springer.com/chapter/10.1007/978-1-62703-200-1_3]
- 75. -Derived Mesenchymal in Ophthalmology... Stem Cell Exosomes [https://pubmed.ncbi.nlm.nih.gov/37111652/]
- 76. -derived Mesenchymal for clinical use stem cell exosomes [https://www.nature.com/articles/s41409-019-0616-z?error=cookies_not_supported&code=5bc1dc60-47fd-4ea8-8092-8459117b759c]
- 77. 人骨髓间充质干细胞分泌的外泌体调控恶性胶质瘤相关巨噬 ... [https://kyxuebao.kmmu.edu.cn/cn/article/doi/10.12259/j.issn.2095-610X.S20210101]
- 78. 国内外泌体领域进展总结(2025年4月) [https://www.cytoniche.com/news/775.html]
- 79. Frontiers | Mesenchymal Stromal Cells and Exosomes : Progress and... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2020.00665/full]
- 80. JEV | 南京医科大学李晶教授团队:创新外泌体大规模亚群分 ... [https://www.exosomemed.com/20217.html]
- 81. Exosomes derived from mesenchymal stem cells alleviate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12447933/]
- 82. Extracellular vesicles derived from GMSCs stimulated with TNF-α and IFN-α promote M2 macrophage polarization via enhanced CD73 and CD5L expression - PubMed [https://pubmed.ncbi.nlm.nih.gov/35922474/]
- 83. Extracellular vesicles derived from GMSCs stimulated with ... [https://www.nature.com/articles/s41598-022-17692-0]
- 84. Identification of CD73 as a Novel Biomarker Encompassing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9688912/]
- 85. Elevation of serum CD5L concentration is correlated with ... [https://www.sciencedirect.com/science/article/abs/pii/S1567576918303515]
- 86. Frontiers | Effects of preconditioning with TNFα and IFNγ in angiogenic potential of mesenchymal stromal cell-derived extracellular vesicles [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2023.1291016/full]
- 87. TNF-α and IFN-γ Participate in Improving the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8431422/]
- 88. Hypoxia in tumor microenvironment regulates exosome ... [https://www.sciencedirect.com/science/article/abs/pii/S0304383520301361]
- 89. Hypoxic exosomes orchestrate tumorigenesis: molecular ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-020-02662-9]
- 90. Hypoxia Induced Changes of Exosome Cargo and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9013908/]
- 91. Can Soluble Immune Checkpoint Molecules on Exosomes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10128092/]
- 92. Role of mesenchymal stem cells in modulating cytokine ... [https://www.sciencedirect.com/science/article/abs/pii/S0304419X25001751]
- 93. Targeting hypoxia signaling pathways in angiogenesis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11079266/]
- 94. Mesenchymal stem cell-derived exosomes regulate the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8422151/]
- 95. Mesenchymal stem cell-derived exosome-educated ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-022-03174-7]
- 96. Macrophage-driven exosomes regulate the progression of ... [https://pubmed.ncbi.nlm.nih.gov/40371346/]
- 97. Exosomal miR-21 secreted by IL-1β-primed-mesenchymal ... [https://www.sciencedirect.com/science/article/abs/pii/S0024320520314119]
- 98. Inhibition of tumor progression and M microglial 2 by... polarization [https://pmc.ncbi.nlm.nih.gov/articles/PMC8490520/]
- 99. Mesenchymal Stem-Cell-Derived Exosomes and MicroRNAs [https://pmc.ncbi.nlm.nih.gov/articles/PMC12249468/]
- 100. Exosomal miRNAs-mediated macrophage polarization and ... [https://www.sciencedirect.com/science/article/abs/pii/S1567576923002254]
- 101. CD73-expressing endometrial regenerative cell-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12131482/]
- 102. CD73: A Team Player Caught in the “AKT” of Wound ... [https://www.roosterbio.com/blog/cd73-a-team-player-caught-in-the-akt-of-wound-healing-cell-survival-via-msc-exosomes/]
- 103. Exosomal miRNA-mediated intercellular ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-023-00964-w]
- 104. Assessment of CD73 activity in breast cancer-derived small ... [https://pubmed.ncbi.nlm.nih.gov/40390761/]
- 105. Mesenchymal stem cell-derived exosome-educated ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9509559/]
- 106. Extracellular vesicles containing GAS6 protect the liver ... [https://www.nature.com/articles/s41420-024-02169-y]
- 107. MSC-Derived Small Extracellular Vesicles Attenuate ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.888949/full]
- 108. In vivo exosome imaging: applications of diverse visualization ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12402689/]
- 109. In vivo exosome imaging: applications of diverse ... [https://www.bmbreports.org/journal/view.html?uid=2155&vmd=Full]
- 110. Emerging strategies for labeling and tracking of ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365920304971]
- 111. Immunomodulatory and Regenerative Functions of MSC ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12383523/]
- 112. Mesenchymal stem cell-derived exosomes: emerging ... [https://www.sciencedirect.com/science/article/pii/S2773041725000253]
- 113. Mesenchymal Stem Cells vs Exosomes: A Comparison ... [https://celltexbank.com/mscs-vs-exosomes/]
- 114. Mesenchymal stem cell-derived small extracellular vesicles ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590144/]
- 115. Comparison of production methods for mesenchymal stem ... [https://www.nature.com/articles/s41598-025-21218-9]
- 116. Exosomes derived from hypoxia-preconditioned M2 ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06808-5]
- 117. Apoptotic vesicles of mesenchymal stem cells promote M2 ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04546-5]
- 118. Comparative Efficacy of Exosomes Derived from Different ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12193399/]
- 119. Adipose-derived stem cell exosomes: emerging roles and ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1637342/full]
- 120. Bone marrow mesenchymal stem cell-derived exosomes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7687785/]
- 121. Adipose-derived stem cell exosomes - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC12181847/]
- 122. Exosome circ-CBLB promotes M1 macrophage ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1627389/full]
- 123. The power of multi-omics | Standard BioTools [https://www.standardbio.com/resources/blog-articles/2024/02/transcriptomics-vs-proteomics]
- 124. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8334347/]
- 125. Multi-Omics Profiling for Health - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10220275/]
- 126. A technical review of multi-omics data integration methods [https://pmc.ncbi.nlm.nih.gov/articles/PMC12315550/]
- 127. Integrated microRNA and proteome analysis of cancer datasets with MoPC - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10956802/]
- 128. Evaluation of commercial micro-flow liquid chromatography ... [https://pubmed.ncbi.nlm.nih.gov/39983561/]
- 129. mitch: multi-contrast pathway enrichment for multi-omics and ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-020-06856-9]
- 130. Adipose-derived stem cell exosomes: multifaceted therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12527763/]
- 131. Mesenchymal stem cell-derived exosomes as a plausible ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1563427/full]
- 132. Molecular and therapeutic effects of mesenchymal stem ... [https://pubmed.ncbi.nlm.nih.gov/40740529/]
- 133. Targeting M2-like tumor-associated macrophages is a ... [https://www.nature.com/articles/s41698-024-00522-z]
- 134. M2 macrophage-derived exosomes for bone regeneration [https://www.sciencedirect.com/science/article/abs/pii/S0003996924001559]
- 135. The role of mesenchymal stem cells and their exosomes in ... [https://www.sciencedirect.com/science/article/abs/pii/S0304416525000686]
- 136. Trends in mesenchymal stem cell-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488731/]
- 137. Exosome circ-CBLB promotes M1 macrophage polarization in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12310475/]
- 138. Exploring the Interaction of Tumor-Derived Exosomes and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11988628/]
- 139. engineering mesenchymal stem cell-derived small ... [https://cancerci.biomedcentral.com/articles/10.1186/s12935-025-03900-0]