Navigating MSC Exosome Heterogeneity: From Biological Complexity to Clinical Standardization
Mesenchymal stem cell (MSC)-derived exosomes and other extracellular vesicles (EVs) represent a promising cell-free therapeutic platform with significant advantages over whole-cell therapies.
Navigating MSC Exosome Heterogeneity: From Biological Complexity to Clinical Standardization
Abstract
Mesenchymal stem cell (MSC)-derived exosomes and other extracellular vesicles (EVs) represent a promising cell-free therapeutic platform with significant advantages over whole-cell therapies. However, their inherent heterogeneity in population and cargo presents a major challenge for clinical translation and reproducible efficacy. This article provides a comprehensive analysis for researchers and drug development professionals, exploring the foundational sources of heterogeneity, methodological advances for its control, strategies for troubleshooting manufacturing challenges, and frameworks for validation and comparative analysis. By synthesizing the latest research and clinical data, this review aims to equip scientists with the knowledge to standardize MSC-exosome products and harness their full therapeutic potential in regenerative medicine and drug delivery.
Decoding the Sources of MSC Exosome Heterogeneity: Vesicles, Cargo, and Biological Drivers
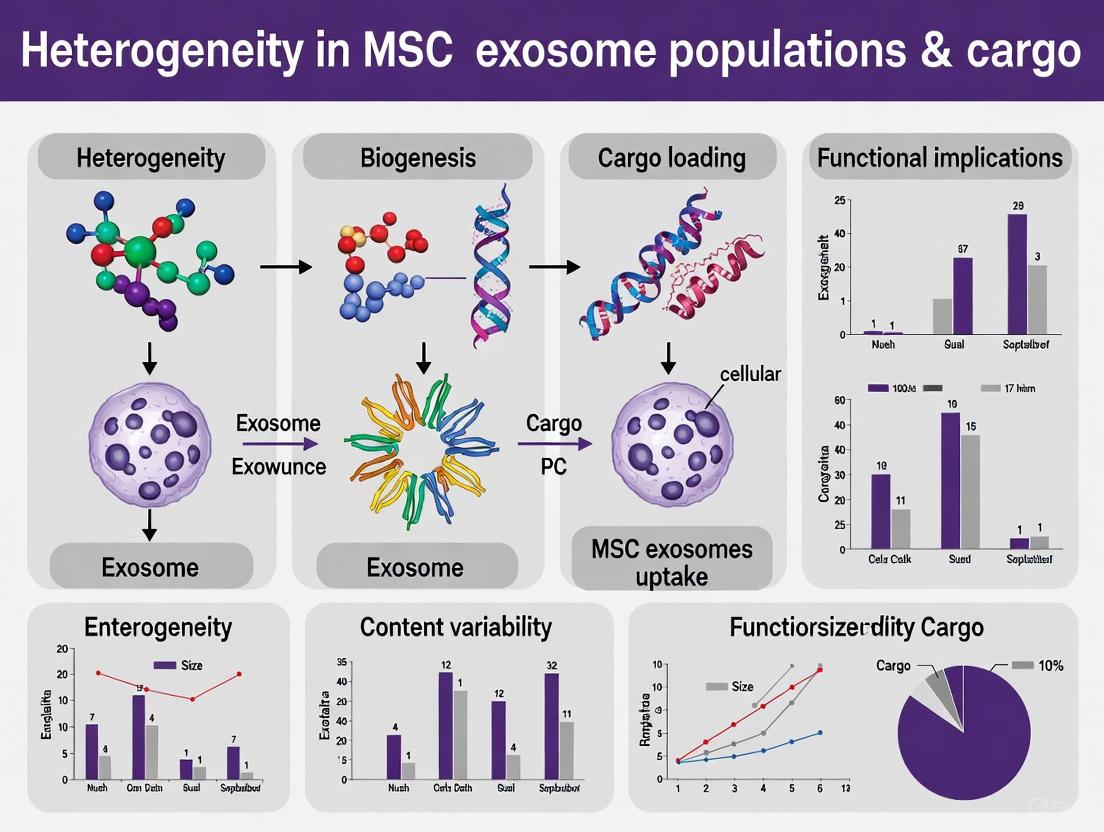
The therapeutic potential of Mesenchymal Stem Cells (MSCs) is now largely attributed to their secretome—the complex mixture of factors they release, which includes various types of extracellular vesicles (EVs). These EVs are nanoscale lipid bilayer-enclosed particles that act as essential messengers in intercellular communication, shuttling bioactive molecules like proteins, lipids, and nucleic acids between cells. The MSC secretome primarily contains three distinct classes of EVs: exosomes, microvesicles, and apoptotic bodies, each with unique origins, sizes, and compositional profiles. Understanding this spectrum is crucial for research and therapeutic development, as the significant heterogeneity within and between these populations directly impacts experimental reproducibility and therapeutic efficacy [1] [2].
This technical support center addresses the key challenges researchers face when working with heterogeneous MSC-EV populations. The following sections provide targeted troubleshooting guides, detailed protocols, and strategic insights to help you isolate, characterize, and functionally analyze these complex vesicle mixtures, thereby advancing your research in regenerative medicine and drug development.
FAQ: Fundamental Concepts of MSC-Derived EVs
Q1: What are the key defining characteristics that differentiate exosomes, microvesicles, and apoptotic bodies?
The three main EV types in MSC secretomes are classified based on their biogenesis, size, and molecular markers.
- Exosomes (30-150 nm) are formed intracellularly within multivesicular bodies (MVBs). When MVBs fuse with the plasma membrane, exosomes are released into the extracellular space. They are characterized by the presence of tetraspanin markers (CD63, CD81, CD9) and ESCRT-related proteins (TSG101, Alix) [1] [3] [2].
- Microvesicles (100-1000 nm) are formed directly through the outward budding and fission of the plasma membrane. Their composition reflects the parent cell's plasma membrane and they are typically enriched with proteins like selectins and integrins [1] [4] [2].
- Apoptotic Bodies (50-5000 nm) are generated during the programmed cell death (apoptosis) of MSCs. They contain cellular debris, such as fragmented organelles and condensed chromatin, and are identified by the presence of phosphatidylserine (PS) on their surface [1] [4] [5].
Table 1: Key Characteristics of Major Extracellular Vesicle Types in MSC Secretomes
| Feature | Exosomes | Microvesicles | Apoptotic Bodies |
|---|---|---|---|
| Biogenesis | Endosomal pathway (MVBs) | Outward budding of plasma membrane | Cell disassembly during apoptosis |
| Size Range | 30 - 150 nm | 100 - 1000 nm | 50 - 5000 nm |
| Key Markers | CD63, CD81, CD9, TSG101, Alix | Selectins, Integins, ARF6 | Phosphatidylserine, Histones |
| Key Cargo | miRNAs, mRNAs, cytoplasmic & membrane proteins | miRNAs, mRNAs, cytoplasmic proteins | Cellular debris, organelle fragments, nuclear parts |
| Primary Function | Targeted intercellular communication | Local cell signaling & communication | Clearance of apoptotic cell debris |
Q2: Why is understanding EV heterogeneity critical for my research outcomes?
EV heterogeneity is a critical variable that can significantly influence your experimental results and their interpretation. This heterogeneity arises from several factors:
- Cell Source: MSCs isolated from different tissues (e.g., bone marrow, adipose tissue, umbilical cord) produce EVs with distinct molecular cargo and functional properties [1] [5]. For instance, bone marrow MSC-EVs highly inhibit inflammatory cell accumulation, while umbilical cord-derived MSC-EVs are particularly effective at suppressing oxidative stress [1].
- Culture Conditions: The medium composition (e.g., fetal bovine serum vs. platelet lysate vs. serum/xeno-free media), 3D vs. 2D culture, use of bioreactors, and exposure to hypoxia can all alter the yield, composition, and biological activity of the EVs produced by MSCs [1] [6].
- Isolation Method: The technique used to isolate EVs (e.g., ultracentrifugation, density gradient, precipitation, size-exclusion chromatography) can selectively enrich for different EV subpopulations, directly affecting the purity and functional profile of your final sample [4] [7].
Failure to account for this heterogeneity can lead to poor reproducibility between experiments and labs, inconsistent therapeutic outcomes, and difficulty in identifying genuine EV-specific biomarkers [1] [5]. Standardizing and reporting these parameters is essential for robust science.
Troubleshooting Guide: Common Experimental Challenges
Challenge 1: Low Yield or Purity of MSC-derived EVs
Problem: The quantity of isolated EVs is insufficient for downstream analysis, or the preparation is contaminated with non-EV components like proteins or lipoprotein particles.
Solutions:
- Optimize Cell Expansion: Ensure MSCs are healthy and expanded under optimal conditions. Using human platelet lysate or defined xeno-free media instead of fetal bovine serum (FBS) can enhance EV production and reduce contaminating bovine EVs [6].
- Select the Appropriate Isolation Technique: Choose an isolation method based on your downstream application. For high purity, consider density gradient centrifugation or size-exclusion chromatography. For large volumes, tangential flow filtration is efficient [4] [8].
- Combine Methods: A common strategy is to use a combination of methods, such as ultrafiltration followed by size-exclusion chromatography (SEC), to increase both yield and purity [4] [7].
- Characterize Rigorously: Always use multiple characterization techniques to assess purity, such as Nanoparticle Tracking Analysis (NTA) for concentration and size, and Western Blot for specific markers (e.g., CD63, CD81) and the absence of negative markers like GM130 (Golgi) or calnexin (ER) [8] [5].
Challenge 2: Inconsistent Functional Results in Target Cell Assays
Problem: EV preparations from the same MSC source yield variable results in functional assays, such as proliferation, migration, or gene expression in recipient cells.
Solutions:
- Control MSC Passage Number: MSC properties can drift with repeated passaging. Use MSCs within a defined, low passage range (e.g., P3-P6) to maintain consistency in EV production [2].
- Standardize the "Cell State": The physiological state of the MSCs at the time of EV collection is critical. Implement serum-starvation during EV production if required, and control for factors like cell confluency and metabolic state [6] [5].
- Functional Titering: Do not simply dose experiments based on EV protein quantity or particle number. Perform a functional titration for your specific assay (e.g., a dose-response curve for angiogenesis or immunomodulation) to find the optimal and reproducible effective dose [1].
- Analyze Cargo: Heterogeneity in EV cargo (e.g., miRNA or protein profiles) due to slight changes in culture conditions can cause functional variation. Use techniques like RNA sequencing or proteomics to correlate cargo with function [1] [6].
Challenge 3: Inefficient Loading of Therapeutic Cargo into EVs
Problem: Low efficiency when loading drugs, nucleic acids (siRNA, miRNA), or proteins into isolated MSC-EVs for drug delivery applications.
Solutions:
- Choose the Right Loading Method: Select a method appropriate for your cargo.
- Electroporation: Common for nucleic acids like siRNA, but can cause EV aggregation and membrane damage [8].
- Sonication: Uses ultrasound to transiently disrupt the EV membrane, allowing cargo entry. Can be more efficient but requires optimization to avoid damaging EVs [8].
- Co-incubation: The simplest method; incubate the cargo with EVs. Efficiency is highly dependent on the cargo's lipophilicity and concentration gradient [8].
- Saponin-Assisted Loading: Uses saponin to create pores in the EV membrane. Can achieve high loading capacity but requires careful control and cleaning steps [8].
- Post-Loading Purification: Always include a purification step (e.g., SEC, ultrafiltration) after loading to remove unencapsulated free cargo, which can confound functional results [8].
- Verify Loading and EV Integrity: Use techniques like NTA to confirm EV size stability post-loading and a functional assay to confirm the loaded cargo is bioactive and protected [8].
Detailed Experimental Protocols
Protocol 1: Isolation of EVs from MSC Conditioned Medium via Ultracentrifugation
This protocol describes the classic "gold standard" method for isolating EVs from MSC-conditioned medium [4] [8].
Research Reagent Solutions:
- Mesenchymal Stem Cells: Source (e.g., bone marrow, adipose). Use at 70-80% confluency.
- Serum-free MSC Medium: To avoid contamination with serum-derived EVs.
- Dulbecco's Phosphate Buffered Saline (DPBS): For washing cells.
- Ultracentrifugation Tubes: Polypropylene tubes compatible with ultracentrifuge.
- Protease Inhibitor Cocktail: Added to conditioned medium to prevent protein degradation.
Method:
- Cell Culture and Conditioning: Culture MSCs to 70-80% confluency. Wash cells three times with DPBS to remove residual serum. Add serum-free medium and incubate for 24-48 hours. Using a defined and consistent conditioning time is crucial for reproducibility.
- Collection and Pre-Clearing: Collect the conditioned medium and centrifuge at 376 × g for 10 minutes at 4°C to remove floating cells.
- Sequential Centrifugation:
- Transfer supernatant to new tubes and centrifuge at 2,000 × g for 20 minutes at 4°C to remove dead cells and large debris.
- Transfer supernatant and centrifuge at 10,000 × g for 30 minutes at 4°C to pellet larger microvesicles and organelles.
- Filter the resulting supernatant through a 0.22 µm pore filter to remove remaining large particles.
- Ultracentrifugation: Transfer the filtered supernatant to ultracentrifuge tubes. Pellet EVs by ultracentrifugation at 100,000 - 120,000 × g for 70-90 minutes at 4°C.
- Washing and Resuspension: Carefully discard the supernatant. Gently wash the pellet (often not visible) with a large volume of DPBS. Perform a second ultracentrifugation under the same conditions (100,000 - 120,000 × g, 70-90 min). Finally, resuspend the final EV pellet in a small volume of DPBS or your chosen storage buffer (e.g., with trehalose). Aliquot and store at -80°C.
Diagram 1: Ultracentrifugation Workflow for EV Isolation
Protocol 2: Characterization of Isolated EVs using Nanoparticle Tracking Analysis (NTA) and Western Blot
This protocol confirms the size, concentration, and presence of EV-specific markers in your isolation [8] [5].
Research Reagent Solutions:
- DPBS, filtered (0.1 µm): For diluting EV samples for NTA.
- RIPA Lysis Buffer: For lysing EVs for protein analysis.
- BCA Assay Kit: For quantifying protein concentration.
- Primary Antibodies: Anti-CD63, anti-CD81, anti-TSG101, anti-Calnexin.
- Secondary Antibodies: HRP-conjugated antibodies.
- SDS-PAGE Gel, PVDF Membrane, ECL Reagent: For Western Blot.
Method: Part A: Nanoparticle Tracking Analysis (NTA)
- Dilution: Thaw an EV aliquot on ice. Dilute the sample 100- to 1000-fold in filtered (0.1 µm) DPBS to achieve an ideal concentration for the NTA instrument (e.g., 10^8 - 10^9 particles/mL).
- Instrument Calibration: Calibrate the NTA instrument according to the manufacturer's instructions using standard beads of known size.
- Measurement: Inject the diluted sample. Capture several 30-60 second videos. Ensure the particle count is within the linear range of the camera.
- Analysis: Use the instrument's software to analyze the videos, which tracks the Brownian motion of individual particles to calculate the hydrodynamic diameter and concentration of the EV population.
Part B: Western Blot Analysis
- EV Lysis: Mix a volume of EV suspension with an equal volume of 2X RIPA buffer containing protease inhibitors. Incubate on ice for 30 minutes.
- Protein Quantification: Use the BCA assay to determine the protein concentration of the lysate.
- Gel Electrophoresis: Load 10-20 µg of EV protein per well on an SDS-PAGE gel. Include a molecular weight marker. Run the gel at constant voltage.
- Membrane Transfer: Transfer the separated proteins from the gel to a PVDF membrane.
- Immunoblotting: Block the membrane with 5% non-fat milk. Incubate with primary antibodies (e.g., CD63, TSG101) overnight at 4°C. Wash and incubate with an HRP-conjugated secondary antibody. Develop the membrane using ECL reagent and image.
- Control for Purity: Probe for negative markers like Calnexin (an endoplasmic reticulum protein), which should be absent or significantly reduced in a pure EV preparation compared to a whole cell lysate.
Table 2: Comparison of Major EV Isolation Techniques
| Method | Principle | Advantages | Disadvantages | Best For |
|---|---|---|---|---|
| Ultracentrifugation (UC) | Sequential centrifugation based on size/density | Considered gold standard; no chemical additives; good for large volumes | Time-consuming; requires expensive equipment; can cause co-precipitation & EV damage | Large-scale prep; initial EV research |
| Size-Exclusion Chromatography (SEC) | Separation by size using porous polymer beads | High purity; preserves EV integrity & function; simple protocol | Sample dilution; limited volume per run; may not separate similar-sized particles | High-purity isolates for functional studies |
| Precipitation (e.g., PEG) | Reduce solubility using polymers | Simple; high yield; no special equipment | Co-precipitation of contaminants (proteins, lipoproteins); difficult downstream analysis | Quick, crude isolation for diagnostics |
| Tangential Flow Filtration (TFF) | Size-based separation with parallel flow | Scalable; high yield; good for processing large volumes | Membrane fouling; requires optimization; initial equipment cost | Industrial-scale manufacturing |
| Immunoaffinity Capture | Antibody-binding to surface markers | High specificity & purity; isolates specific EV subtypes | High cost; low yield; dependent on antibody specificity | Isolating specific EV subpopulations |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for MSC-EV Work
| Reagent / Material | Function / Application | Key Considerations |
|---|---|---|
| Xeno-Free/SFDA-Compliant Cell Media | Expands MSCs for clinical translation | Eliminates bovine EV contaminants; ensures reproducible, GMP-compliant EV production [6]. |
| Human Platelet Lysate (hPL) | Serum-substitute for MSC culture | Enhances MSC proliferation and EV yield; human-derived reduces immunogenicity risks [6]. |
| Size-Exclusion Chromatography (SEC) Columns | High-purity EV isolation | Preserves EV biological activity and morphology; ideal for functional studies [4] [7]. |
| Nanoparticle Tracking Analyzer (NTA) | Measures EV size distribution & concentration | Essential for quantitative quality control of EV preparations [8] [5]. |
| Tetraspanin Antibody Panel (CD63, CD81, CD9) | EV characterization via Western Blot/Flow Cytometry | Confirms the vesicular nature of isolates; standard markers for exosomes [3] [8]. |
| MicroRNA/RNA Extraction Kits (EV-specific) | Isolates RNA cargo from EVs | Designed for low-abundance RNA from small volumes; enables cargo profiling [5]. |
| Trehalose or Sucrose-based Cryoprotectant | Long-term storage of EV samples | Helps maintain EV integrity and function during freeze-thaw cycles [4]. |
Signaling Pathways in MSC-EV Mediated Effects
MSC-derived EVs exert their therapeutic effects by modulating key signaling pathways in recipient cells through the transfer of proteins, miRNAs, and other bioactive molecules. The diagrams below illustrate two critical pathways involved in immune regulation and tissue repair.
Diagram 2: MSC-EV Modulation of T Cell Immune Response
Diagram 3: MSC-EV Mediated Tissue Repair Signaling
Mesenchymal stem cell (MSC)-derived exosomes are increasingly recognized as potent mediators of intercellular communication, capable of transferring diverse bioactive cargoes—including miRNAs, proteins, and lipids—to recipient cells to modulate their function [1] [9]. This cargo dictates the exosomes' targeting specificity and functional roles upon delivery [9]. A central challenge in harnessing these vesicles for therapeutic applications is their inherent heterogeneity; the composition and biological effects of MSC-derived exosomes are profoundly influenced by the MSC source, culture conditions, and isolation methods [1] [10]. This technical support center is designed to help researchers troubleshoot the complexities of analyzing variable exosomal cargo, providing targeted FAQs and detailed guides to ensure reproducible and reliable results within the broader thesis of addressing heterogeneity in MSC exosome research.
Frequently Asked Questions (FAQs)
FAQ 1: Our MSC-exosome preparations show high variability in miRNA cargo yield between batches. What are the primary factors we should investigate?
Batch-to-batch variability in miRNA cargo is often linked to upstream process parameters. You should systematically investigate:
- Cell Source: MSCs from different tissues (e.g., bone marrow, adipose tissue, umbilical cord) inherently produce exosomes with distinct molecular compositions and functional properties [1].
- Culture Conditions: Factors like the use of 3D culture systems, bioreactors, and exposure to hypoxia can significantly alter the cargo profile of the resulting exosomes [1].
- Cell Physiological State: The age of the donor and the passage number of the MSCs in culture can impact their exosomal output. Aging MSCs, for instance, show functional decline and altered cargo secretion [10].
FAQ 2: When analyzing exosomal proteins, we encounter significant contamination from non-vesicular components. How can we improve purity for more accurate cargo analysis?
The isolation method is critical for purity. While differential ultracentrifugation is common, it can co-pellet non-vesicular contaminants. Consider these alternatives:
- Size Exclusion Chromatography (SEC): This method effectively separates exosomes from soluble proteins based on size, resulting in higher purity preparations [11].
- Density Gradient Centrifugation: This technique separates components based on buoyant density, offering a high-purity exosome fraction, though it can be more time-consuming [11]. Always pair your isolation with robust characterization (e.g., NTA, Western Blot for CD63, CD81, TSG101) to confirm the presence of exosomes and assess purity.
FAQ 3: What are the primary biological mechanisms that explain why specific miRNAs and proteins are selectively loaded into MSC-exosomes?
Cargo sorting is a regulated process. The key mechanisms include:
- ESCRT-Dependent Pathway: The Endosomal Sorting Complexes Required for Transport (ESCRT) machinery, comprising complexes ESCRT-0, -I, -II, and -III, works with accessory proteins like ALIX and TSG101 to ubiquitinate and sort cargo into intraluminal vesicles (ILVs) that become exosomes [9].
- ESCRT-Independent Pathways: These rely on lipids like ceramide, which promotes membrane invagination, and tetraspanins (e.g., CD63), which help form protein microdomains for cargo selection [9].
- RNA-Binding Proteins: Proteins such as hnRNPs can bind specific RNA sequences and facilitate their packaging into exosomes [12].
Troubleshooting Guides
Problem 1: Low and Inconsistent Yield of Exosomal RNA
Issue: Inadequate quantity or quality of RNA isolated from MSC-exosomes for downstream sequencing or qPCR analysis.
Investigation and Resolution:
| Investigation Step | Recommended Action | Expected Outcome |
|---|---|---|
| Confirm Exosome Yield | Quantify exosome particles before lysis using Nanoparticle Tracking Analysis (NTA). | Verifies sufficient starting material; if low, revisit cell culture and exosome isolation steps. |
| Check Cell Viability | Ensure >90% viability of MSC cultures before exosome collection. Apoptotic cells release different vesicles (apoptotic bodies) [1]. | Improves consistency of exosome population and its cargo. |
| Optimize Lysis Protocol | Use a commercial exosomal RNA isolation kit with a vigorous lysis step. Include a DNase digest step. | Maximizes RNA recovery and removes genomic DNA contamination. |
| Quality Control | Assess RNA quality with a Bioanalyzer (e.g., RIN >7). | Confirms RNA is intact and suitable for sequencing. |
Detailed Protocol: Isolation of Exosomal RNA for miRNA Sequencing
- Exosome Isolation: Isolate exosomes from 20 mL of MSC-conditioned media using differential ultracentrifugation (2000 × g for 30 min, 10,000 × g for 45 min, followed by 100,000 × g for 70 min) [12].
- Validation: Resuspend the pellet and validate exosome presence via Western Blot for CD63 and CD81 and characterize size by NTA.
- RNA Extraction: Lysate the exosome pellet with a denaturing lysis buffer. Perform RNA extraction using acid-phenol:chloroform, followed by precipitation with isopropanol and glycogen.
- rRNA Depletion: Treat the RNA with a ribosomal RNA depletion kit to enrich for miRNA and other small RNAs.
- Library Prep and Sequencing: Construct sequencing libraries using a small RNA library preparation kit and sequence on an appropriate platform (e.g., Illumina) [12].
Problem 2: Unclear Functional Transfer of Exosomal Cargo to Recipient Cells
Issue: Difficulty in verifying that a specific exosomal cargo (e.g., mRNA, lncRNA) is delivered to a recipient cell and is functionally active.
Investigation and Resolution:
| Investigation Step | Recommended Action | Expected Outcome |
|---|---|---|
| Track Exosomes | Label isolated exosomes with a fluorescent lipid dye (e.g., PKH67) and image uptake in recipient cells over 24h. | Confirms physical transfer of exosomes into target cells. |
| Monitor Functional mRNA Transfer | Isolate exosomes from donor MSCs and co-culture with recipient cells. Use qPCR and Western Blot in recipient cells to detect exosome-derived mRNA and its protein product. | Demonstrates functional delivery, as shown with Rab13 mRNA transfer [12]. |
| Employ CRISPR-based Tracking | For non-coding RNAs, use a CRISPR/Cas9-based RNA-tracking system in donor cells with export signals from secreted RNAs [12]. | Allows direct visualization and confirmation of functional RNA transfer to recipient cells. |
Detailed Protocol: Validating Functional mRNA Transfer
- Exosome Collection: Collect exosomes from donor MSCs (e.g., mutant KRAS cells known to enrich specific mRNAs like Rab13) [12].
- Recipient Cell Treatment: Treat recipient cells (e.g., 50% confluency) with 50 μg/mL of isolated exosomes for 48 hours.
- RNA/Protein Isolation: Harvest recipient cells. Split the sample for parallel RNA and protein extraction.
- Downstream Analysis:
- Perform RT-qPCR on the recipient cell RNA using sequence-specific primers for the gene of interest (e.g., Rab13).
- Perform Western Blot on the recipient cell lysates using an antibody against the corresponding protein (e.g., Rab13).
- A significant increase in both the mRNA and protein levels in recipient cells confirms functional transfer.
Experimental Pathways and Workflows
Diagram: Mechanisms of Cargo Sorting in Exosome Biogenesis
Diagram: Troubleshooting Workflow for Exosomal Cargo Heterogeneity
Research Reagent Solutions
Table: Essential Materials for MSC-Exosome Cargo Analysis
| Item | Function/Application in Research |
|---|---|
| CD63 / CD81 / TSG101 Antibodies | Standard markers for identifying and validating exosomes via Western Blot or flow cytometry [9]. |
| ALIX Antibody | Protein marker associated with the ESCRT pathway and exosome biogenesis; used for validation [9]. |
| rRNA Depletion Kits | Critical for library preparation in RNA-seq to enrich for informative coding and non-coding RNAs over abundant ribosomal RNA [12]. |
| Nanoparticle Tracking Analyzer (NTA) | Instrument for determining the size distribution and concentration of exosome particles in a preparation [11]. |
| Size Exclusion Chromatography (SEC) Columns | Used for high-purity isolation of exosomes away from soluble protein contaminants [11]. |
| PKH67 / PKH26 Fluorescent Cell Linkers | Lipophilic dyes used to fluorescently label the exosome membrane for tracking and uptake studies in recipient cells. |
The therapeutic potential of Mesenchymal Stem Cells (MSCs) is significantly influenced by their tissue of origin. Bone marrow (BM), umbilical cord (UC), and adipose tissue (AT) are the most common sources, each imparting distinct functional and compositional characteristics to the cells and their secreted exosomes. Understanding these differences is critical for experimental design and clinical application. The table below summarizes the key comparative characteristics.
Table 1: Key Characteristics of MSC Tissue Sources
| Feature | Bone Marrow (BM) | Umbilical Cord (UC), notably Wharton's Jelly | Adipose Tissue (AT) |
|---|---|---|---|
| Isolation Invasiveness | Highly invasive (bone marrow aspiration) [13] | Non-invasive; uses medical waste (birth tissue) [14] [13] | Minimally invasive (mini-liposuction) [13] |
| Cell Yield & Proliferative Capacity | Lower yield; proliferation potential decreases significantly with donor age [13] [10] | High volume in Wharton's Jelly; cells from younger donors have higher proliferation rates [14] [13] | Very high yield (~5,000 MSCs/gram of tissue); robust proliferative capacity in vitro [13] |
| Donor Age Impact | High impact; cellular function declines with age [14] [13] | Low impact; sourced from birth, representing a "young" cell population [14] | Moderate impact; more resilient to age-related decline than BM-MSCs [13] |
| Differentiation Potential | High osteogenic (bone) affinity; multipotent [13] | Multipotent; limited heterogeneity and some unique properties [14] | Multipotent; high proliferative and differentiation capacity [13] |
| Immunomodulatory Properties | Potent immunomodulation; first MSC product (Prochymal) for GvHD [14] | Strong immunomodulatory properties; lower immunogenicity of UC-MSC exosomes [14] [1] | Strong immunomodulatory and immunosuppressive effects [10] |
| Key Advantages | Extensive historical data and clinical research track record [13] | No ethical concerns; no risk to donor; ease of isolation [14] | High tissue abundance; simple extraction; suitable for autologous therapy [13] |
| Key Challenges | Painful donor procedure; lower cell numbers [13] | For adults, typically requires allogeneic donation with associated matching needs [13] | Not suitable for individuals with very low body fat [13] |
This heterogeneity directly influences the resulting exosome populations. MSC-derived exosomes from different sources have been shown to possess different molecular cargoes (proteins, miRNAs) and, consequently, varying therapeutic efficacies for specific disease models [1]. For instance, BM-MSC exosomes highly inhibit inflammatory cells, while UC-MSC exosomes are particularly effective at suppressing oxidative stress [1].
Frequently Asked Questions (FAQs) & Troubleshooting
Q1: Our team is getting inconsistent results in our exosome-based angiogenesis assays. Could the MSC tissue source be a factor?
Yes, absolutely. The angiogenic potential of MSC-exosomes varies by source. For example, UC-MSC exosomes have been highlighted for their strong pro-angiogenic effects, promoting blood vessel formation in fracture healing and cardiovascular disease models [1] [15]. BM-MSC exosomes also supply pro-angiogenic factors to damaged tissues [15]. If your assays are inconsistent, first confirm and standardize your MSC source. Furthermore, consider pre-conditioning strategies (e.g., 3D culture or hypoxia) to enhance and standardize the angiogenic cargo of your exosomes [16].
Q2: We are scaling up exosome production but are concerned about donor-related variability. Which source is most suitable?
For large-scale production, source consistency is paramount.
- Umbilical Cord (Wharton's Jelly): Often the preferred choice for allogeneic biobanking because the cells are derived from a young, healthy donor population, reducing age and health-related variability [14] [17].
- Adipose Tissue: Provides a high initial yield of MSCs, which is advantageous for scaling [13]. However, for autologous therapies, variability between individual donors (age, health status) will persist.
- Bone Marrow: Generally less suitable due to lower cell yield and higher sensitivity to donor age [13].
To mitigate variability, implement strict donor screening criteria and use early-passage cells, as cellular aging in long-term culture alters exosome production and functionality [18].
Q3: Why are our intravenously injected MSC-exosomes not homing effectively to the target tissue?
The homing efficiency of MSCs and their exosomes is influenced by multiple factors. The tissue source can affect the expression of homing receptors (e.g., CXCR4), which can be lost during in vitro culture [14]. Furthermore, a significant proportion of intravenously infused MSCs/exosomes can become trapped in capillary networks, particularly in the lungs [14]. Troubleshooting Steps:
- Characterize Homing Markers: Check the expression of key homing-related receptors (like CXCR4) on your parent MSCs.
- Consider Alternative Administration Routes: For localized injuries, direct injection (e.g., intra-arterial, intramyocardial) can dramatically increase the number of exosomes reaching the target site [14].
- Explore Engineering Strategies: Surface modification of exosomes with targeting ligands (e.g., RGD peptides) can improve their specific binding to injured tissues [15] [16].
Detailed Experimental Protocol: Preconditioning MSCs to Modulate Exosome Cargo
A key strategy to address heterogeneity and enhance exosome potency is to precondition MSCs before exosome collection. The following protocol details a method using hypoxic conditioning, which mimics the physiological niche and can enhance the therapeutic properties of secreted exosomes [16].
Protocol: Hypoxic Preconditioning of MSCs to Enhance Angiogenic Exosome Yield
Objective: To increase the production and angiogenic potential of exosomes derived from MSCs.
Materials:
- Cell Source: Mesenchymal Stem Cells (e.g., from BM, UC, or AT) at passage 3-5.
- Basal Medium: DMEM/F-12.
- Supplements: Fetal Bovine Serum (FBS), exosome-depleted. Penicillin-Streptomycin.
- Culture Vessels: T-175 flasks.
- Hypoxia Chamber/Workstation: Capable of maintaining 1-3% O₂, 5% CO₂, and balance N₂.
- Standard Cell Culture Incubator: (Normoxic conditions: 21% O₂, 5% CO₂).
- Other Reagents: PBS, Trypsin-EDTA.
Method:
- Cell Seeding: Seed MSCs at a consistent density (e.g., 5,000 cells/cm²) in T-175 flasks using complete medium supplemented with 10% exosome-depleted FBS.
- Pre-attachment: Incubate the cells under standard normoxic conditions (21% O₂, 5% CO₂) for 24 hours to allow for cell attachment.
- Hypoxic Induction: After 24 hours, carefully move the experimental group of flasks to the hypoxia chamber, pre-set to 1% O₂, 5% CO₂, at 37°C. The control group remains in the normoxic incubator.
- Conditioning Period: Culture the cells under these respective conditions for 48-72 hours.
- Collection of Conditioned Medium: After the conditioning period, carefully collect the culture medium from all flasks.
- Exosome Isolation: Isolate exosomes from the conditioned medium using your standard method (e.g., ultracentrifugation, size-exclusion chromatography). It is critical to process normoxic and hypoxic samples in parallel.
- Characterization: Characterize the isolated exosomes for particle size and concentration (NTA), specific markers (CD63, CD81, TSG101 via western blot), and morphology (TEM). To confirm enhanced efficacy, perform functional assays, such as a tube formation assay using Human Umbilical Vein Endothelial Cells (HUVECs).
Workflow Diagram:
Signaling Pathways: MSC Exosome-Mediated Angiogenesis
The pro-angiogenic effect of MSC-exosomes is a key therapeutic mechanism, particularly in cardiovascular and bone regeneration. The following diagram illustrates the primary signaling pathway by which MSC-exosomes can promote the formation of new blood vessels.
Diagram: MSC Exosome-mediated Angiogenic Signaling Pathway
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for MSC Exosome Research
| Reagent / Material | Function / Application | Key Considerations |
|---|---|---|
| Exosome-Depleted FBS | Essential for cell culture to prevent contamination of isolated exosomes with bovine vesicles [18]. | Validate the depletion efficiency. Alternatives include using serum-free media or human platelet lysate. |
| Isolation Kits (e.g., Precipitation) | Rapid isolation of exosomes from conditioned medium or biofluids [15] [18]. | Can co-precipitate contaminants like proteins and lipoproteins. Not suitable for all downstream applications. |
| Antibodies for Characterization | Identification of exosomal markers (CD63, CD81, CD9, TSG101, Alix) via western blot or flow cytometry [15] [16]. | Always include negative markers (e.g., calnexin, GM130) to confirm exosomal purity. |
| Nanoparticle Tracking Analysis (NTA) | Measures the size distribution and concentration of particles in an exosome preparation [15]. | The gold standard for physical characterization. Tunable Resistive Pulse Sensing (TRPS) is another common alternative. |
| PKH67 / Other Lipophilic Dyes | Fluorescent labeling of exosomes for tracking their uptake by recipient cells in vitro [15]. | Can form dye aggregates that are mistaken for exosomes. Proper controls are critical. |
| 3D Culture Systems (e.g., Bioreactors) | Scalable production of MSCs and exosomes in an environment that more closely mimics the in vivo niche [16]. | Can significantly increase exosome yield and enhance biological activity compared to 2D culture. |
Troubleshooting Guides
FAQ 1: How Does Donor Age Affect the Cargo and Function of MSC-Derived Exosomes?
Problem Statement: Researchers observe inconsistent therapeutic outcomes in experiments using MSC-derived exosomes from different donors and suspect donor age as a contributing factor.
Explanation: Donor age is a critical factor introducing heterogeneity in MSC-derived exosomes. Aging impacts the molecular cargo of exosomes, shifting their functional profile from regenerative to senescent or pro-inflammatory. This occurs because MSCs themselves undergo functional decline with age, which is reflected in their secretome, including exosomes [10] [19].
Solution:
- Action 1: Source MSCs from Younger Donors: When targeting regenerative applications, prioritize MSCs from younger donors, such as umbilical cord tissue (UC-MSCs) or Wharton’s Jelly (WJ-MSC). Studies indicate these sources often exhibit superior regenerative and anti-aging potential compared to those from older adults [10] [19].
- Action 2: Pre-screen Exosome Cargo: For critical experiments, pre-characterize exosome cargo from donors of different ages. Key markers to analyze include:
- Action 3: Consider Pooling Strategies: To mitigate individual age-related variability, create a pool of exosomes isolated from MSCs derived from multiple, age-matched donors. This can help average out extreme phenotypes and produce a more consistent product [19].
Preventive Measures: Establish a standardized donor screening and age-tracking protocol for your cell bank. Clearly document the donor age for every MSC line and its corresponding exosome batch.
FAQ 2: How Do Underlying Health Conditions of the Donor Influence Exosome Function in Disease Models?
Problem Statement: MSC-derived exosomes intended for a disease model (e.g., cancer) show unexpected effects, potentially because the MSCs were isolated from a donor with an unrelated health condition.
Explanation: The health status of the donor directly shapes MSC phenotype and the composition of their exosomes. Cells derived from diseased individuals can produce exosomes with altered molecular profiles that may carry pathological cargo or have impaired therapeutic functionality [22] [19]. For instance, MSCs from diabetic or obese donors may have a different secretome profile compared to those from healthy individuals [19].
Solution:
- Action 1: Implement Rigorous Donor Health Screening: Define strict health exclusion criteria for donors. These should encompass chronic conditions (diabetes, cardiovascular disease, autoimmune disorders), infectious diseases, and recent surgical history [20] [21].
- Action 2: Functional Validation in a Relevant Assay: Do not rely solely on marker expression. Before committing a batch to a large study, functionally test the exosomes in a small-scale, disease-specific assay to confirm the intended biological effect.
- Action 3: Use Disease-Specific Donor Cells with Caution: If studying a specific disease, using MSCs from a diseased donor might be intentional. However, be aware that this introduces a specific and highly variable phenotype. Always compare against a healthy donor control to understand the disease-specific contribution [22].
Preventive Measures: Maintain detailed and annotated donor health records. For the most consistent results in fundamental research, source MSCs from healthy, screened donors whenever possible.
FAQ 3: How Can We Control for Genetic Background and Inter-Individual Variation in Preclinical Studies?
Problem Statement: Significant experiment-to-experiment variability is observed, even when using MSCs from donors of the same age and health status, likely due to inherent genetic and individual differences.
Explanation: Even among demographically similar healthy donors, inherent genetic and epigenetic differences lead to inter-individual variation in MSCs. This results in differences in their proliferation, differentiation potential, and exosome cargo, creating a "batch effect" that is a major challenge in clinical translation [10] [19]. Single-cell RNA sequencing has confirmed that MSCs are a heterogeneous population with distinct functional subpopulations, the balance of which can vary between individuals [19].
Solution:
- Action 1: Standardize the MSC Source: For a given study or project, use MSCs isolated from a single, well-characterized tissue source (e.g., only umbilical cord or only adipose tissue) to minimize source-induced heterogeneity [1] [10].
- Action 2: Characterize Multiple Donor Lines: If possible, establish and characterize several MSC lines from different donors. Test their exosomes for key properties (cargo, uptake, function in a target assay) and select the one with the most desirable and consistent profile for your research [19].
- Action 3: Employ a Pooling Strategy: As with age, pooling exosomes from multiple, genetically diverse but phenotypically screened donors can create a more standardized and reproducible research material, reducing the impact of any single donor's outlier characteristics [19].
Preventive Measures: Build a characterized biobank of MSC lines. Report the specific donor tissue source, passage number, and all characterization data (e.g., surface markers, differentiation potential) for the MSCs used to produce exosomes in any publication, as per ISCT guidelines [10] [19].
Table 1: Summary of Age-Related Changes in MSC-Derived and Plasma Exosomes
| Parameter | Change with Aging | Experimental Evidence | Functional Consequence |
|---|---|---|---|
| Exosome Concentration | Decreases in plasma [23]. MSC source impact is area of active research. | Longitudinal study showed EV concentration in plasma decreased over ~5 years and correlated with age [23]. | May indicate altered intercellular communication and reduced availability of signaling vesicles. |
| Pro-apoptotic Cargo | Increases (e.g., BAX/BCL-2 ratio) [21]. | HSCs treated with exosomes from older donors showed significant upregulation of BAX and downregulation of BCL-2 [21]. | Can promote apoptotic pathways in recipient cells, potentially counteracting regenerative processes. |
| Aging-Related Markers | Increase (e.g., P21 protein) [20]. | HSCs treated with exosomes from older donors showed significantly increased P21 protein expression [20]. | Induces cell cycle arrest and contributes to a senescent phenotype in target cells. |
| Regenerative Signaling | Decreases (e.g., HIF-1α expression) [20]. | HSCs treated with exosomes from younger donors showed increased HIF-1α gene expression, which was decreased with exosomes from older donors [20]. | Impairs cellular responses to hypoxia, a key mechanism in tissue repair and stem cell maintenance. |
Table 2: Impact of Donor Tissue Source on MSC-Exosome Properties
| Tissue Source | Reported Functional Specialization | Key References / Rationale |
|---|---|---|
| Bone Marrow (BM-MSC) | Immunomodulation; B-cell maturation/activation [1]. | Considered the "gold standard" source, widely used for immune-related studies. |
| Umbilical Cord (UC-MSC) | Suppression of oxidative stress; angiogenesis; wound healing [1] [10]. | Younger cell source with high proliferative potential and strong paracrine activity. |
| Adipose Tissue (AD-MSC) | Wound healing; treatment of inflammation and transplantation [1]. | Easily accessible, used prominently in plastic/aesthetic surgery and wound healing research. |
| Pluripotent Stem Cell-Derived (Pluri-MSC) | Treatment of liver, musculoskeletal diseases; low immunogenicity [1]. | Offers a scalable and potentially more uniform source, but requires careful differentiation. |
Experimental Protocols
Protocol 1: Assessing the Functional Impact of Donor Age on Exosomes in a Hematopoietic Stem Cell (HSC) Aging Model
Objective: To evaluate the effect of young vs. old donor-derived plasma exosomes on markers of aging (HIF-1α, P21) in HSCs [20].
Materials:
- Plasma from young (e.g., 25-44 years) and old (e.g., 60-75 years) male donors, health-screened [20] [21].
- Umbilical cord blood for HSC isolation.
- Ficoll-Paque for density gradient centrifugation.
- MACS CD34+ isolation kit and magnetic separator.
- RPMI-1640 culture medium with antibiotics.
- Ultracentrifuge.
- Phosphate Buffered Saline (PBS).
- BCA Protein Assay Kit.
- TEM, DLS/Zeta sizer, and CD63 antibody for exosome characterization.
- qRT-PCR setup for HIF-1α.
- Western Blot setup for P21 protein.
Methodology:
- Exosome Isolation (Ultracentrifugation):
- Thaw plasma and dilute 1:3 in PBS.
- Centrifuge at 17,000 × g for 30 min at 4°C to remove debris and apoptotic bodies.
- Transfer supernatant to ultracentrifuge tubes and centrifuge at 100,000 × g for 75 min at 4°C.
- Resuspend pellet in 2 mL PBS and filter through a 0.22 µm filter.
- Dilute filtered suspension to 13 mL with PBS and repeat ultracentrifugation (100,000 × g, 75 min, 4°C).
- Resuspend final pellet in 0.5-1 mL PBS.
- Quantify exosome protein concentration using BCA assay [20] [21].
- Exosome Characterization:
- HSC Isolation:
- Isolate mononuclear cells from cord blood using Ficoll density gradient centrifugation.
- Isolate CD34+ HSCs using the MACS CD34+ MicroBead kit according to the manufacturer's protocol [20].
- Cell Treatment & Analysis:
- Culture isolated HSCs and treat with exosomes (e.g., 5 µg/mL and 10 µg/mL concentrations) from young and old donors for 24 hours. Include an untreated control.
- Viability Check: Perform MTT assay to confirm non-toxicity of exosome doses.
- Gene Expression: Extract RNA and perform qRT-PCR to analyze HIF-1α mRNA expression.
- Protein Expression: Perform Western Blot on cell lysates to detect P21 protein levels [20].
Expected Outcome: HSCs treated with exosomes from older donors are expected to show decreased HIF-1α mRNA and increased P21 protein, indicating a pro-aging effect, compared to those treated with exosomes from younger donors.
This experimental workflow is outlined in the diagram below:
Protocol 2: Evaluating Pro-Apoptotic Effects of Donor-Derived Exosomes
Objective: To determine if exosomes from older donors can induce a pro-apoptotic shift in the BAX/BCL-2 ratio in recipient HSCs [21].
Materials: (As in Protocol 1, with a focus on apoptosis markers.) Methodology:
- Exosome Isolation & Characterization: Follow steps 1 and 2 from Protocol 1.
- HSC Isolation & Treatment: Follow steps 3 and 4 from Protocol 1.
- Apoptosis Marker Analysis:
- After 24-hour treatment with exosomes, extract RNA from HSCs.
- Perform qRT-PCR using primers for BAX (pro-apoptotic) and BCL-2 (anti-apoptotic) genes.
- Calculate the BAX/BCL-2 mRNA expression ratio, a key indicator of apoptotic tendency [21].
Expected Outcome: HSCs treated with exosomes from older donors are expected to show a significantly higher BAX/BCL-2 ratio compared to the control and young exosome-treated groups.
Signaling Pathways
The molecular mechanisms underlying the aging effects mediated by exosomes involve key signaling pathways. The following diagram integrates findings from the provided research, showing how exosomal cargo from aged donors can influence recipient HSCs, promoting a senescent and pro-apoptotic phenotype.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Studying Donor-Derived Exosome Variability
| Reagent / Material | Function / Application | Key Considerations |
|---|---|---|
| Ficoll-Paque | Density gradient medium for isolation of peripheral blood mononuclear cells (PBMCs) or HSCs from cord blood [20] [21]. | Ensure proper density and osmolarity for the specific cell type being isolated. |
| MACS CD34+ MicroBead Kit | Immunomagnetic positive selection for highly pure CD34+ Hematopoietic Stem Cells [20] [21]. | Purity should be confirmed by flow cytometry (e.g., >90% CD34+CD45-). |
| Ultracentrifuge | Gold-standard equipment for high-speed isolation of exosomes from plasma or cell culture media [20] [21] [24]. | Protocol parameters (g-force, time, temperature) must be strictly standardized for reproducibility. |
| CD63 Antibody | Primary antibody for Western Blot confirmation of exosomal markers, validating successful isolation [20] [21] [24]. | Part of the MISEV guidelines for minimal characterization of extracellular vesicles. |
| Dynamic Light Scattering (DLS) / NTA | Instruments for determining exosome size distribution and particle concentration [20] [23] [24]. | NTA is often preferred for its direct visualization and sizing capabilities. |
| TEM (Transmission Electron Microscopy) | Imaging technique for confirming the classic "cup-shaped" morphology and size of isolated exosomes [20] [24]. | Requires specialized equipment and sample preparation. |
| BCA Protein Assay Kit | Colorimetric quantification of total protein in exosome samples, used for normalizing doses in functional experiments [20] [21]. | A common and sensitive method for protein quantification in dilute samples. |
Frequently Asked Questions (FAQs) on Exosome Heterogeneity
Q1: What is exosome heterogeneity, and why is it a critical consideration in research?
Exosome heterogeneity refers to the existence of distinct subpopulations of exosomes that differ in their biophysical properties, molecular composition, and biological functions. Cells do not release a single, uniform population of exosomes but rather a diverse mixture of vesicles [25] [26]. This heterogeneity arises from variations in biogenesis pathways, the cell's physiological state, and the cell source [1] [27]. Recognizing this is critical because different exosome subpopulations can have unique and sometimes opposing effects on recipient cells. Ignoring this complexity can lead to inconsistent experimental results and misinterpretation of data.
Q2: Are there specific markers that can identify all exosomes or their subpopulations?
Currently, there is no single, universal marker that identifies all exosomes or exclusively defines specific subpopulations [28] [29]. The research community recommends a combination of markers for verification. The most commonly used tetraspanins (CD9, CD63, CD81) are found in many exosome preparations but are not universally present; for example, Jurkat cells release exosomes that are CD9 negative [28]. Other common markers include ESCRT-related proteins like ALIX and TSG101 [26]. It is equally important to test for the absence of contaminants from other cellular compartments using markers for the endoplasmic reticulum (e.g., calnexin), Golgi (e.g., GM130), mitochondria (e.g., cytochrome C), and nucleus (e.g., histones) [28].
Q3: How does the source of Mesenchymal Stem Cells (MSCs) impact exosome heterogeneity?
The tissue source of MSCs is a major determinant of exosome heterogeneity, leading to variations in their molecular cargo and therapeutic functions [1]. For instance:
- Bone marrow MSC-EVs highly inhibit inflammatory and apoptotic cells and affect B-cell maturation [1].
- Umbilical cord MSC-EVs suppress oxidative stress in kidney injury and promote angiogenesis for wound healing [1].
- Pluripotent stem cell-derived MSC-EVs show low immunogenicity and are used for liver and musculoskeletal diseases [1]. This indicates that the choice of MSC source should be tailored to the specific therapeutic application.
Q4: What are the primary challenges in loading therapeutic cargo into exosomes?
Loading cargo into exosomes remains a significant technical challenge. While various strategies exist, including incubation, electroporation, sonication, extrusion, freeze-thaw cycling, and transfection, inadequate loading efficiency is a common problem [30]. Each method has potential drawbacks, such as causing exosome aggregation, damaging the exosome membrane, or being inefficient for certain types of cargo (e.g., small molecules, nucleic acids, or proteins) [30]. The field is actively developing more efficient and gentle loading techniques to enable reliable exosome-based drug delivery.
Q5: How should exosomes be stored to maintain stability?
Exosomes can be stored in PBS with 0.1% BSA [28]. Isolation efficiency is not changed after freezing at -80°C compared to freshly made exosomes. For direct isolation from cell culture media or urine, freezing without cryo-protectants like glycerol is possible [28]. However, standardized protocols for long-term storage are still an area of investigation to ensure functional consistency.
Troubleshooting Common Experimental Issues
Issue 1: Inconsistent Functional Effects in Recipient Cells
Potential Cause: The problem may stem from the unrecognized heterogeneity of your exosome preparation. Your isolated "exosome" sample is likely a mixture of subpopulations, and variations in the relative abundance of these subpopulations between preparations can lead to inconsistent biological outcomes [25] [26].
Solution:
- Characterize Subpopulations: Move beyond basic characterization. Use techniques like density gradient centrifugation to separate and identify distinct subpopulations within your exosome samples [26]. Research shows that low-density exosomes (LD-Exo) and high-density exosomes (HD-Exo) have different protein and RNA contents and elicit differential effects on recipient cells [26].
- Standardize Production: Control upstream process parameters that influence heterogeneity. This includes using a consistent MSC source, culture conditions (2D vs. 3D, bioreactors), and medium composition [1] [31]. One study demonstrated that using a defined system (Hollow Fiber 3D bioreactor with RoosterBio exosome-harvesting system) allowed for stable production of exosome subpopulations over a 28-day culture period, ensuring more consistent bioactive components [31].
Issue 2: Low Yield or Purity of Isolated Exosomes
Potential Cause: The isolation method may be inefficient, may not be suited to your starting material, or may be co-isolating contaminants.
Solution:
- Optimize Isolation Protocol: The choice of isolation method depends on the downstream application and starting material. Ultracentrifugation is common but can lead to vesicle loss and aggregation. Consider alternatives like size-exclusion chromatography (SEC), which preserves exosome integrity and improves purity [26].
- Combine Methods: For complex samples like plasma, a combination of methods is often necessary. A recommended approach is to perform a pre-clearing step using SEC prior to isolation with immunocapture beads (e.g., targeting CD9) [28].
- Validate Purity: Always confirm the purity of your isolates by checking for the absence of organelle-specific markers (e.g., calnexin for ER) [28].
Issue 3: Difficulty in Tracking Exosome Uptake by Recipient Cells
Potential Cause: The mechanisms of exosome uptake and delivery are not fully understood and can be cell-type specific, involving endocytosis, direct fusion, or receptor-ligand interactions [32].
Solution:
- Use Multiple Tracking Methods: Employ a combination of fluorescent lipid dyes to label membranes and fluorescently tagged exosomal cargo (e.g., miRNAs or proteins) to track both the vesicle and its contents [32].
- Investigate Biodistribution: For in vivo studies, use advanced tracking methods. One study successfully used isotopic labeling with Zirconium-89 (89Zr) to trace the biodistribution of intravenously injected MSC exosomes in rats, finding predominant accumulation in the liver [31]. This highlights the importance of administration route selection.
Quantitative Data on Exosome Subpopulations
Table 1: Characteristics of Distinct Exosome Subpopulations Isolated from B16F10 Melanoma Cells [26]
| Subpopulation | Density (g/ml) | Peak Size (nm) | Key Proteomic Features | Functional Impact on Recipient Cells |
|---|---|---|---|---|
| Low-Density Exosomes (LD-Exo) | 1.12 - 1.19 | 117 nm | Enriched in proteins involved in endocytosis, membrane trafficking, and signal transduction. | Mediated distinct alterations in the gene expression programs of recipient cells. |
| High-Density Exosomes (HD-Exo) | 1.26 - 1.29 | 66 nm | Enriched in ribosomal proteins, translation initiation factors, and mitochondrial proteins. | Mediated distinct alterations in the gene expression programs of recipient cells. |
Table 2: Impact of MSC Source on Exosome Function [1]
| MSC Tissue Source | Documented Therapeutic Effects / Specialties |
|---|---|
| Bone Marrow | Inhibition of inflammatory/apoptotic cells; impact on B-cell maturation; most prevalent source in research. |
| Umbilical Cord | Suppression of oxidative stress in kidney injury; promotion of angiogenesis for wound healing. |
| Adipose Tissue | Used for skin, inflammation, and transplantation diseases; less used in cancer or pancreatic diseases. |
| Placenta | Used for a diversity of diseases, except for autoimmune conditions. |
| Pluripotent Stem Cell-Derived | Low immunogenicity; used for liver, inflammation, transplantation, and musculoskeletal diseases. |
Key Experimental Protocols
Protocol 1: Isolation of Exosome Subpopulations via Density Gradient Centrifugation
This protocol is adapted from a key study that demonstrated the existence of functionally distinct exosome subpopulations [26].
Workflow Diagram: Isolation of Exosome Subpopulations
Materials:
- Ultracentrifuge and fixed-angle or swinging-bucket rotors
- Sucrose solutions in deuterium oxide (D2O) or heavy water (e.g., 2.5 M, 2.0 M, 1.3 M, 1.16 M, 0.8 M, 0.5 M, 0.25 M)
- Phosphate-Buffered Saline (PBS)
- Opti-MEM or exosome-depleted FBS medium
Procedure:
- Collect conditioned medium from cells grown in Opti-MEM or medium with exosome-depleted FBS.
- Pre-clear the medium of cells and debris by centrifugation at 2,000 × g for 20 minutes, followed by 10,000 × g for 30 minutes.
- Pellet total exosomes by ultracentrifugation of the supernatant at 110,000 × g for 70 minutes.
- Resuspend the exosome pellet (P110) in a small volume of PBS or gradient base solution.
- Load the resuspended exosomes at the bottom of a discontinuous sucrose density gradient.
- Perform ultracentrifugation in a swinging-bucket rotor at 210,000 × g for 16 hours to allow exosomes to float to their equilibrium density.
- Collect fractions from the top of the gradient. Typically, low-density exosomes (LD-Exo) will be found in fractions with a density of 1.12-1.19 g/ml, and high-density exosomes (HD-Exo) will be found in denser fractions (1.26-1.29 g/ml) [26].
- Analyze fractions for particle concentration (NTA), marker expression (Western Blot), and density.
Protocol 2: Evaluating the Functional Consistency of MSC-Derived Exosomes
This protocol is based on a recent study that established a long-term biomanufacturing workflow for MSC-exosomes [31].
Workflow Diagram: Evaluating Functional Consistency of MSC-Exosomes
Materials:
- Hollow Fiber 3D Bioreactor system
- RoosterBio exosome-harvesting system (or similar defined medium)
- hUC-MSCs (or other MSC sources)
- Characterized exosome purification kit (e.g., based on SEC or precipitation)
- Disease model (e.g., silica-induced mouse silicosis model)
Procedure:
- Expand MSCs in a Hollow Fiber 3D bioreactor integrated with an exosome-promoting system to ensure scalable production.
- Harvest exosomes repeatedly over an extended period (e.g., over 28 days) from the same culture to assess production stability.
- Purify and rigorously characterize the exosomes at each harvest point. Key characterization includes:
- Particle concentration and size (via NTA)
- Marker expression (CD9, CD63, CD81 via Western Blot or flow cytometry)
- Subpopulation profile analysis (e.g., via density gradient) to check for consistency over time.
- Evaluate therapeutic efficacy in a relevant disease model. The cited study used a silica-induced mouse silicosis model.
- Compare administration routes. Importantly, the study found that respiratory delivery significantly improved disease progression, whereas intravenous infusion did not yield notable therapeutic effects in this pulmonary model [31]. This underscores the critical importance of route selection.
- Correlate the consistent subpopulation profile with the stable therapeutic outcome to define an optimal collection window for production.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents and Kits for Exosome Heterogeneity Research
| Reagent / Kit | Function / Application | Example & Notes |
|---|---|---|
| Tetraspanin Antibodies | Detection and validation of exosome markers via Western Blot, Flow Cytometry. | Anti-CD9, CD63, CD81. Note: Not all exosomes express all tetraspanins (e.g., Jurkat exosomes are CD9-). Validate for your system [28]. |
| ESCRT Protein Antibodies | Detection of exosome biogenesis markers. | Anti-ALIX, Anti-TSG101. Commonly used as positive markers for exosomes [26]. |
| Contaminant Antibodies | Assessing purity of exosome isolates by detecting non-exosomal proteins. | Anti-Calnexin (ER), Anti-GM130 (Golgi), Anti-Cytochrome C (Mitochondria), Anti-Histones (Nucleus) [28]. |
| Dynabeads (Immuno-capture) | Isolation of specific exosome subpopulations based on surface markers. | Exosome Human CD9/CD63/CD81 Isolation Reagents. Useful for enriching subpopulations from complex samples like plasma [28]. |
| RoosterBio Exosome System | Scalable production of MSC-exosomes in 3D bioreactors. | Includes culture media and harvesting system. Enables long-term, stable production of exosomes with consistent subpopulations [31]. |
| Sucrose/Density Gradient Media | Separation of exosome subpopulations based on buoyant density. | Sucrose or Nycodenz solutions. Critical for resolving low-density and high-density exosome subpopulations [26]. |
Strategies and Engineering Solutions to Control and Harness Exosome Heterogeneity
The production of mesenchymal stem cell (MSC)-derived exosomes begins with upstream bioprocessing, which encompasses all initial steps from cell culture to the point of harvest. This phase is critical in biopharmaceutical production as it establishes the foundation for the yield, quality, and heterogeneity of the final exosome product [33] [34]. Upstream process optimization focuses on cultivating cells to produce therapeutic extracellular vesicles (EVs), including exosomes, by providing a meticulously controlled environment for cell growth [33].
For MSC-derived exosomes, upstream processing presents a unique challenge: the heterogeneity of the final product is intrinsically linked to the conditions in which the parent cells are cultivated [1]. The composition of exosomes—their cargo of proteins, RNA, and lipids—is largely determined by the cell source and its physiological state [1]. Furthermore, process parameters such as the culture system (2D vs. 3D), medium composition, the use of bioreactors, and exposure to hypoxia can crucially affect the resulting therapeutic properties and biological functions of the exosomes [1]. Therefore, optimizing upstream processes is not merely about increasing yield; it is about controlling and directing the inherent variability of MSC-derived exosomes for specific therapeutic applications, such as heart repair, immunomodulation, and drug delivery [1] [35].
Key Challenges in Upstream Optimization
Researchers face several interconnected challenges when optimizing upstream processes for MSC exosome production. The table below summarizes the primary hurdles and their direct impacts on exosome yield and quality.
Table 1: Key Challenges in Upstream Process Optimization for MSC Exosomes
| Challenge Category | Specific Challenge | Impact on MSC Exosomes |
|---|---|---|
| Culture System | Transitioning from 2D to 3D culture systems [36] | Alters exosomal RNA content, secretion efficiency, and therapeutic efficacy [37] [35]. |
| Process Control | Maintaining precise environmental control (pH, O₂, temperature) [33] | Deviations decrease cell growth and product yield, affecting exosome cargo [33]. |
| Cell Source & Health | Managing MSC source (bone marrow, umbilical cord, adipose) and health [1] | Different sources produce exosomes with varying molecular composition and functional specificity [1]. |
| Scalability | Scaling up from laboratory to industrial production [33] | Fluid dynamics and mass transfer changes can alter exosome characteristics and yield [33]. |
| Heterogeneity | Controlling exosome population and cargo diversity [1] [11] | Influences batch-to-batch consistency, therapeutic reproducibility, and functional reliability [1]. |
The Scientist's Toolkit: Essential Research Reagents & Materials
Successful upstream optimization relies on a foundation of high-quality materials and reagents. The following table details essential components for experiments aimed at controlling MSC exosome heterogeneity.
Table 2: Essential Research Reagents and Materials for Upstream Process Optimization
| Item | Function/Application | Key Considerations |
|---|---|---|
| Chemically Defined Media | Provides essential nutrients, vitamins, and growth factors without introducing unknown variables from serum [34]. | Enables process consistency; critical for optimizing nutrients like amino acids and trace elements for specific exosome cargo [1] [34]. |
| 3D Scaffolds & Matrices | Provides an ECM-mimicking 3D environment for cell growth (e.g., HYDROX, hydrogels) [36]. | Influences cell morphology, differentiation, and the yield and molecular content of secreted exosomes [1] [36]. |
| Microcarriers | Beads that provide a high surface-to-volume ratio for 3D cell culture in stirred-tank bioreactors [36]. | Facilitates the scale-up of MSC cultures for large-volume exosome production [36]. |
| Bioreactor Systems | Provides a controlled environment (temperature, pH, O₂, agitation) for cell cultivation at various scales [33] [36]. | Perfusion bioreactors enable high cell densities and improved productivity; dynamic mechanical stimulation affects cell behavior and exosome output [33] [36]. |
| Cell Lines | Source of exosomes (e.g., Bone Marrow MSCs, Umbilical Cord MSCs, Adipose-derived MSCs) [1]. | The choice of MSC source dictates the baseline protein and RNA footprint of the derived exosomes, affecting their therapeutic function [1]. |
Experimental Protocols for Key Upstream Processes
Protocol: Establishing a 3D Spheroid Culture for Enhanced Exosome Yield
Objective: To create a 3D culture environment that increases the yield and modifies the cargo of MSC-derived exosomes compared to traditional 2D monolayer culture [37] [35].
Materials:
- Mesenchymal Stem Cells (e.g., from bone marrow or umbilical cord)
- Low-attachment U-bottom 96-well plates or hanging drop plates
- Complete cell culture medium (e.g., DMEM/F12 supplemented with specific growth factors)
- Centrifuge
- Phosphate Buffered Saline (PBS)
Methodology:
- Cell Preparation: Harvest and count MSCs from a 2D culture. Create a single-cell suspension with a viability of >95%.
- Seeding for Spheroid Formation:
- For U-bottom plates: Prepare a cell suspension at a concentration of 1x10⁵ cells/mL. Aliquot 100 µL per well (10,000 cells/well) into the low-attachment plate.
- For Hanging drop plates: Place a 20 µL drop of cell suspension (at a higher density, e.g., 2.5x10⁴ cells/drop) onto the lid of a culture dish, which is then inverted.
- Incubation: Culture the plates at 37°C with 5% CO₂ for 72-96 hours. Monitor daily for spheroid formation. Compact, spherical structures should form within this period.
- Harvesting Spheroids and Conditioned Media: After spheroid formation, carefully collect the culture medium (conditioned media) containing the secreted exosomes by centrifugation at 300 x g for 5 minutes to remove any loose cells or debris.
- Exosome Isolation: Proceed with standard exosome isolation techniques (e.g., ultracentrifugation, size-exclusion chromatography) from the collected conditioned media.
Data Interpretation: The success of spheroid formation is confirmed by visual inspection under a microscope. A successful protocol will demonstrate a higher yield of exosomes and potentially different miRNA or protein profiles compared to 2D culture-derived exosomes, which can be verified by nanoparticle tracking analysis and western blotting, respectively [37] [35].
Protocol: Optimizing a Perfusion Bioreactor for Continuous Production
Objective: To utilize a perfusion bioreactor system to maintain high cell densities over an extended period for continuous harvest of MSC-derived exosomes [33] [34].
Materials:
- Stirred-tank bioreactor system with perfusion capabilities (including cell retention device)
- MSC seed train culture
- Bioreactor-specific culture medium
- Peristaltic pumps for media addition and harvest
- pH, dissolved oxygen (DO), and temperature probes and controllers
Methodology:
- Bioreactor Setup and Sterilization: Assemble the bioreactor vessel and all fluid paths. Sterilize in-place (SIP) or autoclave components as per manufacturer's instructions.
- Inoculation: Transfer the expanded MSC culture from the seed train into the bioreactor vessel to achieve a target initial cell density (e.g., 2.0x10⁵ cells/mL).
- Parameter Control: Set and maintain critical process parameters (CPPs):
- Temperature: 37°C
- pH: 7.2 (controlled through CO₂ sparging and base addition)
- Dissolved Oxygen (DO): 40% air saturation (controlled by adjusting air/O₂ sparging and agitation speed)
- Agitation: Use a low-shear impeller to prevent cell damage while ensuring homogeneity.
- Initiating Perfusion: Once the cell density reaches a pre-defined threshold (e.g., 2.0x10⁶ cells/mL), initiate the perfusion process.
- Start adding fresh medium at a defined perfusion rate (e.g., 1 vessel volume per day).
- Simultaneously, harvest spent medium through the cell retention device (e.g., acoustic settler, tangential flow filtration) to keep the working volume constant.
- Continuous Monitoring and Harvest: Monitor cell density, viability, and nutrient/metabolite levels (e.g., glucose, lactate) daily. The harvested spent medium from the perfusion process is the source for continuous exosome isolation.
Data Interpretation: A stable perfusion process will maintain high cell viability (>90%) and a steady-state cell density for several weeks. The exosome yield per day from the harvest stream will be significantly higher and more consistent than from a batch or fed-batch process [33].
Diagram 1: Upstream process parameter influence on exosome output.
Troubleshooting Guides & FAQs
Frequently Asked Questions
Q1: Why is my 3D MSC spheroid culture producing exosomes with different cargo profiles than my 2D culture? The 3D culture environment more accurately mimics the in vivo physiological conditions, altering cell-to-cell and cell-to-matrix interactions. This change in the cellular microenvironment directly influences the molecular sorting mechanisms that load cargo into exosomes. Consequently, exosomes from 3D cultures often have RNA and protein profiles that are more representative of in vivo conditions and can exhibit enhanced therapeutic efficacy [37] [36].
Q2: How can I increase the yield of exosomes from my MSC cultures without scaling up vessel size? Transitioning to a 3D culture system, such as using microcarriers in a bioreactor or forming spheroids, can significantly increase cell density and thus exosome yield per unit volume compared to 2D monolayers [36] [35]. Furthermore, implementing a perfusion bioreactor system allows for continuous nutrient supply and waste removal, supporting very high cell densities over prolonged periods and enabling the continuous harvest of exosomes from the spent media [33] [34].
Q3: My bioreactor parameters (pH, DO) are deviating from setpoints. What is the immediate impact on my exosome product? Deviations in critical process parameters like pH and dissolved oxygen (DO) can cause immediate cellular stress. This stress alters the metabolic state of the MSCs, which in turn can change the cargo (e.g., stress-related miRNAs, proteins) packaged into exosomes and potentially affect their yield. Consistent deviation can lead to increased batch-to-batch heterogeneity, compromising product consistency and therapeutic reproducibility [33] [1].
Q4: Does the source of my MSCs (e.g., bone marrow vs. umbilical cord) matter for the resulting exosomes? Yes, the source is a primary determinant of exosome heterogeneity. MSCs from different tissues (bone marrow, umbilical cord, adipose) have distinct molecular and functional identities. These differences are reflected in their exosomes, which will have varying protein, lipid, and RNA compositions. This means exosomes from different sources may have preferential efficacy for specific therapeutic applications (e.g., bone marrow for immunomodulation, umbilical cord for angiogenesis) [1].
Troubleshooting Common Problems
Table 3: Troubleshooting Common Upstream Process Issues
| Problem | Potential Causes | Solutions & Recommendations |
|---|---|---|
| Low Exosome Yield | 1. Suboptimal cell viability/density.2. Nutrient depletion in media.3. Using 2D instead of 3D culture. | 1. Monitor cell health and optimize medium formulation [33].2. Switch to fed-batch or perfusion modes [34].3. Transition to a 3D culture system (spheroids, bioreactors) [35]. |
| High Heterogeneity Between Batches | 1. Inconsistent culture conditions.2. Uncontrolled MSC differentiation.3. Variations in serum lots (if used). | 1. Strictly control CPPs (pH, DO, temp) using bioreactors [33].2. Monitor MSC surface markers and limit passages.3. Use chemically defined, serum-free media [1] [34]. |
| Contamination in Bioreactor | 1. Failure in sterilization procedures.2. Leak in seals or tubing. | 1. Validate sterilization protocols (e.g., SIP).2. Perform pre-culture leak tests and integrity checks. |
| Poor Cell Growth in 3D System | 1. Excessive shear stress in bioreactor.2. Inadequate nutrient diffusion in spheroids.3. Incorrect matrix/scaffold choice. | 1. Optimize agitation speed; use low-shear impellers [36].2. Control spheroid size to prevent necrotic cores.3. Screen different 3D matrices for your MSC type [1]. |
Diagram 2: Troubleshooting high exosome heterogeneity.
Mesenchymal stem cell-derived exosomes (MSC-exosomes) have emerged as a promising cell-free therapeutic platform, offering the regenerative and immunomodulatory benefits of their parent cells without the risks associated with live cell transplantation [16] [38]. However, a significant challenge hindering their clinical translation is heterogeneity—variations in exosome yield, molecular cargo, and subsequent biological activity [1]. This heterogeneity stems from differences in MSC sources, culture conditions, and the physiological state of the cells [1].
Preconditioning strategies involve exposing MSCs to specific environmental cues before collecting their exosomes. This process is not merely a stress response; it is a method to deliberately steer the MSC phenotype, thereby tailoring the content and enhancing the functional consistency and efficacy of the resulting exosomes [16] [39] [40]. By controlling these variables, researchers can actively combat heterogeneity and produce exosome populations with more predictable and potent therapeutic profiles for applications in regenerative medicine, immunomodulation, and drug delivery [16] [41].
Frequently Asked Questions (FAQs) on Preconditioning
Q1: How does preconditioning specifically address the problem of heterogeneity in MSC-exosome research? Preconditioning addresses heterogeneity by providing a defined and controlled stimulus to the parent MSCs. This stimulus creates a more uniform cell population that, in turn, secretes exosomes with a more consistent and enriched cargo profile. For instance, hypoxia preconditioning consistently upregulates pro-angiogenic miRNAs like miR-126 and miR-210-3p across different MSC sources [42] [39] [43]. This standardization reduces batch-to-batch variability and enhances the reliability of experimental and therapeutic outcomes.
Q2: What are the key signaling pathways activated by hypoxia preconditioning, and how do they alter exosome cargo? Hypoxia preconditioning primarily activates the HIF-1α (Hypoxia-Inducible Factor 1-alpha) signaling pathway [42]. HIF-1α acts as a master regulator, leading to specific changes in the exosomal cargo:
- miRNA Enrichment: Upregulates miRNAs such as miR-125a-5p (targeting RTEF-1 to protect endothelial cells [43]), miR-210-3p (promoting angiogenesis [42]), and miR-486-5p (inhibiting MMP19 to enhance angiogenesis [41]).
- Protein Enrichment: Increases the load of pro-angiogenic factors like VEGF (Vascular Endothelial Growth Factor) and angiopoietin-1 [42]. The mechanism also involves upstream regulators like HMGB1, which activates the JNK pathway to induce HIF-1α/VEGF expression [42].
Q3: Can inflammatory preconditioning make exosomes too immunosuppressive, potentially promoting cancer growth? The dual role of MSC-exosomes in cancer is a critical consideration. While preconditioning with factors like TNF-α or IFN-γ enhances immunomodulatory miRNAs (e.g., miR-146a and miR-21-5p) for anti-inflammatory therapy [39], there is a theoretical risk that this could suppress immune surveillance in an oncological context. The effect is highly dependent on the specific cytokine, dose, MSC source, and tumor microenvironment [44]. Therefore, thorough safety and efficacy testing in relevant disease models is mandatory before clinical application.
Q4: How do I choose between different preconditioning strategies for my specific research application? The choice of preconditioning strategy should be directly aligned with your desired therapeutic outcome. The following table provides a guideline:
| Desired Therapeutic Outcome | Recommended Preconditioning Strategy | Key Mediators in Exosomes |
|---|---|---|
| Angiogenesis & Vascular Repair | Hypoxia (1-5% O₂) [42] [41] | miR-125a-5p, miR-210-3p, miR-486-5p, VEGF [42] [43] [41] |
| Anti-inflammatory & Immunomodulation | Inflammatory Cytokines (e.g., IFN-γ, TNF-α, IL-1β) [39] [41] | miR-146a, miR-21-5p, miR-181a [39] |
| Anti-apoptosis & Cell Survival | Hypoxia [42]; Drugs (e.g., Atorvastatin [41]) | miR-125a-5p, miR-21, Bcl-2/Bcl-xL proteins [42] [43] |
| Chondrogenic Differentiation | Chemical Agents (e.g., Kartogenin/KGN [41]) | Chondrogenic inducing factors (specific miRNAs/proteins under investigation) |
| Scalable Production | 3D Culture [16] | Varies based on 3D system and MSC source |
Troubleshooting Common Preconditioning Experiments
Problem 1: Low Exosome Yield After Preconditioning
- Potential Cause: The preconditioning stimulus (e.g., severe hypoxia, high cytokine dose) is cytotoxic, reducing MSC viability and proliferation.
- Solutions:
- Titrate the stimulus: Systemically test different intensities (e.g., 1% vs. 5% O₂ for hypoxia [42]) and durations to find the optimal window that enhances cargo without killing cells.
- Switch to 3D Culture: Implement 3D culture systems, such as hollow fiber bioreactors or scaffold-based cultures, which can significantly increase both MSC biomass and exosome production (up to 19.4-fold compared to 2D culture [16]).
- Monitor Cell Health: Always assess cell viability and apoptosis (e.g., via flow cytometry) after preconditioning to ensure the regimen is not overly toxic.
Problem 2: Preconditioned Exosomes Do Not Show Enhanced Therapeutic Efficacy
- Potential Cause 1: Inefficient uptake of exosomes by the target cells.
- Solutions:
- Use fluorescent lipophilic dyes (e.g., PKH67, DiR) to label exosomes and confirm their internalization by recipient cells via fluorescence microscopy or flow cytometry.
- Potential Cause 2: The cargo loading was insufficient or not relevant to the disease model.
- Solutions:
- Validate Cargo Enrichment: After preconditioning, use qRT-PCR or RNA-Seq to confirm the upregulation of target miRNAs (e.g., check for miR-146a after TNF-α treatment [39]) in the isolated exosomes. Do not assume enrichment has occurred.
- Confirm Mechanism: Use inhibitor or knockout experiments to validate the functional role of a specific miRNA. For example, transfecting a miR-125a-5p inhibitor into recipient cells can block the beneficial effects of H-EXO [43].
Problem 3: Inconsistent Results Between Batches
- Potential Cause: Uncontrolled heterogeneity in starting materials or preconditioning parameters.
- Solutions:
- Standardize MSC Sources: Characterize MSCs thoroughly (ISCT standards) and use low-passage cells to avoid replicative senescence [16] [1].
- Control the Environment: Use specialized hypoxia workstations or incubators for precise, continuous O₂ control instead of chemical hypoxia mimetics, which can introduce variability [42].
- Document Rigorously: Meticulously record all parameters: passage number, preconditioning agent concentration, duration, and cell confluence at the time of treatment.
Detailed Experimental Protocols
Protocol 1: Hypoxia Preconditioning for Pro-angiogenic Exosomes
This protocol is designed to enhance the angiogenic potential of MSC-exosomes.
Workflow Overview:
Key Reagent Solutions:
| Item | Function in Protocol | Example & Note |
|---|---|---|
| Hypoxia Chamber/Workstation | Maintains precise, low oxygen tension | Use a tri-gas incubator for precise control of O₂, CO₂, and N₂. |
| Serum-Free Medium | Prevents contamination with bovine exosomes from FBS | Use exosome-depleted FBS if serum is absolutely required. |
| Ultracentrifugation | Gold-standard for exosome isolation | Alternative: Size-Exclusion Chromatography (SEC) for higher purity [1]. |
| CD63/CD81/TSG101 Antibodies | Confirms exosome identity via Western Blot | Use a combination of positive markers. |
| miR-210-3p Assay | Validates functional cargo enrichment | Key miRNA for hypoxia response [42]. |
Step-by-Step Method:
- Cell Preparation: Culture human MSCs (e.g., from bone marrow or umbilical cord) to 70-80% confluence in standard culture flasks.
- Medium Change: Gently wash cells with PBS and replace the medium with serum-free basal medium. This is critical to avoid contaminating the sample with exogenous vesicles from fetal bovine serum (FBS).
- Hypoxic Exposure: Place the flasks in a pre-equilibrated hypoxia chamber or workstation set to 1-5% O₂, 5% CO₂, balance N₂, at 37°C. A 24 to 48-hour incubation is commonly used [42] [41].
- Conditioned Medium Collection: After incubation, collect the conditioned medium and centrifuge it at 300 × g for 10 min to remove cells, followed by 2,000 × g for 20 min to remove dead cells and debris.
- Exosome Isolation: Isolve exosomes from the supernatant using ultracentrifugation (100,000-120,000 × g for 70 min) or size-exclusion chromatography (SEC) for a purer preparation [1].
- Validation: Resuspend the exosome pellet and characterize using:
- Nanoparticle Tracking Analysis (NTA): For size and concentration.
- Transmission Electron Microscopy (TEM): For morphology.
- Western Blot: For positive exosome markers (CD63, CD81, TSG101) and negative markers (e.g., Calnexin).
- Functional Cargo Check: Perform qRT-PCR to confirm the enrichment of hypoxia-specific miRNAs like miR-210-3p or miR-125a-5p [42] [43].
Protocol 2: Inflammatory Preconditioning with TNF-α for Immunomodulatory Exosomes
This protocol enhances the anti-inflammatory properties of MSC-exosomes.
Workflow Overview:
Key Reagent Solutions:
| Item | Function in Protocol | Example & Note |
|---|---|---|
| Recombinant Human TNF-α | The inflammatory preconditioning agent | Aliquot to avoid freeze-thaw cycles; test dose response (10-20 ng/mL [39]). |
| Control IgG Antibody | Isotype control for functional assays | - |
| anti-miR-146a Inhibitor | Validates mechanistic role | Transfect into recipient cells to block exosome effect [39]. |
| Macrophage Cell Line | Functional validation of immunomodulation | e.g., THP-1 or primary human monocytes. |
Step-by-Step Method:
- Cell Preparation: Seed MSCs and allow them to reach 60-70% confluence.
- Cytokine Stimulation: Add recombinant human TNF-α to the culture medium at a final concentration of 10-20 ng/mL [39]. Include an untreated control group.
- Incubation: Incubate the cells under normoxic conditions (21% O₂, 5% CO₂, 37°C) for 24-48 hours.
- Collection and Isolation: Collect the conditioned medium and isolate exosomes following the same centrifugation and isolation steps described in Protocol 1.
- Validation: Characterize the isolated exosomes using NTA, TEM, and Western Blot.
- Cargo and Functional Validation:
- qRT-PCR: Confirm the upregulation of miR-146a in the treated exosomes compared to controls [39].
- Functional Assay: Co-culture the preconditioned exosomes with lipopolysaccharide (LPS)-stimulated macrophages. Assess the induction of M2 polarization (e.g., via CD206 staining by flow cytometry) and the reduction of pro-inflammatory cytokines (e.g., TNF-α, IL-6) by ELISA.
Essential Research Reagent Solutions
The following table lists key reagents for implementing and validating preconditioning strategies.
| Category | Reagent / Tool | Primary Function in Preconditioning Research |
|---|---|---|
| Preconditioning Agents | Tri-Gas Hypoxia Incubator | Provides precise, physiological (1-5% O₂) low-oxygen environment [42]. |
| Recombinant Human Cytokines (IFN-γ, TNF-α, IL-1β) | Precondition MSCs to enhance immunomodulatory exosome cargo [39] [41]. | |
| Lipopolysaccharide (LPS) | Bacterial endotoxin used to mimic inflammatory stress at low doses (0.1-1 μg/mL) [39]. | |
| Kartogenin (KGN) | Small molecule to precondition MSCs for enhanced chondrogenic exosomes [41]. | |
| Exosome Isolation & Analysis | Ultracentrifuge | Gold-standard instrument for pelleting exosomes from conditioned medium. |
| Size-Exclusion Chromatography (SEC) Columns | Purifies exosomes with high specificity, reducing protein contaminants [1]. | |
| Antibodies: CD63, CD81, TSG101 | Western Blot validation of exosome identity (positive markers) [1]. | |
| Nanoparticle Tracking Analyzer (NTA) | Measures exosome size distribution and concentration. | |
| Functional Validation | miRNA Assays (qRT-PCR) | Quantifies specific miRNA enrichment (e.g., miR-146a, miR-21, miR-125a) [39] [43]. |
| miRNA Mimics/Inhibitors | Validates the causal role of specific exosomal miRNAs in recipient cell effects [43]. | |
| Human Umbilical Vein Endothelial Cells (HUVECs) | Standard cell model for in vitro angiogenesis (tube formation) assays [41]. | |
| Macrophage Cell Line (e.g., THP-1) | Model for testing exosome-induced immunomodulation and M2 polarization [39] [41]. |
FAQs: Core Concepts and Strategic Planning
Q1: What is the primary advantage of genetically engineering parent MSCs over directly modifying the isolated exosomes? Genetically engineering the parent Mesenchymal Stem Cells (MSCs) is a biological modification strategy that leverages the cell's own natural machinery to load specific therapeutic cargo (proteins, nucleic acids) into exosomes during their biogenesis. This endogenous loading method often results in higher encapsulation efficiency and better preservation of cargo bioactivity compared to many passive loading techniques applied to already-isolated exosomes. Furthermore, this approach can be used to modify the exosome surface with targeting ligands (e.g., by expressing fusion proteins like CXCR4), enhancing their homing capability to specific tissues [45].
Q2: Which genetic modification tools are most suitable for engineering parent MSCs? The choice depends on the desired outcome and experimental constraints:
- Viral Transduction (e.g., Lentivirus, Adenovirus): Highly efficient for stable, long-term gene expression in MSCs. This is ideal for endogenously overexpressing proteins, miRNAs, or targeting ligands in MSC-derived exosomes [45].
- CRISPR/Cas9: Best for precise gene knockout (e.g., knocking in a gene of interest) or modifying endogenous genes to alter exosome cargo or biogenesis. Delivery into MSCs can be achieved via viral vectors or non-viral methods like lipid nanoparticles (LNPs) [46] [47].
- Non-Viral Methods (e.g., Electroporation, Lipofection): Useful for transient transfection with plasmid DNA or siRNA. While often lower in efficiency and persistence than viral methods, they pose fewer safety concerns related to insertional mutagenesis [48] [45].
Q3: How can I address the challenge of low yield of engineered exosomes? Scalable production remains a key hurdle. Strategies include:
- 3D Bioreactor Culture: Transitioning from 2D flasks to 3D culture systems, such as hollow fiber bioreactors, can increase exosome yield by up to 20-fold while maintaining vesicle quality and biological activity [49].
- Cell Preconditioning: Stimulating MSCs with specific agents like thrombin or bioactive glass (45S5 Bioglass) can enhance paracrine secretion, potentially increasing exosome production by more than four times [49].
- Immortalization: Transfecting MSCs with genes like MYC can create immortalized clones that proliferate indefinitely, ensuring a consistent and expandable source of engineered exosomes. However, oncogenic safety concerns must be carefully evaluated [49].
Troubleshooting Guides: Common Experimental Issues
Issue 1: Low Cargo Loading Efficiency into Exosomes
| Potential Cause | Diagnostic Steps | Recommended Solution |
|---|---|---|
| Inefficient MSC Transduction/Transfection | Measure transfection efficiency via FACS (for reporter genes) or qPCR in parent MSCs. | Optimize viral titer (MOI) or non-viral reagent-to-DNA ratio. Use high-efficiency systems like lentivirus for stable expression. |
| Cargo Not Directed to Exosome Biogenesis Pathway | Analyze parent cell lysates vs. isolated exosomes via Western blot for cargo presence. | Fuse cargo gene to exosome-enriched protein tags (e.g., CD63, CD9, CD81, Lamp2b) to actively shuttle it into intraluminal vesicles [32] [45]. |
| Cargo Size or Structure Disrupts Exosome Formation | Perform nanoparticle tracking (NTA) and TEM on isolated exosomes; check for altered size/morphology. | Consider using smaller cargo variants (e.g., engineered Cas proteins) or split-protein systems if the cargo is too large. |
Issue 2: Inadequate Target Cell Specificity of Engineered Exosomes
| Potential Cause | Diagnostic Steps | Recommended Solution |
|---|---|---|
| Lack of Specific Targeting Ligands | Validate exosome surface expression of the engineered ligand via flow cytometry or immuno-EM. | Genetically engineer parent MSCs to express exosome surface proteins fused with targeting moieties (e.g., CXCR4 for tumor homing, or RGD peptides for angiogenesis sites) [45]. |
| Non-Specific Uptake by Mononuclear Phagocyte System | Track biodistribution of labeled exosomes in vivo in animal models. | Modify the parent MSC culture conditions to enrich exosomes with "self" markers (e.g., CD47) to evade phagocytic clearance. |
Issue 3: Heterogeneity and Impurity of Isolated Engineered Exosomes
| Potential Cause | Diagnostic Steps | Recommended Solution |
|---|---|---|
| Suboptimal Isolation Technique | Characterize preparations with multiple methods (NTA, TEM, Western blot for positive CD63/CD81 and negative calnexin/GM130 markers). | Combine isolation techniques. Use density gradient centrifugation as a "gold standard" to follow concentration steps like tangential flow filtration (TFF) for higher purity [50]. |
| Inherent Heterogeneity of Parent MSCs | Use single-vesicle analysis techniques to characterize subpopulations. | Use low-passage MSCs and standardize culture conditions. Implement clonal selection of parent MSCs after genetic engineering to ensure a uniform starting population [51] [52]. |
Experimental Protocols for Key Workflows
Protocol: Genetic Modification of Parent MSCs using Lentiviral Vectors
This protocol outlines the process for generating MSCs that stably express an exosome-targeted therapeutic cargo.
Workflow Diagram: Lentiviral Engineering of MSCs
Materials:
- Parent MSCs (e.g., Bone Marrow or Umbilical Cord derived, low passage)
- Lentiviral Transfer Plasmid (e.g., pLVX with GOI fused to CD63/Lamp2b)
- Lentiviral Packaging Mix (e.g., psPAX2, pMD2.G)
- Transfection Reagent (e.g., PEI or commercial lipofectamine)
- HEK293T cells for virus production
- Polybrene (to enhance transduction)
- Appropriate Antibiotic (e.g., Puromycin) for selection
Step-by-Step Method:
- Virus Production: Co-transfect HEK293T cells with the transfer plasmid and packaging plasmids using PEI. Harvest the virus-containing supernatant at 48 and 72 hours post-transfection. Concentrate the supernatant using ultracentrifugation or PEG-it virus precipitation solution.
- MSC Transduction: Plate MSCs at 60-70% confluence. Replace the medium with fresh medium containing the concentrated lentivirus and polybrene (e.g., 8 µg/mL). Spinoculate by centrifuging plates at 800-1000 x g for 30-60 minutes at 32°C. Incubate for 24 hours before replacing with fresh medium.
- Selection and Cloning: 48 hours post-transduction, begin selection with the appropriate antibiotic. Maintain selection pressure for at least 1-2 weeks, changing the medium every 3-4 days, until distinct resistant colonies form. Use limited dilution or FACS to isolate single-cell clones.
- Validation: Expand clonal populations. Validate the expression of the GOI at the mRNA level (qRT-PCR) and protein level (Western blot of cell lysates). Confirm that the protein is successfully packaged into isolated exosomes.
Protocol: Isolation and Purification of Engineered Small Extracellular Vesicles (sEVs)
This protocol describes a standardized method for isolating high-purity sEVs from conditioned media of engineered MSCs.
Workflow Diagram: sEV Isolation via Ultracentrifugation
Materials:
- Conditioned Media from engineered MSCs (serum-free, centrifuged)
- Ultracentrifuge with fixed-angle or swinging-bucket rotors
- Polycarbonate Bottles or Thin-Walled Tubes compatible with ultracentrifugation
- Phosphate-Buffered Saline (PBS), sterile and cold
- 0.22 µm PVPF syringe filter
Step-by-Step Method:
- Pre-Clearing: Centrifuge the conditioned media at 300 × g for 10 minutes to remove floating cells. Transfer supernatant to a new tube and centrifuge at 2,000 × g for 20 minutes to remove dead cells and debris. Filter the supernatant through a 0.22 µm filter.
- Ultracentrifugation (UC): Transfer the filtered supernatant to ultracentrifuge tubes. Balance tubes precisely. Pellet sEVs by ultracentrifugation at 100,000 × g for 90 minutes at 4°C.
- Washing: Carefully discard the supernatant. Resuspend the pellet in a large volume of cold PBS to remove contaminating proteins. Perform a second ultracentrifugation step under the same conditions (100,000 × g, 90 minutes).
- Final Resuspension: Discard the supernatant and resuspend the final, purified sEV pellet in a small volume (e.g., 50-200 µL) of PBS or an appropriate storage buffer. Aliquot and store at -80°C.
The Scientist's Toolkit: Essential Research Reagents
| Reagent / Material | Function in Experiment | Key Considerations |
|---|---|---|
| Lentiviral Packaging System (psPAX2, pMD2.G) | Production of replication-incompetent lentiviral particles for stable gene expression in MSCs. | Use 3rd generation systems for enhanced safety. Always handle in BSL-2 containment [45]. |
| Polybrene | A polycation that reduces charge repulsion between virions and the cell membrane, increasing transduction efficiency. | Titrate for optimal performance (typical range 4-8 µg/mL); can be cytotoxic at high concentrations. |
| Puromycin Dihydrochloride | Antibiotic selection agent for cells transduced with a puromycin resistance gene. | Perform a kill curve on untransduced MSCs to determine the minimal effective concentration (typically 1-5 µg/mL). |
| Polyethylenimine (PEI), linear | High-efficiency, low-cost transfection reagent for plasmid DNA, used for producing lentivirus in HEK293T cells. | PEI/DNA complexes are formed in serum-free medium before addition to cells [45]. |
| Dulbecco's Phosphate Buffered Saline (DPBS) | Washing cells and diluting reagents; used as a resuspension buffer for the final sEV pellet. | Always use calcium- and magnesium-free PBS for cell washing and trypsin neutralization. |
| Serum-Free Mesenchymal Stem Cell Medium | For producing conditioned media free of bovine exosome contaminants. | Essential for all steps during sEV production and harvest to avoid FBS-derived vesicle contamination. |
| Protease and Phosphatase Inhibitor Cocktails | Added to lysis buffers and sometimes to PBS during sEV isolation to prevent degradation of cargo proteins and phosphoproteins. | Use a broad-spectrum, ready-to-use solution. |
| RIPA Lysis Buffer | For efficient lysis of parent MSCs and isolated sEVs to extract total protein for Western blot validation. | Contains strong detergents (SDS, Triton X-100) for complete solubilization. |
| Antibodies for Characterization: Anti-CD63, Anti-CD81, Anti-TSG101, Anti-Calnexin | Western blot analysis to confirm sEV enrichment (CD63, CD81, TSG101) and the absence of cellular contaminants (Calnexin, a negative marker). | Use a combination of positive and negative markers to ensure sEV purity [50] [32]. |
| PKH67/PKH26 Lipophilic Dyes | For fluorescently labeling the membrane of isolated sEVs to track their uptake in vitro or biodistribution in vivo. | Dye can form micelles; always include a control with dye alone and use extensive post-labeling purification [32]. |
Table 1: Reported Efficacy of Different Genetic Cargo in MSC-sEVs
| Cargo Type | Target Disease (Model) | Engineering Method | Key Outcome Metric | Reported Result | Citation |
|---|---|---|---|---|---|
| miR-29a-3p (Overexpression) | Glioma (in vivo) | Lentiviral transduction of parent MSCs | Inhibition of migration & vasculogenic mimicry | Significant inhibition of tumor growth and VM formation | [45] |
| miR-124a (Overexpression) | Glioblastoma (GBM) (Mouse model) | Transfection of parent MSCs | Tumor growth inhibition & survival | Improved survival rates in mouse models | [45] |
| Pigment Epithelium-Derived Factor (PEDF) (Overexpression) | Cerebral Ischemia-Reperfusion Injury (in vivo/vitro) | Overexpression in Ad-MSCs | Reduction in brain injury | Mitigated injury via apoptosis/autophagy modulation | [45] |
| CXCR4 (Overexpression) + si-Survivin (Loaded) | Tumor Targeting (Experimental) | Engineering MSCs for CXCR4high sEVs + Electroporation | Targeted delivery & gene silencing | Created a novel targeted gene-drug delivery system | [45] |
Table 2: Comparison of sEV Isolation Techniques
| Method | Principle | Average Yield | Pros | Cons | Suitability for Engineered sEVs |
|---|---|---|---|---|---|
| Differential Ultracentrifugation (dUC) | Sequential centrifugation based on size/density | Medium | "Gold standard," high purity, scalable | Time-consuming, requires specialized equipment, potential vesicle damage | High - Widely used for research-scale prep [50] |
| Density Gradient Centrifugation | Separation based on buoyant density | Low | Very high purity, separates sEVs from contaminants | Complex, low yield, time-consuming | High - Ideal for final purification step [50] |
| Tangential Flow Filtration (TFF) | Size-based separation using filters | High | Gentle process, highly scalable, good for large volumes | Requires specialized equipment, lower purity than dUC | High - Excellent for pre-concentration and scalable processing [53] [49] |
| Size-Exclusion Chromatography (SEC) | Size-based separation in a column | Low-Medium | Good purity, preserves vesicle integrity, simple | Diluted samples, low capacity | Medium - Good for final polishing step of concentrated samples [50] |
| Polymer-Based Precipitation | Reduction of solubility using polymers | High | Simple, fast, no special equipment | Co-precipitation of contaminants (e.g., proteins), lower purity | Low - Not recommended for therapeutic-grade sEVs due to impurity issues [49] |
What is the primary goal of direct exosome surface engineering? The primary goal is to enhance the targeting specificity and therapeutic efficacy of exosomes by deliberately modifying their surface to display functional ligands, peptides, or antibodies that direct them to particular cell types or tissues, thereby minimizing off-target effects [54].
How does direct surface engineering fit into the broader context of addressing heterogeneity in MSC exosome research? Mesenchymal Stem Cell (MSC) exosomes inherently exhibit significant heterogeneity in their population and cargo, which can lead to inconsistent experimental and therapeutic outcomes [55]. Direct surface engineering provides a strategy to impose a layer of uniformity and control. By consistently equipping a proportion of MSC exosomes with a defined targeting moiety, researchers can potentially enrich for a subpopulation that performs a desired function, thereby reducing the variable impact of heterogeneity on targeting efficiency [56] [54].
Troubleshooting Guides
Low Efficiency in Chemical Conjugation
Problem: Low yield of ligand attachment to the exosome surface using chemical methods.
| Possible Cause | Verification Experiment | Solution |
|---|---|---|
| Insufficient reactive groups | Measure the concentration of surface amines/thiols via colorimetric assays (e.g., Ellman's reagent for thiols). | Increase the molar ratio of ligand-to-exosome during reaction; consider lipid insertion with pre-functionalized lipids [54]. |
| Reduced exosome integrity | Perform nanoparticle tracking analysis (NTA) pre- and post-reaction; check for degradation of marker proteins (CD63, CD81) via Western blot [57] [58]. | Optimize reaction conditions (pH, temperature, solvent); use milder catalysts or shift to physical modification methods like extrusion [54]. |
| Improper purification post-conjugation | Use size-exclusion chromatography (SEC) to separate conjugated exosomes from free ligand and analyze fractions [58]. | Implement a robust purification protocol (e.g., SEC, ultrafiltration) to effectively remove unreacted ligands [59]. |
Poor Targeting Specificity
Problem: Engineered exosomes show insufficient specificity for target cells in vitro or in vivo.
| Possible Cause | Verification Experiment | Solution |
|---|---|---|
| Loss of ligand functionality | Use ELISA or surface plasmon resonance (SPR) to confirm the binding affinity of the conjugated ligand. | Employ a different conjugation chemistry that does not impair the ligand's active site; use a flexible PEG spacer [56] [54]. |
| Incomplete characterization of target receptor | Perform flow cytometry or immunofluorescence on the target cell line to confirm receptor expression levels. | Select a target receptor with higher and more specific expression on the target cell population [54]. |
| Non-specific uptake by off-target cells | Use imaging (e.g., confocal microscopy) to track the cellular uptake of labeled exosomes in a co-culture system. | Re-engineer exosomes with "do not eat me" signals (e.g., CD47) to reduce non-specific phagocytosis [56]. |
Reduced Exosome Integrity Post-Engineering
Problem: Engineered exosomes exhibit aggregation, fusion, or cargo leakage.
| Possible Cause | Verification Experiment | Solution |
|---|---|---|
| Harsh modification conditions | Analyze exosome size and polydispersity index (PDI) via NTA; use cryo-electron microscopy for morphology [58]. | Switch from sonication or extrusion to gentler methods like incubation or freeze-thaw cycles for certain cargoes [57]. |
| Membrane disruption from hydrophobic insertion | Conduct a cargo retention assay (e.g., measure encapsulated dye release) before and after modification. | Titrate the amount of inserted lipid-conjugated ligand to find a balance between functionalization and membrane integrity [54]. |
Frequently Asked Questions (FAQs)
Q1: What are the main strategies for directly engineering the exosome surface? The three main strategies are:
- Genetic Engineering: Modifying parent cells to express targeting ligands fused to exosomal membrane proteins (e.g., Lamp2b, CD63). This is a pre-isolation method [59] [54].
- Chemical Conjugation: Using chemical linkers (e.g., Click chemistry, NHS-PEG-Maleimide) to covalently attach ligands to amine or sulfhydryl groups on exosome surface proteins post-isolation [56] [54].
- Physical Modification: Utilizing hydrophobic insertion, electrostatic interactions, or membrane fusion techniques (e.g., extrusion, freeze-thaw) to attach ligands to the exosome membrane post-isolation [57] [54].
Q2: How do I choose the best surface modification strategy for my target? The choice depends on the ligand and experimental goals, as summarized in the table below.
| Strategy | Best For | Advantages | Limitations |
|---|---|---|---|
| Genetic Engineering | Proteinaceous ligands (e.g., peptides, scFv); stable, long-term expression. | Stable, homogeneous display; high reproducibility [54]. | Requires cell transfection/transduction; not suitable for non-biological ligands (e.g., chemicals, polymers); potential for altered cell/exosome biology [59]. |
| Chemical Conjugation | Broad range of ligands (peptides, antibodies, sugars); precise control over ligand density. | High modularity and flexibility; applicable to pre-isolated exosomes [56] [54]. | Risk of damaging exosome membrane or ligand; requires purification steps; potential batch-to-batch variability [54]. |
| Physical Modification | Lipophilic ligands (e.g., DSPE-PEG-anchor); quick and simple protocols. | Relatively simple and rapid; no harsh chemicals needed [57] [54]. | Ligand attachment can be unstable; may lead to exosome aggregation; lower control over ligand orientation [54]. |
Q3: What are the critical quality control checkpoints after surface engineering? Essential quality control steps include:
- Size and Concentration: Use Nanoparticle Tracking Analysis (NTA) to confirm that engineering did not cause significant aggregation or a shift in size distribution [57] [58].
- Surface Marker Expression: Use Western blot or flow cytometry (Exo-FCM) to confirm the presence of both the engineered ligand and canonical exosome markers (e.g., CD63, CD81) [57].
- Ligand Presence and Function: Use techniques like ELISA, immuno-gold EM, or surface plasmon resonance to verify the presence and binding functionality of the conjugated ligand [58] [54].
- Membrane Integrity: Assess using cryo-electron microscopy or cargo retention assays to ensure the lipid bilayer remains intact [58].
Q4: Our engineered exosomes are internalized by target cells but show inadequate therapeutic effect. What could be wrong? This points to an issue with endosomal entrapment. Even after successful internalization, exosomes can remain trapped in endosomes and degrade without releasing their therapeutic cargo into the cytoplasm. Consider strategies to enhance endosomal escape, such as co-engineering exosomes with endosomolytic peptides (e.g., L17E) or fusogenic lipids [58] [54].
Experimental Protocols
Detailed Protocol: Post-Isonation Ligand Conjugation via Click Chemistry
This protocol describes a reliable method for conjugating azide-modified ligands to DBCO-modified exosomes, known for its high efficiency and bioorthogonality [56] [54].
1. Principle: Strain-promoted azide-alkyne cycloaddition (SPAAC) between dibenzocyclooctyne (DBCO) groups on the exosome surface and azide groups on the targeting ligand allows for covalent linkage without cytotoxic copper catalysts.
2. Reagents and Equipment:
- Purified exosomes (e.g., from MSC culture medium)
- DBCO-PEG4-NHS Ester (commercial reagent)
- Azide-functionalized targeting ligand
- Size-Exclusion Chromatography (SEC) columns (e.g., qEVoriginal)
- Phosphate Buffered Saline (PBS), pH 7.4
- Ultracentrifugation equipment or ultrafiltration devices
3. Step-by-Step Procedure: Step 1: Exosome Surface Modification with DBCO.
- Dilute purified exosomes in PBS (pH ~7.4).
- Prepare a fresh stock of DBCO-PEG4-NHS ester in DMSO.
- Add the DBCO reagent to the exosome solution at a predetermined molar ratio (requires optimization, e.g., 1000:1 reagent-to-exosome ratio). Incubate for 2 hours at room temperature with gentle agitation.
- Purify the DBCO-modified exosomes from unreacted reagent using a SEC column or ultrafiltration. Collect the exosome-containing fractions.
Step 2: Conjugation with Azide-Modified Ligand.
- Add the azide-functionalized ligand to the DBCO-exosome solution. The ratio should be optimized based on the ligand.
- Incubate the reaction mixture for 2-4 hours at room temperature or overnight at 4°C with gentle mixing.
Step 3: Purification of Conjugated Exosomes.
- Pass the final reaction mixture through a SEC column to separate conjugated exosomes from free ligand and reaction by-products.
- Concentrate the purified, conjugated exosomes if necessary. Aliquot and store at -80°C.
4. Key Calculations and Data Interpretation:
- Ligand Coupling Efficiency: Determine using a BCA assay for protein ligands or HPLC for small molecules by comparing the amount of free ligand pre- and post-purification.
- Functional Validation: The success of conjugation must be confirmed by a binding assay (e.g., ELISA, flow cytometry) against the target receptor.
Experimental Workflow Visualization
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function/Application in Direct Exosome Engineering |
|---|---|
| DSPE-PEG-Maleimide | An amphiphilic polymer used for chemical conjugation. The DSPE moiety inserts into the exosome lipid bilayer, while the PEG spacer and Maleimide group allow covalent conjugation to thiol-containing ligands [54]. |
| DBCO-PEG-NHS Ester | A key reagent for copper-free "Click Chemistry." The NHS ester reacts with surface amines on exosomes, displaying DBCO groups that subsequently react with azide-modified ligands [56]. |
| Lentiviral Vectors | Used for genetic engineering of parent MSCs to stably express fusion proteins (e.g., target peptide-Lamp2b) on the exosome surface [54]. |
| Size-Exclusion Chromatography (SEC) Columns | Critical for gentle purification of engineered exosomes, effectively separating them from unreacted dyes, ligands, and protein aggregates while preserving vesicle integrity [58]. |
| Anti-Tetraspanin Antibodies (CD63, CD81, CD9) | Essential for quality control via Western Blot or flow cytometry to confirm the presence of exosomal markers after engineering steps [57]. |
| Lipid-Anchored Peptides | Pre-synthesized constructs where the targeting peptide is linked to a hydrophobic anchor (e.g., cholesterol). These can be inserted into exosomes via simple incubation or extrusion [54]. |
Quality Control Framework Visualization
Frequently Asked Questions (FAQs)
Q1: What is the primary advantage of using MSC-derived exosomes over whole MSC therapies? MSC-derived exosomes offer several key advantages over whole cell therapies. They have lower immunogenicity because they lack replicative function and express lower levels of major histocompatibility complex (MHC) molecules, significantly reducing the risk of immune rejection and carcinogenesis [38] [1]. As acellular nanoparticles, they exhibit enhanced safety profiles and a superior ability to cross biological barriers, such as the blood-brain barrier, without causing embolism [38]. Furthermore, they provide logistical benefits, including easier storage and stability at -80°C for extended periods without losing biological activity, and they can be administered through various routes (topical, intravenous, oral) [38].
Q2: How does heterogeneity in MSC exosome populations affect my therapeutic outcomes? Heterogeneity is a double-edged sword. Exosomes vary in size, cargo (proteins, RNA, lipids), and functional effects based on the MSC source (bone marrow, umbilical cord, adipose tissue), culture conditions (2D vs. 3D, hypoxia), and isolation methods [1]. This inherent variability can lead to inconsistent experimental results and therapeutic efficacy. However, this heterogeneity can also be controlled and leveraged. By systematically optimizing upstream process parameters, researchers can steer the exosome population toward a desired cargo profile and functional output, transforming heterogeneity from a challenge into a tool for precision medicine [1] [60].
Q3: What are the most critical factors to control for producing consistent MSC exosome batches? Producing consistent batches requires strict control over several upstream and downstream processes. The most critical factors are:
- Cell Source: The tissue origin of the MSCs (e.g., bone marrow, umbilical cord) fundamentally defines the baseline exosome characteristics [1].
- Culture Conditions: Parameters like the use of 3D dynamic culture systems, bioreactors, oxygen tension (hypoxia), and medium composition drastically influence exosome yield and cargo [38] [1].
- Isolation and Purification: The choice of method (e.g., ultracentrifugation, size-exclusion chromatography, precipitation) affects the purity, size distribution, and ultimately, the functionality of the final exosome preparation [1] [60].
Q4: Can MSC exosomes be engineered for better targeting? Yes, MSC exosomes can be engineered to enhance their targeting specificity. Surface functionalization techniques allow for the attachment of targeting ligands (e.g., peptides, antibodies) to direct exosomes to specific cell types or tissues [38] [61]. Furthermore, genetic engineering of the parent MSCs can be used to express targeting motifs or enriched therapeutic cargo directly on or within the secreted exosomes [38]. These strategies are central to developing them from general "injectable regenerative factors" into sophisticated "programmable nanomedicines" [38].
Q5: What are the key challenges in scaling up MSC exosome production for clinical use? The transition from lab-scale to clinical-scale manufacturing faces several hurdles [60] [62]. Scalable Production requires moving from flask-based cultures to controlled bioreactor systems that can consistently produce high yields of exosomes with the desired critical quality attributes (CQAs) [62]. Potency and Characterization are challenging due to the complex, multimodal mechanisms of action of exosomes and the lack of standardized, robust assays to define their therapeutic potency [60]. Finally, establishing Good Manufacturing Practice (GMP)-compliant processes for consistent, sterile, and well-characterized exosome products is a significant but essential regulatory challenge [62].
Troubleshooting Common Experimental Issues
Issue 1: Low Exosome Yield from MSC Cultures
Problem: Inadequate quantity of exosomes isolated for downstream experiments or characterization.
Possible Causes and Solutions:
- Cause 1: Suboptimal Cell Culture Health.
- Solution: Ensure MSCs are healthy and in their logarithmic growth phase. Avoid using over-confluent or senescent cells. Regularly monitor cell viability and morphology.
- Cause 2: Inefficient Induction of Exosome Biogenesis.
- Cause 3: Inefficient Isolation Protocol.
- Solution: Validate and potentially switch the isolation method. While ultracentrifugation is common, it can have variable recovery. Consider alternative or complementary methods like tangential flow filtration (TFF) for better scalability and yield [1].
Issue 2: Inconsistent Functional Results in Target Cell Assays
Problem: The same nominal exosome preparation produces variable effects in functional assays (e.g., proliferation, migration, gene expression) between experiments.
Possible Causes and Solutions:
- Cause 1: Uncharacterized Heterogeneity in Exosome Batches.
- Solution: Implement rigorous batch-to-batch characterization. Go beyond simple particle counting (NTA) and protein quantification. Include cargo profiling (e.g., miRNA sequencing, proteomics) and establish a functional potency assay relevant to your therapeutic goal (e.g., T-cell suppression for immunomodulation) to ensure consistency [1] [60].
- Cause 2: Inefficient Uptake by Target Cells.
- Solution: Confirm exosome uptake. Use fluorescently labeled exosomes (e.g., with PKH67 or CellMask dyes) and confirm internalization into your specific target cell line via flow cytometry or confocal microscopy. If uptake is low, consider engineering exosomes for improved targeting [61].
- Cause 3: Misunderstanding the Mechanism of Action (MoA).
- Solution: Consider alternative MoA models. The therapeutic effect may not require direct internalization. The Extracellular Modulation of Cells by EVs (EMCEV) model proposes that exosomes can activate surface receptors and modulate the extracellular environment, affecting many cells without being taken up [60]. Design experiments to test for surface-level signaling events.
Issue 3: Poor Loading Efficiency of Therapeutic Cargo
Problem: Low incorporation of desired drug, nucleic acid, or protein into the isolated exosomes.
Possible Causes and Solutions:
- Cause 1: Suboptimal Loading Method for the Cargo Type.
- Solution: Match the loading technique to the cargo. The table below summarizes standard methods [61].
- Cause 2: Inadequate Purity of Pre-Loaded Exosomes.
- Cause 3: Cargo is Damaged During Loading.
- Solution: For sensitive cargo like RNA, optimize harsh parameters like electroporation voltage and buffer composition. Alternatively, use gentler methods like sonication or saponin-assisted loading. The endogenous loading strategy, where the parent MSCs are transfected or pre-treated to package the cargo during exosome biogenesis, can often yield superior and more natural loading [61].
Table 1: Overview of Exosome Cargo Loading Strategies
| Loading Method | Principle | Best For | Considerations |
|---|---|---|---|
| Electroporation | Uses electrical pulses to create temporary pores in the exosome membrane. | Nucleic acids (siRNA, miRNA), small hydrophilic drugs. | Can cause cargo aggregation or exosome aggregation; optimization required. |
| Sonication | Uses ultrasonic energy to disrupt the exosome membrane. | Hydrophobic and hydrophilic drugs, proteins. | May damage exosome membrane integrity if overdone. |
| Co-incubation | Passive diffusion of cargo across the membrane. | Small, lipophilic molecules. | Simple but often has very low efficiency. |
| Endogenous Loading | Engineering parent MSCs to produce and package the desired cargo. | Proteins, nucleic acids. | Considered more natural; cargo is packaged during biogenesis. |
Experimental Protocols & Workflows
Protocol 1: Standardized Production and Isolation of MSC Exosomes
Objective: To reliably produce and isolate MSC-exosomes with controlled heterogeneity for in vitro functional assays.
Materials:
- Cell Source: Human bone marrow-derived MSCs (passage 3-5).
- Culture Medium: Serum-free MSC medium, supplemented as required.
- Equipment: CO2 incubator, ultracentrifuge, nanoparticle tracking analyzer (NTA), BCA protein assay kit.
Procedure:
- Cell Culture & Conditioning: Grow MSCs to 70-80% confluence. Replace standard growth medium with serum-free medium. For enhanced yield, use a bioreactor or implement a 3D culture system [1]. Culture for 48-72 hours.
- Collection of Conditioned Medium: Collect the conditioned medium and perform sequential centrifugation steps:
- 300 × g for 10 min (remove cells)
- 2,000 × g for 20 min (remove dead cells)
- 10,000 × g for 30 min (remove cell debris and large vesicles)
- Exosome Isolation (Ultracentrifugation): Ultracentrifuge the supernatant at 100,000 × g for 70 minutes at 4°C. Carefully discard the supernatant. Resuspend the pellet (containing exosomes) in sterile PBS.
- Purification: Perform a second ultracentrifugation step (wash) with PBS at 100,000 × g for 70 minutes. Resuspend the final pellet in a small volume of PBS or storage buffer.
- Characterization:
- Concentration & Size: Use NTA to determine particle size distribution and concentration.
- Protein Content: Use BCA assay to quantify total protein.
- Marker Expression: Confirm presence of exosomal markers (CD63, CD81, TSG101) and absence of negative markers (calnexin) via western blot.
Diagram: MSC Exosome Isolation Workflow. This standard protocol outlines the key steps for isolating exosomes from mesenchymal stem cell culture medium via differential ultracentrifugation.
Protocol 2: Engineering Exosomes for Targeted Drug Delivery
Objective: To load a small molecule drug (e.g., Doxorubicin) into MSC-exosomes and functionalize their surface with a targeting peptide (e.g., RGD).
Materials:
- Isolated MSC-exosomes (from Protocol 1).
- Therapeutic drug (e.g., Doxorubicin).
- Targeting peptide (e.g., RGD) with a NHS-PEG-DBCO linker.
- Equipment: Sonication bath, desalting column, incubator.
Procedure:
- Drug Loading via Sonication:
- Mix the exosome preparation with the drug at an optimal ratio (requires optimization).
- Subject the mixture to sonication in a water bath sonicator at 30-50 W for 2-6 cycles (30s sonication, 30s rest on ice).
- Incubate the mixture at 37°C for 30-60 minutes to allow membrane recovery.
- Purification of Loaded Exosomes:
- Use a desalting column (e.g., Sephadex G-25) or size-exclusion chromatography to separate drug-loaded exosomes from unencapsulated free drug.
- Surface Functionalization:
- Click Chemistry Approach: First, incubate exosomes with the NHS-PEG-DBCO linker to conjugate DBCO groups to surface amines.
- Purify the DBCO-labeled exosomes via SEC.
- Incubate with the azide-functionalized RGD peptide to allow covalent conjugation via click reaction.
- Validation:
- Confirm drug loading efficiency via HPLC.
- Verify surface conjugation using flow cytometry (e.g., via a fluorescently tagged peptide) or western blot for the peptide tag.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents and Materials for MSC Exosome Research
| Reagent/Material | Function/Application | Key Considerations |
|---|---|---|
| Serum-Free, Xeno-Free Media | For MSC culture and exosome production. Prevents contamination with bovine exosomes from FBS. | Essential for producing clinically relevant exosomes and for accurate cargo profiling. |
| 3D Culture Scaffolds/Bioreactors | Upstream culture systems to enhance exosome yield and modulate cargo. | Bioreactors provide better control over the microenvironment (pH, O2, nutrients) compared to static 2D culture [38] [1]. |
| Size-Exclusion Chromatography (SEC) Columns | Downstream isolation and purification of exosomes. | Provides high-purity exosome preparations with retained biological activity; superior to precipitation kits for functional studies [1]. |
| Nanoparticle Tracking Analyzer (NTA) | Characterizing particle concentration and size distribution. | A key tool for establishing basic physical attributes of your exosome preparation. |
| Click Chemistry Conjugation Kits | For surface engineering and functionalization of exosomes. | Enables robust and specific attachment of targeting ligands (peptides, antibodies) to the exosome surface [61]. |
| CRISPR-Cas9 Systems | For genetic engineering of parent MSCs to endogenously load protein or RNA cargo. | Allows for precise manipulation of the exosome cargo during biogenesis [61]. |
| Tetraspanin Antibody Kits (CD63/CD81/CD9) | Standard markers for exosome characterization via western blot, flow cytometry, or ELISA. | Critical for confirming the vesicular identity of your isolate. |
| Lipid-Binding Dyes (e.g., PKH67) | For labeling and tracking exosome uptake in recipient cells. | Vital for mechanistic studies to visualize and quantify exosome-cell interactions. |
Overcoming Manufacturing Hurdles and Standardization Challenges in MSC Exosome Production
Core CQA Concepts and Definitions
What are Critical Quality Attributes (CQAs) and why are they essential for MSC exosome therapeutics?
Critical Quality Attributes (CQAs) are "physical, chemical, biological, or microbiological property or characteristic that should be within an appropriate limit, range, or distribution to ensure the desired product quality" according to FDA definitions [63]. For MSC-derived exosome therapeutics, establishing well-defined CQAs is particularly challenging due to the inherent biological variability of both the parent cells and the secreted exosomes, their complex mechanisms of action, and the current lack of standardized characterization protocols [64] [60]. The transition from cell therapies to exosome-based therapies offers clinical advantages including reduced challenges with cell viability, storage, and administration, but manufacturing faces significant challenges in defining CQAs for consistent identity and potency [60].
How do regulatory frameworks classify CQAs for biological products?
The Code of Federal Regulations (21CFR610) defines four fundamental CQAs for biological products: Safety, Purity, Identity, and Potency [64]. These categories form the foundation for quality assessment of MSC exosome products, though specific implementation requires adaptation to account for their unique biological nature and heterogeneity.
Table: Fundamental CQA Categories for MSC Exosome Products
| CQA Category | Regulatory Definition | Primary Concern for MSC Exosomes |
|---|---|---|
| Safety | "Relative freedom from harmful effect when prudently administered" [63] | Sterility, endotoxin levels, absence of tumorigenic potential [63] [38] |
| Identity | Characteristics that "distinguish one product from another produced in the same facility" [63] | Specific surface markers, size distribution, and cellular origin [63] [31] |
| Purity | Measure of "impurities in the final product from the manufacturing process" [63] | Residual process contaminants, non-exosome particulate matter [63] |
| Potency | "The specific ability or capacity of the product to effect a given result" [63] | Biological activity relevant to the intended mechanism of action [63] [64] |
Troubleshooting Common CQA Challenges
FAQ: How can we address the inherent heterogeneity in MSC exosome populations when defining identity attributes?
Challenge: MSC exosomes exhibit substantial heterogeneity in size, composition, and function depending on their tissue source, culture conditions, and isolation methods [53] [31]. This natural variability complicates the establishment of consistent identity criteria across production batches.
Solutions:
- Implement multi-parameter characterization: Instead of relying on single markers, use a combination of physical (size, concentration), biochemical (surface proteins), and functional attributes to define identity [31].
- Establish subpopulation consistency: Monitor exosome subpopulations throughout production. Recent research demonstrates that certain subpopulations can remain stable over extended culture periods (e.g., 28 days in 3D bioreactors), providing a window for consistent harvest [31].
- Standardize source materials: Begin with well-characterized MSC sources that meet International Society for Cellular Therapy (ISCT) minimal criteria [64].
- Control manufacturing processes: Maintain consistent culture conditions, expansion protocols, and harvesting timelines to minimize introduced variability [64].
Table: Technical Solutions for Heterogeneity Challenges
| Challenge Area | Technical Approach | Expected Outcome |
|---|---|---|
| Source Variability | Implement strict donor screening criteria; use standardized MSC characterization panels [64] | Reduced batch-to-batch variability originating from cellular source differences |
| Process-Induced Heterogeneity | Control critical process parameters (CPPs) including culture duration, bioreactor conditions, and feeding schedules [31] [64] | Consistent exosome subpopulation profiles across manufacturing lots |
| Analytical Variability | Use orthogonal characterization methods (NTA, flow cytometry, electron microscopy) with standardized protocols [65] [31] | Reproducible identity measurements across different laboratories and operators |
FAQ: What strategies can improve potency assay development for MSC exosomes with complex modes of action?
Challenge: MSC exosomes exhibit multimodal mechanisms of action rather than single defined pathways, making traditional potency assays insufficient [60]. The field lacks standardized assays that correlate with clinical efficacy, as highlighted by regulatory feedback on MSC products requesting "further scientific rationale to demonstrate the relationship of potency measurements to the product's biologic activity" [64].
Solutions:
- Implement matrix assays: Evaluate multiple characteristics collectively, where the cumulative assessment provides a measure of potency [63].
- Focus on mechanism-specific bioactivity: Develop assays targeting specific pathways relevant to the therapeutic indication (e.g., immunomodulation, angiogenesis, tissue repair) [64].
- Consider emerging models: The Extracellular Modulation of Cells by EVs (EMCEV) model suggests MSC exosomes may exert effects by modulating the extracellular environment rather than through direct cellular internalization, which may require new assay approaches [60].
- Correlate with clinical outcomes: Establish bridges between in vitro potency measures and in vivo efficacy during clinical development, recognizing that assays may need refinement as clinical experience accumulates [64].
Experimental Protocols for CQA Assessment
Detailed Methodology: Comprehensive Identity Profiling of MSC Exosomes
This protocol provides a standardized approach for establishing identity attributes through orthogonal characterization methods, adapted from recent studies addressing exosome heterogeneity [31].
Materials and Equipment:
- Purified MSC exosome sample
- Nanoparticle Tracking Analysis (NTA) system (e.g., Malvern Nanosight)
- Transmission Electron Microscope (TEM)
- Flow cytometer with high-sensitivity configuration
- Western blot apparatus
- Specific antibodies for exosome markers (CD9, CD63, CD81) and MSC markers
- Phosphate buffered saline (PBS)
- Exosome-free ultracentrifuged fetal bovine serum
Procedure:
- Sample Preparation: Dilute purified exosomes in PBS to appropriate concentration for each analysis method. Prepare aliquots to avoid freeze-thaw cycles.
Physical Characterization:
- Size and Concentration Analysis (NTA): Dilute sample to 10^7-10^9 particles/mL in filtered PBS. Acquire five 30-second videos using standardized camera level and detection threshold. Analyze particle size distribution and concentration using integrated software.
- Morphological Assessment (TEM): Apply 10μL of exosome suspension to formvar/carbon-coated grids. Negative stain with 2% uranyl acetate for 1 minute. Image using TEM at 80-100kV accelerating voltage.
Biochemical Characterization:
- Surface Marker Analysis (Flow Cytometry): Incubate exosomes with antibody-coupled beads for 1 hour. Wash and incubate with fluorescent-labeled detection antibodies against CD9, CD63, CD81. Include isotype controls. Analyze using high-sensitivity flow cytometer with appropriate gating for vesicle populations.
- Protein Marker Confirmation (Western Blot): Lyse exosomes in RIPA buffer. Separate proteins by SDS-PAGE, transfer to membrane, and probe with antibodies against tetraspanins (CD9, CD63, CD81) and MSC-associated markers. Use appropriate secondary antibodies and detection system.
Data Analysis and Acceptance Criteria:
- Establish historical ranges for size distribution (typically 30-150nm for exosomes)
- Define expected protein marker profile (positive for at least two tetraspanins)
- Document percentage of particles falling within expected size and marker parameters
Troubleshooting Notes:
- If NTA concentration appears artificially high, check for salt crystals or protein aggregates and consider additional purification steps.
- If flow cytometry shows high background, ensure thorough washing and use of exosome-free buffers.
- For western blot with weak signal, confirm antibody specificity and consider concentrating exosome sample.
Identity Assessment Workflow
Advanced CQA Measurement Techniques
Detailed Methodology: Potency Assessment Through Functional Bioassays
This protocol outlines a matrix approach to potency measurement that addresses the multimodal functionality of MSC exosomes, recognizing that no single assay may fully capture therapeutic potential [63] [64].
Materials and Equipment:
- Isolated MSC exosomes with known particle concentration
- Target cell lines relevant to mechanism of action (e.g., immune cells for immunomodulation, endothelial cells for angiogenesis)
- Cell culture facilities and appropriate media
- ELISA kits for cytokine detection (e.g., IFN-γ, TNF-α, IL-10)
- Endothelial tube formation assay kit (e.g., Matrigel-based)
- qPCR system and reagents
- Migration assay chambers (e.g., Transwell system)
Procedure:
- Immunomodulatory Potency Assay:
- Isolate peripheral blood mononuclear cells (PBMCs) from healthy donors.
- Activate PBMCs with mitogens (e.g., PHA-L) in the presence of varying concentrations of MSC exosomes.
- After 48-72 hours, collect supernatants for cytokine analysis by ELISA.
- Analyze suppression of pro-inflammatory cytokines (IFN-γ, TNF-α) and induction of anti-inflammatory cytokines (IL-10).
- Calculate IC50 values for inhibition of activation.
Angiogenic Potential Assessment:
- Seed endothelial cells (HUVECs) on Matrigel-coated plates.
- Treat with MSC exosomes at multiple concentrations.
- After 4-8 hours, quantify tube formation by measuring total tube length, number of branches, and junction points.
- Compare to positive (VEGF) and negative controls.
Gene Expression Modulation:
- Treat relevant target cells with MSC exosomes for 24 hours.
- Extract RNA and perform qPCR for genes associated with the proposed mechanism of action.
- Analyze expression changes relative to untreated controls.
Data Interpretation and Potency Assignment:
- Establish dose-response curves for each assay system.
- Assign relative potency units based on comparison to a reference standard.
- Apply multivariate analysis to combine results from multiple assay systems.
Troubleshooting Notes:
- If assay variability is high, ensure consistent exosome storage conditions and avoid freeze-thaw cycles.
- If dose-response is not observed, verify exosome integrity and consider functional assessment of parent MSCs.
- Include reference standards in each assay to control for inter-assay variability.
Potency Assessment Strategy
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table: Key Research Reagents for MSC Exosome CQA Assessment
| Reagent/Category | Specific Examples | Function in CQA Assessment |
|---|---|---|
| Bioreactor Systems | Hollow Fiber 3D Bioreactors [31] | Scalable production of consistent exosome populations with stable subdistribution |
| Exosome Enhancement | RoosterBio exosome-promoting system [31] | Increased exosome yield while maintaining critical quality attributes |
| Characterization Instruments | Nanoparticle Tracking Analysis (NTA) [31] | Physical characterization of size distribution and particle concentration |
| Surface Marker Analysis | High-sensitivity flow cytometry [31] [64] | Detection of tetraspanins (CD9, CD63, CD81) and MSC-specific markers |
| Functional Assay Tools | Endothelial tube formation assays [64] | Measurement of angiogenic potential for potency assessment |
| Immunomodulation Assays | Mixed lymphocyte reaction, cytokine secretion [63] | Quantification of immunomodulatory capacity for potency |
| Reference Materials | Standardized beads, control exosomes [65] | Calibration of instruments and cross-experiment standardization |
| Cell Culture Media | Defined, xeno-free MSC media [64] | Consistent expansion of parent MSCs with maintained characteristics |
Regulatory and Standardization Considerations
FAQ: How should we approach CQA development for early-phase clinical trials?
Guidance: During early product development, focus on identifying measurable attributes that correlate with biological activity, even if complete validation isn't yet possible [65]. The time to design and undertake CQA testing is during original product development, when transferring technology, and whenever the manufacturing process changes [65]. Develop and validate assays for CQAs as early as possible in the pre-clinical product development process to enable better decision making at each step and build confidence that observed effects are reproducible in the clinical phase [65].
Key Considerations:
- Establish acceptance criteria using known reference and patient samples before clinical trials [65]
- Identify critical reagents early to streamline the manufacturing process [65]
- Implement comparability protocols to manage process changes during development
- Engage with regulatory agencies early regarding CQA strategy, especially for complex attributes like potency
FAQ: What are emerging solutions for the "product is the process" challenge in MSC exosome therapeutics?
Emerging Approaches: The field is gradually moving from a complete "product is the process" paradigm toward more defined CQA-based specifications through several advanced strategies:
- Advanced process analytics: Implementing in-line or at-line monitoring of Critical Process Parameters (CPPs) that influence CQAs [64]
- Bioengineering solutions: Using genetic engineering of parent MSCs to produce more consistent exosome populations [38]
- 3D culture systems: Implementing bioreactor technologies that maintain more stable exosome subpopulations over extended culture periods [31]
- Computational modeling: Developing models that predict in vivo performance based on multifaceted CQA assessment
The progression toward Quality by Design approaches where processes operate within defined ranges and CQAs truly define the product represents the future state for the field, though this remains aspirational for most MSC exosome products currently in development [64].
Within mesenchymal stem cell (MSC) research, exosomes have emerged as critical mediators of therapeutic effects, offering a promising cell-free alternative for regenerative medicine, immunotherapy, and drug delivery [1]. However, the inherent heterogeneity of MSC-derived exosome populations and their cargo presents a significant challenge for research reproducibility and therapeutic development [1]. This heterogeneity is profoundly influenced by the methods used to isolate exosomes from conditioned media or complex biofluids. Selecting an appropriate isolation technique is therefore not merely a procedural step but a critical decision that directly affects the yield, purity, and biological functionality of the resulting exosome preparations, ultimately impacting the validity of downstream experimental results [66] [67]. This guide addresses common pitfalls and troubleshooting approaches for three widely used isolation methods: Ultracentrifugation, Size Exclusion Chromatography (SEC), and Precipitation.
Quantitative Method Comparison
The choice of isolation method directly impacts critical physical and functional characteristics of the isolated exosomes. The table below summarizes a comparative analysis of three common techniques.
Table 1: Comparative Analysis of Exosome Isolation Methods from Cell Culture Media
| Isolation Method | Average Particle Size (nm) | Particle / Protein Ratio (Purity Indicator) | Impact on Cell Viability (Hypoxic Model) | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Ultracentrifugation (UC) | ~60 nm [66] | Varies; can be low due to co-pelleted proteins [67] | Increased live cell content by ~20% [66] | Considered a "gold standard"; cost-effective for consumables [68] | Low isolation efficiency (~10-25%); long processing time; requires specialized equipment; can damage exosomes [69] |
| Size Exclusion Chromatography (SEC) | N/A (Separates by size) | Can be low (e.g., 1.12 × 10⁷) but yields high tetraspanin positivity [67] | Functional particles maintained [67] | Gentle process; retains bioactivity; good separation from soluble proteins [67] [69] | Sample dilution; requires a concentration step; pressure can cause damage [67] [69] |
| Polymer-Based Precipitation | ~89 nm [66] | Generally low due to high co-precipitation of contaminants [67] | Increased live cell content by ~15% [66] | Simple protocol; no specialist equipment; high yield of particles [66] [67] | High co-precipitation of non-EV proteins (e.g., albumin); potential reagent immunogenicity [67] |
Troubleshooting Common Isolation Pitfalls
Ultracentrifugation (UC)
Problem: Low Yield and Poor Exosome Functionality
- Potential Cause: Excessive g-force or prolonged centrifugation times can damage exosomes, disrupting their membrane integrity and compromising their biological function [69].
- Solution: Optimize the centrifugation protocol. Avoid excessively long run times and high speeds beyond what is necessary. A typical protocol for pelleting exosomes involves a final ultracentrifugation step at 100,000–110,000×g for 70-120 minutes [66]. Using a softer resuspension buffer and allowing the pellet to rehydrate at 4°C for 30 minutes before gentle pipetting can also help preserve integrity.
Problem: Low Purity and High Contaminant Protein
- Potential Cause: Co-pelletion of protein aggregates and non-exosomal lipoproteins, particularly from complex biofluids like serum [70] [67].
- Solution: Incorporate a density gradient centrifugation step following differential centrifugation. This separates particles based on buoyant density, effectively isolating exosomes from most contaminants [70] [68]. Although this adds time and complexity, it significantly enhances purity. Always ensure complete removal of the supernatant after each centrifugation wash step without disturbing the pellet.
Size Exclusion Chromatography (SEC)
Problem: Highly Diluted Sample
- Potential Cause: The fundamental principle of SEC separates particles by size, which inherently leads to sample dilution as the exosomes are eluted in a relatively large volume of buffer [69].
- Solution: Follow SEC with a gentle concentration step. Ultrafiltration using centrifugal filters with an appropriate molecular weight cut-off (e.g., 10 kDa or 100 kDa) is commonly used to concentrate the exosome-containing fractions [67] [70]. Ensure the filter material is chosen to minimize exosome adhesion and shear stress.
Problem: Incomplete Separation from Lipoproteins
- Potential Cause: While SEC effectively separates exosomes from most soluble proteins, some lipoprotein particles, particularly HDL, have a size and density similar to small exosomes, leading to co-elution [70].
- Solution: For studies requiring extremely high purity from serum/plasma, use SEC in combination with a prior purification step. Pre-processing the sample with ultracentrifugation or tangential flow filtration to remove the majority of lipoproteins and other large particles can significantly improve the purity of the final SEC product [70] [67].
Polymer-Based Precipitation
Problem: Significant Contamination with Non-Vesicular Material
- Potential Cause: Precipitation reagents (e.g., PEG) co-precipitate any particles within a similar size range, including non-exosomal proteins, protein aggregates, and nucleic acids, leading to low sample purity [67] [66].
- Solution: Implement rigorous post-precipitation washing steps. After the initial precipitation and low-speed centrifugation, resuspend the pellet in a sufficient volume of PBS and re-centrifuge to remove soluble contaminants. For higher purity, combining precipitation with a subsequent SEC clean-up step can effectively remove polymeric reagents and co-precipitated proteins [67].
Problem: Reagent Interference with Downstream Analysis
- Potential Cause: Residual polymer from precipitation kits can inhibit downstream enzymatic reactions (e.g., in PCR or proteomic analyses) and interfere with protein quantification assays [67].
- Solution: Ensure thorough washing of the precipitate, as mentioned above. If interference persists, consider switching isolation methods for specific applications. SEC is generally preferred for downstream 'omics' analyses due to its minimal chemical interference [67].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Exosome Isolation and Characterization
| Reagent / Kit | Primary Function | Isolation Context |
|---|---|---|
| Polyethylene Glycol (PEG) | Volume-excluding polymer that reduces exosome solubility, leading to precipitation [66] [67]. | Used in polymer-based precipitation methods. |
| Iodixanol | Density gradient medium for separating particles based on buoyant density [70]. | Used in density gradient centrifugation to achieve high-purity exosome isolation. |
| Sepharose/ Agarose Beads | Porous stationary phase for chromatographic separation based on hydrodynamic volume [67]. | The packing material used in Size Exclusion Chromatography (SEC) columns. |
| Antibodies (CD9, CD63, CD81) | Immunoaffinity capture of vesicles displaying specific surface tetraspanins [66] [71]. | Used for highly specific isolation of exosome subpopulations or for characterization via ELISA/flow cytometry. |
| Protease Inhibitors | Prevent degradation of protein cargo by endogenous proteases. | Added to isolation buffers throughout the process to preserve exosome integrity for cargo analysis. |
| PBS (Phosphate-Buffered Saline) | Isotonic buffer for washing and resuspending exosome pellets. | Used universally across all methods for dilution, washing, and final resuspension. |
Isolation Workflow and Decision Pathway
The following diagram illustrates a generalized isolation workflow, while the subsequent decision tree aids in selecting the most appropriate method based on research priorities.
General Exosome Isolation Workflow
Isolation Method Selection Guide
Frequently Asked Questions (FAQs)
Q1: My exosome yield from ultracentrifugation is consistently low. What are the main factors I should check? A1: Low yield in UC can stem from several factors:
- Rotor Calibration: Ensure your ultracentrifuge rotor is properly calibrated. An incorrect k-factor will affect pelleting efficiency.
- Pelleting Efficiency: The fixed-angle rotors are most common. Be aware that the pellet will form on the side of the tube, not the bottom, and may be invisible. Always orient the tube the same way in the rotor and carefully aspirate the supernatant from the opposite side.
- Resuspension Technique: The pellet can be difficult to resuspend. After removing the supernatant, let the pellet sit in your resuspension buffer (e.g., PBS) for 30 minutes on ice before using gentle pipetting to avoid fragmentation.
Q2: How can I effectively assess the purity of my exosome preparation? A2: Purity assessment requires more than just particle concentration [71]. The gold standard is the particle-to-protein (PtP) ratio, which compares the total particle count (e.g., via NTA) to the total protein concentration (e.g., via BCA assay) [70] [67]. A higher PtP ratio indicates a preparation with more vesicles relative to soluble protein or aggregates. Furthermore, techniques like high-resolution flow cytometry or Western blotting for positive markers (CD9, CD63, CD81) and negative markers (e.g., APOB/APOE for lipoproteins) provide a more specific purity evaluation [70] [67].
Q3: Why is there so much heterogeneity in my isolated MSC-exosome population, and how does the isolation method contribute? A3: Heterogeneity is inherent to MSC-exosomes due to different cellular origins, multivesicular body states, and environmental cues [1]. The isolation method acts as a filter that can select for certain subpopulations. For instance:
- Ultracentrifugation may preferentially pellet larger vesicles and miss smaller ones.
- SEC will isolate all particles above a certain size threshold without distinction.
- Precipitation may capture a broad range of particles but with many non-vesicular contaminants. Therefore, the observed heterogeneity is a combination of the natural diversity of vesicles and the selective bias of your chosen isolation technique.
FAQs: Scaling and GMP Compliance in MSC-sEV Production
What are the most significant challenges in transitioning from lab-scale to commercial-scale production for cell and gene therapies like MSC-sEVs?
The most significant challenge is often not just the science itself, but managing and translating knowledge between Research & Development (R&D) and Good Manufacturing Practice (GMP) environments. Lab teams may develop an elegant process, but upon moving to GMP, questions about scalability, validation, and documentation arise [72]. For autologous therapies and complex products like MSC-sEVs, the main hurdle can be the regulatory burden and product release testing, which must be completed within compressed timeframes due to short shelf lives [72]. Furthermore, for MSC-sEVs, defining Critical Quality Attributes (CQAs) for consistent identity and potency is complicated by variability in cell sources, culture conditions, and the inherent heterogeneity of MSCs [60].
How can companies mitigate scale-up and compliance challenges during early development?
Mitigating these challenges requires a proactive approach focusing on three key areas [72]:
- Knowledge Management: Implement AI-enabled systems to help organize and connect knowledge across the product lifecycle, from early development to commercial tech transfer.
- Cross-Functional Teams: Utilize agile MSAT (Manufacturation, Science, and Technology) teams that act as a bridge between R&D and manufacturing, spotting potential gaps early.
- Real-Time Analytics: Accelerate the implementation of real-time analytics and rapid-release testing to align with critical timelines.
What are the best practices for ensuring GMP compliance without stifling innovation during process design?
The key is integrating compliance into innovation from the start. Think of GMP not as a constraint, but as a design input from day one [72]. Fostering direct communication is crucial; facilitate interactions between manufacturing and scientific teams so each understands the other's environment and constraints [72]. Additionally, building structured, digital knowledge-sharing systems that track decisions and learnings throughout development helps maintain continuity and compliance [72].
Why is defining "potency" particularly challenging for MSC-sEV products, and what strategies can help?
Defining potency for MSC-sEVs is complex due to their multimodal mechanisms of action. They impact various cell types and processes through diverse mechanisms, making it difficult to link a single attribute to biological effect [60]. A pragmatic strategy is to focus on identifying key potency-related CQAs based on specific mechanisms of action relevant to the therapy's intended purpose, while recognizing that for such products, "the process defines the product" [60].
How should a Computer System Validation (CSV) strategy evolve as a biotech facility scales?
A CSV strategy should align with the facility's growth phases [73]:
- Early Phase (Clinical Supply): Focus on risk-based validation for critical systems like LIMS, using frameworks like GAMP 5.
- Mid Phase (Pre-commercial): Standardize validation templates and begin defining core requirements (e.g., audit trails) across new systems like MES (Manufacturing Execution System) and eQMS (electronic Quality Management System).
- Commercial Phase: Implement a full CSV governance framework with periodic reviews, change control procedures, and data integrity monitoring to manage higher regulatory scrutiny [73].
Troubleshooting Guides for MSC-sEV Process Development
Guide 1: Troubleshooting Heterogeneity and Potency
| Problem | Potential Causes | Recommended Solutions & Investigations |
|---|---|---|
| High batch-to-batch variability in sEV potency | • Inconsistent cell source (donor, passage number)• Fluctuations in culture conditions (media, supplements)• Uncontrolled enrichment/purification steps [60] | • Standardize cell banking and establish strict cell passage limits• Implement process controls and define Critical Process Parameters (CPPs)• Use clonal MSC lines (while monitoring for heterogeneity re-emergence) [60] |
| Inconsistent sEV cargo profile | • Uncontrolled bioreactor parameters (e.g., pH, dissolved oxygen)• Lack of in-process controls for CMAs and CPPs [74] | • Move to a controlled bioreactor system• Adopt a QbD approach to identify and control parameters impacting CQAs [74] |
| Difficulty defining a potency CQA | • Complex, multimodal mechanism of action [60] | • Shift focus from traditional "internalization" models to the EMCEV model• Identify key mechanism-based potency assays (e.g., immunomodulation, angiogenesis) |
Guide 2: Troubleshooting Scale-Up and GMP Transitions
| Problem | Potential Causes | Recommended Solutions & Investigations |
|---|---|---|
| Process performs well at lab-scale but fails in pilot-scale bioreactors | • Inefficient knowledge transfer from R&D to manufacturing teams• Scale-up effects not adequately considered in process design [72] | • Engage MSAT early to bridge R&D and GMP• Conduct engineering runs at pilot scale to identify and resolve scale-up issues before GMP production [74] |
| Failed audit due to data integrity issues in electronic systems | • Lack of robust Computer System Validation (CSV)• Inadequate control over system interfaces and data flows [73] | • Implement a scalable CSV lifecycle with risk assessment• Validate data transfer accuracy and error handling between integrated systems (e.g., LIMS to MES) [73] |
| Lengthy product release times due to analytics | • Reliance on traditional, time-consuming release assays [72] | • Develop and validate rapid release tests and real-time analytics• Implement Process Analytical Technology (PAT) for immediate feedback and control [72] [74] |
The Scientist's Toolkit: Essential Research Reagent Solutions
| Item / Category | Function & Rationale |
|---|---|
| Defined Cell Culture Media | To ensure consistency in MSC growth and sEV production, reducing variability introduced by serum-derived components. |
| Bioreactor Systems | To provide a controlled, scalable environment for consistent MSC expansion, replacing flasks and allowing monitoring of CPPs like pH and dissolved oxygen [74]. |
| Chromatography Resins | For the purification and recovery of sEVs during downstream processing, enabling separation based on size, charge, or affinity [74]. |
| Process Analytical Technology (PAT) | Sensors and analytical tools for real-time monitoring of process parameters and product quality, enabling better control and faster release [74]. |
| Reference Standard sEVs | A well-characterized sEV preparation used as a benchmark for comparing identity, purity, and potency across different production batches. |
Experimental Protocols & Workflows
Protocol 1: QbD-Driven Process Development for MSC-sEVs
Aim: To establish a scalable and reproducible manufacturing process for MSC-sEVs by identifying and controlling critical factors impacting quality.
Methodology:
- Define Quality Target Product Profile (QTPP): Outline the desired quality characteristics of the final sEV product (e.g., size, key cargo markers, potency titre).
- Identify Critical Quality Attributes (CQAs): Determine the physical, chemical, biological properties that should be within appropriate limits to ensure product quality. Examples include:
- Identity: Presence of specific surface markers (e.g., CD63, CD81).
- Purity: Ratio of particle count to protein content.
- Potency: Measured by a relevant biological activity assay (e.g., T-cell suppression, angiogenesis).
- Risk Assessment & DOE: Conduct a risk assessment to link Material Attributes and Process Parameters to CQAs. Use Design of Experiments (DOE) to systematically evaluate the impact of Critical Process Parameters (CPPs)—such as harvest time, purification method—on CQAs [74].
- Establish a Control Strategy: Define the validated ranges for CPPs and implement in-process testing and real-time monitoring (PAT) to ensure consistent quality.
Protocol 2: Tech Transfer and Scale-Up Validation
Aim: To successfully transfer a lab-scale MSC-sEV process to a GMP-compliant pilot or commercial manufacturing facility.
Methodology:
- Knowledge Transfer: Compile a comprehensive technology transfer package including all process documentation, analytical methods, and known process constraints [72].
- Facility Fit Assessment: Evaluate the receiving facility's equipment, systems, and capabilities against process requirements. Key systems to review include [73]:
- MES (Manufacturing Execution System): For electronic batch records.
- LIMS (Laboratory Information Management System): For managing sample and test data.
- Environmental Monitoring System: For ensuring air and surface quality.
- Engineering Run: Execute a non-GMP engineering run at the new scale to de-risk the process, troubleshoot potential issues, and confirm CPPs [74].
- Process Performance Qualification (PPQ): Execute a minimum of three consecutive GMP batches at the commercial scale to demonstrate that the process is reproducible and effective.
- Ongoing Monitoring: Implement a continued process verification plan to monitor the process performance over its lifecycle.
Signaling Pathway: The EMCEV Model for MSC-sEV Action
The traditional view of sEV action involves direct internalization by target cells. However, the Extracellular Modulation of Cells by EVs (EMCEV) model proposes that MSC-sEVs exert their effects by modulating the extracellular environment, enabling a "one EV to many cells" interaction [60]. This model is particularly relevant for understanding the multimodal potency of MSC-sEVs.
Technical Support & Troubleshooting Hub
This section provides targeted guidance for common experimental challenges in proteomic and miRNA analysis of MSC exosomes, helping researchers ensure data robustness and reproducibility.
Frequently Asked Questions (FAQs)
Q1: My proteomic data shows high technical variation between replicate MSC-EV samples. What are the primary sources and how can I minimize them?
A: High variation often stems from these key process conditions and can be mitigated as follows:
- EV Isolation Method: Different isolation techniques (e.g., UC, SEC, precipitation) co-isolate varying levels of non-EV contaminants and select for different EV subpopulations, directly impacting protein cargo profiles [75] [76].
- Cell Culture Conditions: The type of culture medium (e.g., classical vs. undefined commercial) and the use of EV-depleted serum significantly influence the observed EV cargo [76]. For consistent results, standardize media and use EV-depleted serum to reduce background contamination.
- Analytical Variability: The LC-MS instrumentation and protein search algorithm contribute to variance [75]. Implement a consistent analytical pipeline and use batch-effect correction methods.
Q2: When performing miRNA sequencing from MSC exosomes, a large portion of my reads do not map to the miRBase database. What could be the cause?
A: This is a common challenge due to several factors:
- Multi-mapping: The short length of miRNAs (18-30 nt) increases the probability that reads will map to multiple locations in the genome [77]. Use aligners like Bowtie2 or STAR with parameters optimized for small RNA to improve accuracy [77].
- Diverse RNA Sources: Biofluids and EV preparations contain a diverse small RNA population, including tRNA fragments, siRNAs, piRNAs, and non-human (e.g., microbial) RNAs [77]. Map reads sequentially to human and microbial genomes to account for this.
- Post-transcriptional Modifications: miRNAs can undergo modifications like 2'-O-methylation, which can affect library preparation and mapping efficiency [77].
Q3: How do I choose the best statistical test for identifying differentially expressed proteins in my MSC exosome experiment?
A: The choice of test depends on your data structure. In proteomics, measurement variance can be unique to each peptide, violating the assumption of uniform variance in standard tests like the t-test [78]. It is recommended to:
- Select tests based on enrichment of known targets. For miRNA target identification, tests can be evaluated by their ability to enrich for top-ranked DEPs that are also reported by orthogonal methods like PAR-CLIP [78].
- Use variance estimation methods that weigh peptide measurements appropriately to account for the complex structure of LC-MS/MS data [78].
Q4: Should I correct for batch effects in my proteomic data at the precursor, peptide, or protein level?
A: A 2025 benchmarking study recommends performing batch-effect correction at the protein level for maximum robustness in MS-based proteomics [79]. While corrections can be applied at the precursor or peptide level, the process of quantifying and aggregating data into protein-level expressions can reintroduce batch-related noise. Protein-level correction has been shown to be the most robust strategy for integrating multi-batch data in large cohort studies [79].
Troubleshooting Guides
Problem: Inconsistent miRNA Quantification Results with TaqMan Assays
- Symptom: Low amplification signal or high Ct values in qPCR.
- Potential Cause & Solution:
- Low Abundance Target: For lowly abundant miRNAs, the standard 1-10 ng total RNA input may be insufficient.
- Troubleshooting Step: Titrate the input total RNA up to 250 ng. Alternatively, double the amount of reverse transcriptase enzyme to 6.6 U/µL to improve cDNA synthesis efficiency [80].
- Symptom: Amplification in No-Template Control (NTC) wells (Ct < 38).
- Potential Cause & Solution:
- Contamination: Contamination from plasmids or other artificial templates in the lab environment.
- Troubleshooting Step: Change all reagents. Use a DNA degradation solution (e.g., DNAZap) to clean surfaces and equipment. If possible, perform the assay in a separate location with dedicated pipets [80].
Problem: High Background Noise in SYBR Green-based miRNA Detection
- Symptom: Multiple peaks in the melt curve analysis.
- Potential Causes & Solutions:
Quantitative Data on Variability
This section compiles empirical data on the key factors contributing to cargo variability in MSC exosomes, providing a quantitative basis for experimental planning and data interpretation.
Table 1: Quantitative Impact of Process Conditions on EV Proteomic Heterogeneity
| Factor Category | Specific Example | Quantitative Impact on Variability | Proposed Mitigation Strategy |
|---|---|---|---|
| EV Source | MSC Source (e.g., Bone Marrow vs. Umbilical Cord) | MSC-derived EVs contained the highest number of unique proteins and a greater fraction of proteins in wound-healing pathways [75] [76]. | Standardize the MSC tissue source and donor criteria (age, sex) [19]. |
| Isolation Method | Ultracentrifugation (UC) vs. Size-Exclusion Chromatography (SEC) | Combined with other factors (source, medium, LC-MS), isolation method contributes to 25-60% of unexplained variance in protein cargo [75] [76]. | Adopt a single, validated isolation method (e.g., following MISEV guidelines) for all comparative studies [75]. |
| Cell Culture Medium | Classical (DMEM) vs. Undefined Commercial Media | ~50% of studies used classical media, while 35% used undefined commercial media, a known source of cargo variability [76]. | Use a defined, consistent medium formulation, and use EV-depleted serum to reduce contaminating background EVs [76]. |
| Cross-Study Consistency | Protein Identification Across 52 Proteomic Studies | 40% of the ~13,000 observed proteins were identified in only a single study, highlighting extreme inter-study heterogeneity [75] [76]. | Improve reproducibility by adopting standardized reporting (MISEV guidelines) and data sharing practices [75]. |
Table 2: Common Batch-Effect Correction Methods for Omics Data
| Method | Principle | Best For | Key Considerations |
|---|---|---|---|
| ComBat [81] [79] | Empirical Bayes framework to adjust for known batch variables. | Bulk proteomic or transcriptomic data with known, additive batch effects. | Requires known batch information; may over-correct if batches are confounded with biology [81]. |
| SVA (Surrogate Variable Analysis) [81] | Estimates and removes hidden sources of variation (unknown batch effects). | Scenarios where batch variables are unknown or partially observed. | Risk of removing biological signal; requires careful modeling [81]. |
| limma removeBatchEffect [81] | Linear modeling-based correction for known batch effects. | Integrating with differential expression analysis workflows in R. | Assumes batch effects are additive and known [81]. |
| Harmony [79] | Iteratively clusters cells and corrects in a shared embedding space. | Single-cell RNA-seq or complex datasets where biological groups need to be aligned across batches. | Effective for preserving biological variation while integrating data [79]. |
| Ratio [79] | Scales feature intensities based on concurrently profiled universal reference samples. | Large-scale proteomic studies, especially when batch effects are confounded with biological groups. | Demonstrated superior performance in large-scale plasma proteomics for prediction tasks [79]. |
Experimental Protocols for Robust Cargo Analysis
Detailed Protocol: Quantitative Proteomic Approach for miRNA Target Identification in MSC Exosomes
This protocol, adapted from a study on KSHV miR targets, outlines an integrated pipeline for confidently identifying miRNA targets by quantifying proteomic changes in recipient cells [78].
1. Sample Preparation and Labeling:
- Transfection & Culture: Transfect human embryonic kidney (HEK293T) or relevant recipient cells with the MSC exosome-derived miRNA of interest (test) or an empty vector (control). Culture for 48 hours to allow for miRNA-mediated protein regulation [78].
- Protein Digestion with 18O Labeling:
- Harvest and digest the protein lysate from the test sample with trypsin in H218O water.
- Digest the control sample with trypsin in normal H216O water.
- Note: 18O/16O labeling is flexible, does not require cell culturing with labels, and has a wide dynamic range [78].
- Sample Mixing & Fractionation: Mix the heavy (18O, test) and light (16O, control) labeled peptide samples together. Reduce complexity by separating peptides using Strong Cation Exchange (SCX) chromatography into 20 fractions. Further, analyze each SCX fraction in three technical LC-MS replicates to reduce quantification variation [78].
2. Mass Spectrometry Data Acquisition:
- Quantification: Use Liquid Chromatography-Mass Spectrometry (LC-MS) for peptide quantification. The relative abundance of heavy vs. light peptides provides the fold-change.
- Identification: Use Tandem Mass Spectrometry (MS/MS) for peptide sequence identification [78].
3. Data Processing and Analysis:
- Peptide Feature Alignment: Apply an alignment algorithm (e.g., SCFIA) across technical replicates to increase the number of quantifications per peptide, thereby reducing measurement variance [78].
- Peptide Quantification & Variance Estimation: Use a dedicated quantification algorithm to remove interference and random suppression effects. Estimate variance for unique and non-unique peptides within their fractions to properly weight measurements [78].
- Statistical Testing for DEPs: Employ statistical tests to identify Differentially Expressed Proteins (DEPs), focusing on downregulated proteins. Select the best-performing test by examining the enrichment of predicted targets (e.g., from PAR-CLIP data) among the top-ranked DEPs [78].
- Target Filtering & Validation: Filter the final list of DEPs by requiring that potential targets also show downregulated gene expression and are reported by a prediction algorithm (e.g., SVMcrio) and PAR-CLIP. Validate novel targets using Western blotting and Luciferase reporter assays [78].
Detailed Protocol: miRNA Sequencing and Analysis from MSC Exosomes
This protocol outlines a specialized workflow for analyzing miRNA sequencing data, addressing challenges like multi-mapping and diverse RNA sources [77].
1. Preprocessing of Raw Reads:
- Adapter Trimming: Use specialized tools like Cutadapt or Trimmomatic to effectively remove adapter sequences, which can constitute a significant portion of the short miRNA reads. Use vendor-recommended commands for optimal results [77].
2. Mapping and Annotation:
- Alignment: Use an aligner optimized for short reads, such as Bowtie2, with parameters precisely controlled for small RNA data [77].
- Handling Multi-mapping: To manage reads that map to multiple locations, use tools like the STAR aligner with adjusted parameters or follow a sequential mapping strategy [77].
- Comprehensive Annotation: For a full analysis, use a comprehensive pipeline like exceRpt. Alternatively, use a tool like miRDeep2 to accurately annotate miRNAs and separate them from other small RNAs and degradation products [77].
3. Quantitation and Normalization:
- Quantification: Quantify miRNA expression, accounting for miRNA family members and isomiRs (sequence variants). Tools like isomiRage or seqBuster can be used for this purpose [77].
- Normalization: Avoid using simple normalization like Reads Per Million (RPM), which may not account for compositional biases. Choose a method appropriate for your data to ensure accurate expression comparisons [77].
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents and Kits for MSC Exosome Cargo Analysis
| Item Name | Function / Application | Technical Notes |
|---|---|---|
| TaqMan MicroRNA Assays [80] | Sensitive and specific detection and quantification of mature miRNAs via RT-qPCR. | Input can be titrated up to 250 ng total RNA for low-abundance targets. Can be used for absolute quantitation with a synthetic miRNA standard curve [80]. |
| NCode VILO miRNA cDNA Synthesis Kit [80] | Generation of cDNA specifically optimized for miRNA SYBR Green-based qPCR detection. | Contains a universal primer for qPCR. The primer sequence is proprietary and not provided [80]. |
| Megaplex RT and PreAmp Primers [80] | Highly multiplexed pools for streamlined profiling of hundreds of miRNA targets in a single experiment. | A small subset of assays may be semi-quantitative; validate significant findings with individual TaqMan Assays [80]. |
| Trypsin (for 18O/16O labeling) [78] | Proteolytic enzyme used to digest proteins into peptides for bottom-up proteomics. Incorporates 18O into the C-terminus of peptides during digestion in H218O. | Enables relative quantification of proteins from two conditions without metabolic labeling. Requires careful control of labeling efficiency [78]. |
| Size-Exclusion Chromatography (SEC) Columns [75] [76] | Isolation of small extracellular vesicles (sEVs/exosomes) with minimal protein contamination and high vesicle purity. | Preferred over precipitation methods for functional studies due to better preservation of vesicle integrity and reduced co-isolation of contaminants [75]. |
| EV-Depleted Fetal Bovine Serum (FBS) [76] | Serum supplement for cell culture that has been processed to remove the majority of bovine EVs, reducing background in EV cargo analysis. | Critical for distinguishing bona fide MSC-EV cargo from artifacts introduced by the culture medium [76]. |
This technical support center article is framed within the broader thesis on Addressing heterogeneity in MSC exosome populations and cargo research.
Troubleshooting Guides
Guide 1: Addressing Loss of Exosome Integrity After Storage
Problem: After storage and thawing, your MSC exosome samples show signs of aggregation, decreased particle concentration, or reduced biological activity.
Investigation and Solution:
| Observation | Possible Cause | Recommended Action |
|---|---|---|
| Increased particle size and aggregation on NTA | Sub-optimal storage temperature; multiple freeze-thaw cycles | Avoid storage at -20°C; limit freeze-thaw cycles; use single-use aliquots [82]. |
| Reduced RNA content and impaired bioactivity | Vesicle rupture and cargo leakage due to ice crystal formation | Switch to rapid freezing methods; add cryoprotectants like trehalose for cargo stability [82]. |
| Membrane deformation or vesicle fusion on EM images | Damage from slow freezing or inappropriate cryoprotectants | Optimize freezing protocol for fast cooling; store at a constant -80°C [82]. |
| Low yield from a previously used sample | Degradation from repeated temperature fluctuations | Store in native biofluids if possible, as they offer better stability than purified EVs in buffers [82]. |
Guide 2: Managing Heterogeneity in Stored MSC-sEV Batches
Problem: Inconsistent functional outcomes between different batches of stored MSC-small Extracellular Vesicles (MSC-sEVs), complicating the correlation of potency with specific cargo.
Investigation and Solution:
| Observation | Possible Cause | Recommended Action |
|---|---|---|
| Variable therapeutic efficacy in bioassays | Inherent heterogeneity of MSC-sEVs amplified by storage | Adopt a "process defines the product" mindset; rigorously control cell source, culture, and storage conditions to define CQAs [60]. |
| Difficulty defining Critical Quality Attributes (CQAs) for potency | Complex, multimodal mechanisms of action (e.g., EMCEV model) | Focus stability testing on key potency-related CQAs (e.g., specific surface proteins or miRNAs) linked to your intended mechanism [60]. |
| Inconsistent results in animal models | Uncontrolled variability from parent MSCs | Use well-characterized, clonal MSC lines and monitor for epigenetic or genetic drift over passages [60]. |
Frequently Asked Questions (FAQs)
FAQ 1: What is the single most important factor for preserving MSC exosome functionality during long-term storage?
The most critical factor is maintaining a constant subzero temperature of -80°C. Storage at -20°C leads to significant particle aggregation and size increase, while liquid nitrogen (-196°C) has been associated with membrane disruption and is less commonly recommended. Storing exosomes at -80°C best preserves their uniform size, integrity, RNA content, and bioactivity [82].
FAQ 2: How do multiple freeze-thaw cycles impact MSC exosomes, and how can I avoid this?
Multiple freeze-thaw cycles are detrimental. They lead to a decrease in particle concentration, loss of RNA content, impaired bioactivity, and an increase in exosome size and aggregation [82]. The best practice is to divide exosome preparations into single-use aliquots to avoid repeated thawing of the main stock.
FAQ 3: Are there any additives that can help stabilize exosomes during freezing?
Yes, adding cryoprotectants like trehalose can help exosomes maintain their integrity during freezing and thawing by stabilizing the lipid bilayer and preventing ice crystal formation. Evidence also suggests that storing exosomes in their native biofluid (e.g., conditioned cell culture media) offers improved stability over purified exosomes resuspended in simple buffers [82].
FAQ 4: How does storage stability relate to the challenge of heterogeneity in MSC exosome research?
Storage conditions can directly amplify pre-existing heterogeneity. Different subpopulations of MSC exosomes may have varying stability, leading to a shift in the overall composition and function of the sample after freeze-thaw. Therefore, a standardized and optimized storage protocol is not just for preservation, but is also a critical tool for controlling heterogeneity and ensuring that the product you test is the same as the product you stored [60].
FAQ 5: What analytical methods are essential for characterizing the stability of my stored exosomes?
A combination of techniques is required to assess different aspects of stability [83]:
- Concentration and Size: Use Nanoparticle Tracking Analysis (NTA) or Tunable Resistive Pulse Sensing (TRPS).
- Morphology: Use Transmission Electron Microscopy (TEM) or Scanning Electron Microscopy (SEM) to visualize vesicle shape and membrane integrity.
- Cargo Integrity: Use Western Blot, ELISA, or flow cytometry to monitor specific protein markers (e.g., CD63, CD81, CD9, TSG101, Alix).
- Bioactivity: Use relevant cell-based functional assays (e.g., immunomodulation or angiogenesis assays) to confirm therapeutic potency is retained.
The table below summarizes quantitative data on the effects of different storage conditions on exosome integrity, synthesized from a systematic review of the literature [82].
Table 1: Impact of Storage Conditions on Exosome Integrity
| Storage Condition | Impact on Particle Concentration | Impact on Size & Morphology | Impact on Cargo (e.g., RNA) | Impact on Bioactivity |
|---|---|---|---|---|
| -80°C (Constant) | Minimal decrease | Preserves uniform size and integrity; minimal aggregation | Best preservation of RNA and protein content | Maintains biological functionality |
| -20°C | Significant decrease | Significant aggregation and size increase | Reduced stability and potential degradation | Likely impaired |
| Liquid Nitrogen (-196°C) | Less optimal than -80°C in some studies | Potential for membrane disruption and size reduction | Data limited; may be variable | Data limited |
| Multiple Freeze-Thaw Cycles | Decreases with each cycle | Increases aggregation and size | Decreases RNA content and integrity | Significantly impaired |
Experimental Protocols
Protocol 1: Standardized Storage and Thawing of MSC Exosomes
This protocol is designed to maximize the recovery of functional exosomes after freezing.
Key Research Reagent Solutions:
| Item | Function & Brief Explanation |
|---|---|
| Trehalose | A cryoprotectant that stabilizes the exosome lipid bilayer, preventing ice crystal damage during freezing. |
| Phosphate-Buffered Saline (PBS) | A common physiological buffer for resuspending purified exosome pellets. |
| Cryogenic Vials | Specially designed tubes that can withstand ultra-low temperatures without cracking. |
| Bovine Serum Albumin (BSA) | Can be used as a protein carrier to reduce surface adsorption and stabilize exosomes in solution. |
Procedure:
- Post-Isolation Resuspension: After isolation (e.g., by ultracentrifugation or size-exclusion chromatography), gently resuspend the final exosome pellet in a suitable buffer. Pure PBS is common, but consider adding 0.5-1% trehalose or 0.1% BSA for enhanced stability [82].
- Aliquoting: Immediately aliquot the exosome suspension into single-use cryogenic vials. The volume should be tailored to the needs of your downstream assays to avoid repeated thawing.
- Freezing: Flash-freeze the aliquots by placing them directly in a -80°C freezer. Alternatively, use a pre-chilled freezing container filled with isopropanol to ensure a rapid and consistent freezing rate, which helps preserve vesicle integrity.
- Storage: Store the aliquots at a constant -80°C. Avoid storing samples in freezer doors or areas prone to temperature fluctuations.
- Thawing: When needed, rapidly thaw a single aliquot by placing it in a 37°C water bath with gentle agitation. Once just thawed, immediately place the vial on ice.
- Post-Thaw Handling: Gently mix the thawed exosomes by flicking the tube or pipetting slowly. Avoid vortexing, as this can cause shear stress and damage the vesicles. Use immediately for experiments.
Protocol 2: Post-Storage Integrity Check
Before using a stored batch of MSC exosomes in critical experiments, perform this multi-parameter quality control check.
Procedure:
- Concentration and Size Distribution: Dilute a small volume of the thawed exosomes in sterile, particle-free PBS. Analyze the sample using Nanoparticle Tracking Analysis (NTA) to determine the particle concentration (particles/mL) and size distribution profile. Compare this to the pre-freeze data [83].
- Morphological Assessment: Use Transmission Electron Microscopy (TEM) to visualize the exosomes. Look for intact, cup-shaped vesicles. The absence of extensive debris, aggregation, or ruptured membranes indicates good preservation [83].
- Marker Profiling: Validate the presence of positive exosomal protein markers (e.g., CD63, CD81, TSG101) and the absence of negative markers (e.g., calnexin) via Western Blot. This confirms the identity of the vesicles and the absence of significant co-isolated contaminants [84] [83].
- Potency Assay (If applicable): Perform a relevant bioactivity assay. For immunomodulatory MSC exosomes, this could be a T-cell proliferation suppression assay or a macrophage polarization assay. This functional readout is the ultimate test of stability [60].
Workflow and Relationship Diagrams
Post-Storage Exosome QC
Key Stability Factors
Benchmarks, Clinical Translation, and Comparative Efficacy of MSC Exosome Products
Mesenchymal stem cell (MSC)-derived exosomes represent a promising frontier in regenerative medicine, drug delivery, and therapeutic applications. However, their profound heterogeneity, influenced by the MSC tissue source, culture conditions, and isolation methods, presents significant challenges for their analytical characterization [85]. Accurate assessment using a combination of tools is crucial for understanding this diversity and developing reproducible, pharmaceutical-grade exosome products. This technical support center provides targeted troubleshooting guides and FAQs to help researchers address common pitfalls in characterizing these complex nanoparticles.
Troubleshooting Guides for Key Characterization Techniques
Nanoparticle Tracking Analysis (NTA)
NTA is vital for determining the size distribution and concentration of MSC exosomes in a suspension. The following table outlines common issues and their solutions.
| Observation | Possible Cause | Solution |
|---|---|---|
| High background noise | Contamination with particles/dust or sub-optimal camera level setting. | Filter buffers (e.g., 0.02 µm) and clean work area. Adjust camera level until particles are clearly visible without background speckling. |
| Multiple peaks in size distribution | Genuine heterogeneity of exosome population or presence of protein aggregates [85]. | Incorporate a purification step (e.g., density gradient) to separate exosomes from non-vesicular particles. |
| Low particle concentration | Dilution factor too high or instrument focus is incorrect. | Re-optimize sample dilution. Carefully adjust the focus on the laser beam while monitoring the live video feed. |
Western Blotting
Western blotting is used to detect specific exosomal protein markers (e.g., CD63, CD81, TSG101) and assess sample purity. The tables below summarize common problems.
General Western Blot Issues
| Observation | Possible Cause | Solution |
|---|---|---|
| High background | Antibody concentration too high or insufficient blocking [86] [87]. | Titrate primary and secondary antibodies to find optimal concentration. Ensure blocking with 3-5% BSA or milk for at least 1 hour at room temperature. |
| Weak or no signal | Insufficient antigen (exosomal protein) or inefficient transfer to membrane [86] [87]. | Load more exosomal protein (10-20 µg). Confirm transfer efficiency by staining the membrane with reversible stains like Ponceau S. |
| Multiple non-specific bands | Antibody cross-reactivity or protein degradation [86]. | Use monospecific/affinity-purified antibodies. Add fresh protease inhibitors to lysis buffers during exosome preparation. |
Protein Gel Electrophoresis Issues
| Observation | Possible Cause | Solution |
|---|---|---|
| Streaking or distorted bands | Too much protein loaded per lane or excess salt in the sample [86]. | Reduce the amount of exosomal protein loaded per lane. Dialyze or desalt samples to ensure salt concentration does not exceed 100 mM. |
| Viscous samples, smeared lanes | Genomic DNA contamination from lysed cells during exosome isolation [86]. | Treat the exosome lysate with a nuclease (e.g., DNase I) to shear genomic DNA before adding sample buffer. |
Flow Cytometry
Flow cytometry is employed for the immunophenotyping of exosomes, often using beads to capture the vesicles for analysis.
| Observation | Possible Cause | Solution |
|---|---|---|
| High background/noise | Non-specific antibody binding or antibody aggregates. | Include an isotype control for every antibody. Ultracentrifuge antibodies before use to remove aggregates. |
| Low signal for specific markers | Antigen heterogeneity—the marker may not be present on all exosomes [85]. | Use a cocktail of antibodies against different exosomal surface markers (e.g., CD9, CD63, CD81) to account for population diversity. |
| Unreducible high sample pressure | Large particles or aggregates clogging the fluidic system. | Filter the exosome sample through a 0.2 µm filter before analysis to remove large aggregates. |
Electron Microscopy (EM)
EM, particularly Transmission EM (TEM), is the gold standard for visualizing the morphology and bilayer structure of exosomes.
| Observation | Possible Cause | Solution |
|---|---|---|
| Clumping of exosomes | Improper sample preparation or staining. | Use hydrophilic support films and ensure adequate negative staining with uranyl acetate. Avoid drying artifacts by using a freeze-plunging method. |
| Broken or deformed vesicles | Damage from harsh chemical fixation or dehydration. | Use a gentler fixation protocol, such as a combination of glutaraldehyde and paraformaldehyde, and ensure critical point drying. |
| Low contrast | Insufficient heavy metal staining. | Optimize the concentration and incubation time of the negative stain (e.g., 2% uranyl acetate for 1-2 minutes). |
Frequently Asked Questions (FAQs)
1. Why is a multi-method approach essential for characterizing MSC-derived exosomes? A single technique cannot capture the full complexity of MSC-exosomes due to their heterogeneous nature [85]. Each method provides complementary information: NTA for size/concentration, Western Blot for specific protein markers, EM for morphology, and Flow Cytometry for surface antigen profiling. Using them together is critical for comprehensive batch-to-batch quality control and validating the identity of your exosome preparation.
2. Our Western Blots for exosomal markers (like CD63) show weak signal even with high protein load. What could be wrong? This is a common challenge. First, confirm your isolation method is yielding sufficient and pure exosomes. Second, the heterogeneity of MSC-exosomes means that a specific marker like CD63 may not be equally abundant in all vesicles [85]. Try probing for a panel of markers (CD9, CD81, TSG101, Alix) instead of relying on a single one. Also, ensure complete lysis of the exosomal membrane with a strong RIPA buffer containing SDS to release all proteins for detection [86].
3. How does the heterogeneity of MSC-exosomes impact their analysis by NTA and Flow Cytometry? Heterogeneity directly affects the results. NTA will often show a polydisperse size distribution rather than a single, sharp peak, which is normal but must be carefully interpreted to distinguish genuine exosome subpopulations from technical artifacts like aggregates [85]. In Flow Cytometry, heterogeneity means that not all captured exosomes will express the same surface markers at the same levels. This necessitates the use of multiple antibodies and careful gating strategies to identify specific exosome subpopulations.
4. What are the critical reagent solutions for standardizing MSC-exosome characterization? Key materials include:
- Antibodies: Validate antibodies for exosomal markers (CD9, CD63, CD81, TSG101, Alix) and negative markers (e.g., Calnexin) using knockout controls if possible.
- Reference Materials: Use standardized polystyrene beads (e.g., 100 nm) for calibrating NTA and flow cytometers.
- Protease Inhibitors: Essential in all buffers during exosome isolation to prevent protein degradation for Western Blot analysis.
- Blocking Buffers: Optimize blocking buffers (e.g., BSA vs. non-fat milk) to minimize background in Western Blots, especially when detecting phosphoproteins [86].
Experimental Workflows and Relationships
The following diagram illustrates the typical workflow for the isolation and multi-method characterization of MSC-derived exosomes, highlighting how heterogeneity influences each step.
The next diagram outlines a logical troubleshooting pathway for a common problem—inconsistent Western blot results—when analyzing heterogeneous MSC-exosome samples.
Frequently Asked Questions (FAQs)
What is the primary challenge in linking exosome cargo to a specific biological function? The primary challenge is the significant heterogeneity of MSC-derived exosomes. Their protein and miRNA cargo is highly variable and is influenced by the MSC tissue source (e.g., bone marrow vs. umbilical cord), culture conditions, and the methods used for isolation and characterization. This variability makes it difficult to consistently correlate a specific cargo profile with a defined biological outcome [1] [75].
Why is an in vitro potency assay preferable to an in vivo assay for product release? In vitro potency assays offer significant benefits over in vivo assays, including:
- Reduced Ethical Concerns: They align with the 3Rs principles (Replacement, Reduction, and Refinement) by minimizing animal use [88].
- Lower Variability: In vitro assays demonstrate much lower variability (%CV often below 10%) compared to in vivo assays (%CV can range from 34% to 125%) [88].
- Faster and More Efficient: They provide faster turnaround times and are less resource-intensive [88].
- Greater Sensitivity: In some cases, in vitro assays can be more sensitive in detecting product degradation than in vivo immunogenicity tests [89].
My in vitro and in vivo potency results are inconsistent. What could be the cause? Inconsistencies often arise because an in vitro assay may not fully capture the complex immune response of a living organism. The in vitro assay might be measuring a single mechanism of action (e.g., antigen expression), while the in vivo immunogenicity is a net result of multiple biological processes. Ensuring that your in vitro assay is based on a biologically relevant mechanism and conducting correlation studies with degraded samples can help bridge this gap [89].
Troubleshooting Guides
Issue 1: High Variability in Exosome Cargo and Function
| Potential Cause | Investigation | Recommended Solution |
|---|---|---|
| Diverse MSC Sources | Characterize exosomes from different sources (e.g., bone marrow, adipose tissue). | Select and standardize the MSC tissue source based on the desired therapeutic function [1]. |
| Inconsistent Culture Conditions | Analyze exosome cargo from MSCs cultured in different media or under different conditions (e.g., 3D vs. 2D, hypoxia). | Implement strict, standardized protocols for cell culture medium and conditions [1]. |
| Uncontrolled Isolation Methods | Compare exosome yield, purity, and cargo using different isolation techniques (e.g., ultracentrifugation vs. precipitation). | Choose an isolation method that balances yield, purity, and efficiency for your application, and apply it consistently [90] [75]. |
Issue 2: Failed Correlation Between In Vitro and In Vivo Potency
| Potential Cause | Investigation | Recommended Solution |
|---|---|---|
| In vitro assay not biologically relevant | Review if the assay measures a key mechanism of action (MOA) linked to the in vivo effect. | Develop a cell-based in vitro assay that mimics the key biological step, such as antigen expression for mRNA vaccines [89]. |
| Insufficient data range for correlation | Test a narrow range of product potencies. | Create samples with a wide potency range (e.g., 0-100%) using controlled stress conditions (thermal, photo) to establish a robust correlation [89]. |
| In vivo model not suitable | Evaluate if the animal species shows a similar immune response to humans. | Investigate alternative animal models or endpoints; consider that some products may show correlation in non-human primates but not in mice [89]. |
Table 1: Variability of Different Potency Assay Types
| Assay Type | Typical Variability (%CV) | Key Sources of Variability |
|---|---|---|
| In Vivo Potency Assay | 34% - 125% | Individual animal physiology, complex biological systems [88]. |
| In Vitro Potency Assay | < 10% | Reagent stability, operator technique, equipment calibration [88]. |
| EV Proteomic Analysis | High (75% of variance unaccounted for) | Cell source, culture medium, isolation method, analytical platform [75]. |
Table 2: Functional Differences of MSC-Derived Exosomes by Tissue Source
| MSC Tissue Source | Associated Biological Functions (from literature) |
|---|---|
| Bone Marrow (BM) | Inhibition of inflammatory and apoptotic cells; maturation, proliferation, and activation of B-cells [1]. |
| Umbilical Cord (UC) | Suppression of oxidative stress; promotion of angiogenesis; improvement in proliferation and migration of skin cells for wound healing [1]. |
| Adipose Tissue (AD) | Used in treatments for skin, inflammation, and transplantation diseases [1]. |
Experimental Protocols
Protocol 1: Establishing In Vitro - In Vivo Potency Correlation
Objective: To develop and validate an in vitro potency assay that is predictive of in vivo immunogenicity.
Materials:
- Test articles (e.g., mRNA-LNP vaccine, MSC-derived exosomes)
- Relevant cell line (e.g., HepG2 for mRNA translation [89])
- Culture medium and reagents
- Antigen-specific antibodies (for immunoassay)
- Laboratory animals (e.g., mice)
- Equipment for cell culture, ELISA, PCR, etc.
Methodology:
- Generate Samples of Varying Potency: Subject your product to controlled stress conditions (e.g., thermal stress at 25°C, 37°C) for different time periods (0, 1, 3, 7 days) to create a series of samples with degraded quality [89].
- Measure In Vitro Potency:
- For mRNA vaccines: Transfert cells (e.g., HepG2) with the stressed samples and quantify antigen expression using a validated immunoassay (e.g., with fluorescently labeled antibodies). Calculate relative potency (EC50) [89].
- For exosomes: Use a relevant cell-based assay that measures a key function (e.g., modulation of immune cell proliferation).
- Measure In Vivo Immunogenicity:
- Immunize groups of animals (e.g., mice, n=5-10/group) with the same set of stressed samples.
- After a set period, collect serum and measure antigen-specific antibody titers (total IgG) and/or virus-neutralizing antibody potency (ED50) [89].
- Statistical Correlation:
- Plot the in vitro relative potency against the in vivo antibody titers or neutralization potency.
- Perform regression analysis to determine if a statistically significant correlation exists.
Protocol 2: Isolating and Characterizing MSC-Derived Exosomes
Objective: To isolate exosomes from MSC-conditioned medium and perform basic characterization.
Materials:
- Conditioned medium from MSCs
- Phosphate-buffered saline (PBS)
- Exosome isolation kit (precipitation-based)
- Ultracentrifuge (if using ultracentrifugation method)
- Transmission Electron Microscope (TEM)
- Nanoparticle Tracking Analysis (NTA) instrument
- Antibodies for flow cytometry (CD9, CD63, CD81, CD90, CD105)
Methodology:
- Isolation (Precipitation Method):
- Centrifuge conditioned medium at 3,000 × g for 15 min to remove cells and debris.
- Mix the supernatant with a precipitation solution and incubate overnight at 4°C.
- Centrifuge at 1,500 × g for 30 min to pellet the exosomes. Resuspend the pellet in PBS [90].
- Characterization:
- Size and Concentration: Use Nanoparticle Tracking Analysis (NTA) to determine the size distribution and particle concentration [90].
- Morphology: Use Transmission Electron Microscopy (TEM) to visualize the cup-shaped morphology of intact exosomes [90].
- Surface Markers: Confirm the presence of exosome-specific markers (CD9, CD63, CD81) and MSC markers (CD90, CD105) using flow cytometry, and confirm the absence of negative markers [90].
Experimental Workflows and Pathways
Diagram: Workflow for Potency Correlation Study
Diagram: Factors Influencing Exosome Heterogeneity
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Exosome and Potency Research
| Item | Function/Benefit |
|---|---|
| Selective Monoclonal Antibodies | Essential for in vitro immunoassays. They recognize conformational antigenic epitopes linked to virus-neutralizing immune response, forming the basis of potency assays [89]. |
| HepG2 Cell Line | A mammalian cell line frequently used in cell-based potency assays for mRNA vaccines due to its efficient protein expression and translatability to in vivo outcomes [89]. |
| Precipitation-based Isolation Kits | Provide a simple, efficient, and equipment-friendly method for isolating exosomes from conditioned medium, offering a practical alternative to ultracentrifugation [90]. |
| Defined MSC Culture Media | Standardized, serum-free media help reduce heterogeneity in exosome cargo by providing consistent growth conditions, improving experimental reproducibility [1] [75]. |
The field of clinical research involving Mesenchymal Stem Cell-derived exosomes (MSC-Exos) has experienced significant growth over the past decade, positioning these extracellular vesicles as promising cell-free therapeutic agents. Their appeal in regenerative medicine stems from their inherent immunomodulatory and regenerative properties, low immunogenicity, and ability to cross biological barriers like the blood-brain barrier [53] [50]. However, the clinical translation of MSC-exosome therapies is confronted with a fundamental challenge: significant heterogeneity in exosome populations and their molecular cargo. This variability influences their biological functions, therapeutic efficacy, and ultimately, the consistency and interpretability of clinical trial results [53] [60] [76].
Understanding this landscape requires analyzing the current state of registered clinical trials, the sources of heterogeneity, and the evolving regulatory framework. This technical support center article provides researchers with a structured guide to navigate these complexities, offering troubleshooting advice and clear protocols to enhance the reproducibility and reliability of their preclinical and clinical work.
A systematic review of global clinical trial registries (including ClinicalTrials.gov, the Chinese Clinical Trial Registry, and the Cochrane Register of Studies) up to February 2024 identified 66 eligible trials investigating MSC-EVs and Exos registered between 2014 and 2024 [53]. The data reveal trends in administration routes, disease targets, and sources.
Table 1: Analysis of Administration Routes in MSC-Exosome Clinical Trials
| Administration Route | Prevalence | Common Disease Targets | Notable Findings |
|---|---|---|---|
| Intravenous Infusion | Predominant method | Various systemic conditions | Requires higher doses compared to local administration |
| Aerosolized Inhalation | Predominant method, especially for respiratory diseases | COVID-19, ARDS, lung injuries | Achieves therapeutic effects at significantly lower doses (approx. 10^8 particles) [53] |
| Local Injection | Less common | Joint disorders, localized wounds | Enables high local concentration, potentially lowering systemic exposure |
Table 2: Common Mesenchymal Stem Cell Sources for Exosome Production in Clinical Trials
| MSC Source | Prevalence in Trials | Therapeutic Associations |
|---|---|---|
| Bone Marrow (BM) | High (Approx. 50% of "Old" MSC sources) | Widely studied; established history [53] [76] |
| Umbilical Cord (UC) | High (Approx. 33% of "Young" MSC sources) | Sought after for prolific output and therapeutic potential [53] [76] |
| Adipose Tissue (AD) | Common | Readily accessible, subject to donor-related variability [76] |
The analysis of these trials highlights a critical and often underappreciated gap: the dose-response relationship. Therapeutic efficacy is highly route-dependent, with nebulization achieving effects at doses around 10^8 particles, which is significantly lower than required for intravenous routes. This underscores a narrow effective dose window and the urgent need for standardized dosing frameworks and potency assays [53].
Critical Challenges: Heterogeneity in MSC Exosome Populations and Cargo
FAQs on Heterogeneity and Its Impact
Q1: What are the primary factors contributing to heterogeneity in MSC-exosome preparations? A1: Heterogeneity arises from multiple variables, which can be categorized as follows [53] [60] [76]:
- Biological Source: The tissue origin of the MSCs (e.g., bone marrow vs. umbilical cord), donor age, and health status significantly influence exosome characteristics [53] [76].
- Process Conditions: Culture conditions (e.g., media composition, use of 2D vs. 3D bioreactors), and the specific methods used for isolation and purification (e.g., ultracentrifugation, tangential flow filtration) introduce substantial variability [53] [60].
- Inherent Biological Diversity: Even clonal MSC lines can exhibit heterogeneity due to factors like epigenetic modifications and genetic drift, leading to a mixed population of vesicles with different cargo and functions [60].
Q2: How does heterogeneity impact the reliability of clinical trial data and regulatory approval? A2: Heterogeneity directly challenges the establishment of Critical Quality Attributes (CQAs), which are essential for defining a product's identity, purity, potency, and safety. Inconsistent exosome populations lead to [60] [76]:
- Variable therapeutic outcomes, making it difficult to determine a true dose-response.
- Difficulty in replicating results across different research sites and manufacturing batches.
- Challenges in satisfying regulatory requirements for product consistency and potency, potentially delaying or preventing approval.
Q3: What does the proteomic data reveal about exosome heterogeneity? A3: A recent analysis of 52 proteomic studies highlighted the extreme variability in exosome cargo. Across 13,000 observed proteins, 40% were identified in only a single study. Statistical models could only account for 25-60% of the variance, even when considering factors like EV source, culture medium, and isolation method. This underscores that a large portion of variability remains unaddressed by current standardization efforts [76].
Troubleshooting Guide: Addressing Heterogeneity in Your Research
Problem: Inconsistent functional outcomes in cell-based assays.
- Potential Cause: Variability in exosome cargo (e.g., protein or miRNA content) between batches.
- Solution:
- Implement robust characterization protocols beyond just particle size and concentration. This includes:
- Western Blot or Flow Cytometry for surface markers (e.g., CD9, CD63, CD81).
- Proteomic analysis to track key functional proteins in different batches [76].
- Develop a functional potency assay relevant to your therapeutic mechanism (e.g., T-cell modulation for immunotherapies) and use it to qualify each batch [60].
- Adhere closely to the Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines for reporting to improve reproducibility [76].
- Implement robust characterization protocols beyond just particle size and concentration. This includes:
Problem: Low yield during exosome isolation.
- Potential Cause: Inefficiency or inconsistency of the isolation method.
- Solution:
- Compare isolation methods for your specific MSC source and medium. Common methods include:
- Differential Ultracentrifugation: The "gold standard" but can cause vesicle damage [50].
- Density Gradient Centrifugation: Higher purity but lower yield and complex operation [50].
- Ultrafiltration: Simpler and avoids chemical contaminants, but shear force may damage EVs [50].
- Size-Exclusion Chromatography: Good for preserving vesicle integrity and function [91].
- Carefully control and document all process parameters (e.g., g-forces, filtration pore sizes, buffer compositions).
- Compare isolation methods for your specific MSC source and medium. Common methods include:
Problem: Difficulty in scaling up production for clinical trials.
- Potential Cause: Laboratory-scale isolation methods are not translatable to large-scale manufacturing.
- Solution:
Emerging Safety and Regulatory Considerations
Monitoring Safety and Adverse Events
While MSC-exosomes are generally considered to have a high biosafety profile due to their low immunogenicity and lack of a nucleus (preventing tumorigenesis), safety monitoring remains paramount [50]. Researchers and clinicians should be aware of emerging frameworks for tracking potential risks.
The FDA Adverse Event Reporting System (FAERS) is a database that contains information on adverse event and medication error reports submitted to the FDA. The FDA regularly screens this database to identify potential signals of serious risks. It is critical to note that the appearance of a therapy on a FAERS report does not mean the FDA has concluded the drug causes the risk, but rather that a potential safety issue has been identified and requires further evaluation [92].
For clinical trial sponsors, compliance with safety reporting is mandatory. The FDAAA 801 Final Rule, updated in 2025, has introduced stricter timelines and transparency requirements. Key updates include [93]:
- Shortened Timelines: Results of applicable clinical trials must now be submitted to ClinicalTrials.gov within 9 months (previously 12 months) of the primary completion date.
- Enhanced Enforcement: There are now real-time public notifications for non-compliance and significantly higher penalties, which can reach $15,000 per day for ongoing violations.
- Increased Transparency: Mandatory posting of informed consent documents (redacted) is now required.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials and Reagents for MSC-Exosome Research
| Reagent/Material | Primary Function | Key Considerations for Use |
|---|---|---|
| CD9, CD63, CD81 Antibodies | Characterization of exosomes via Western Blot or Flow Cytometry | Tetraspanins are common exosome markers; use a combination for reliable identification [76] [91]. |
| Annexin V / Propidium Iodide | Assessment of apoptotic bodies and cell viability during production | Distinguishes exosomes from larger apoptotic bodies (500-800 nm) released by dying cells [50]. |
| Sucrose or Iodixanol | Formulation of density gradients for high-purity isolation | Crucial for density gradient centrifugation; requires careful removal post-isolation [50]. |
| PBS (Exosome-Depleted) | Resuspension and buffer for isolated exosomes | Standard PBS must be ultracentrifuged or filtered to remove bovine exosomes from serum-containing media [76]. |
| RNA/DNA Extraction Kits | Analysis of nucleic acid cargo (miRNA, mRNA) | Select kits optimized for small volumes and low concentrations of nucleic acids [94]. |
| Nanoparticle Tracking Analysis (NTA) | Determining particle size distribution and concentration | Complements protein-based quantification; provides a physical particle count [53] [50]. |
Experimental Protocols and Workflows
Detailed Protocol: Standardized Isolation and Characterization of MSC-Exosomes
This protocol focuses on a common, reproducible workflow for laboratory-scale production.
Step 1: MSC Culture and Conditioned Media Collection
- Culture MSCs from a defined source (e.g., Bone Marrow) in a standardized, xeno-free medium to minimize variability [76].
- At 70-80% confluence, replace the growth medium with a serum-free, exosome-depleted medium.
- After 48 hours, collect the conditioned media and perform a low-speed centrifugation (e.g., 2,000 × g for 30 minutes) to remove cells and large debris.
Step 2: Exosome Isolation via Ultracentrifugation
- Transfer the supernatant to ultracentrifugation tubes.
- Centrifuge at 10,000 × g for 30 minutes at 4°C to remove larger vesicles and organelles.
- Carefully transfer the supernatant to fresh tubes and ultracentrifuge at 100,000 × g for 70 minutes at 4°C to pellet the exosomes [50].
- Discard the supernatant and resuspend the pellet in a large volume of sterile, phosphate-buffered saline (PBS).
- Perform a second ultracentrifugation at 100,000 × g for 70 minutes at 4°C (washing step).
- Finally, resuspend the purified exosome pellet in a small volume of PBS and aliquot for storage at -80°C.
Step 3: Characterization (The "Identity Triad")
- Particle Concentration and Size: Use Nanoparticle Tracking Analysis (NTA) to confirm a size profile of 30-200 nm and determine particle concentration [53] [50].
- Protein Marker Profile: Confirm the presence of positive markers (e.g., CD9, CD63, CD81) and absence of negative markers (e.g., GM130) via Western Blot [76] [91].
- Morphology: Use Transmission Electron Microscopy (TEM) to visualize the classic "cup-shaped" morphology of exosomes.
Diagram 1: Workflow for MSC-Exosome Isolation and Characterization
Visualizing the Impact of Process Conditions on Heterogeneity
The following diagram synthesizes how different factors influence the final therapeutic exosome product, contributing to the heterogeneity challenge.
Diagram 2: Factors Driving Exosome Heterogeneity
What are the core differences between MSC exosomes, whole-cell therapies, and synthetic nanoparticles? The choice of therapeutic platform significantly impacts the strategy for drug delivery and regenerative medicine. The table below compares the core characteristics of these three platforms.
Table: Core Characteristics of Therapeutic Platforms
| Feature | MSC Exosomes | Whole MSC Therapy | Synthetic Nanoparticles |
|---|---|---|---|
| Nature | Cell-derived, natural nanovesicles (40-150 nm) [95] [32] | Living, intact cells [52] | Engineered particles (e.g., liposomes, polymeric NPs) [32] |
| Mechanism | Paracrine signaling; cargo delivery (proteins, nucleic acids) [96] [32] | Direct differentiation & potent paracrine signaling [52] | Encapsulation and controlled release of drugs [32] |
| Immunogenicity | Low (low MHC content) [1] | Moderate (risk of immune rejection) [1] | Variable (can trigger adverse reactions) [32] |
| Tumorigenicity Risk | Low (cell-free, cannot proliferate) [1] | Potential risk [97] | Not applicable |
| Targeting | Innate homing ability; can be engineered [96] | Limited, uncontrolled migration [98] | Requires surface functionalization [32] |
| Production Scalability | Challenging (isolation and purification) [1] [98] | Challenging (costs, culture expansion) [95] | Highly scalable [32] |
| Storage & Stability | Sensitive; requires -80°C; prone to cargo degradation [98] | Requires complex cryopreservation [98] | Generally high stability [32] |
Efficacy and Functional Comparisons
How does the therapeutic efficacy of MSC exosomes compare to whole MSCs in preclinical models? Evidence from umbrella reviews of preclinical meta-analyses demonstrates that MSC-derived extracellular vesicles (MSC-EVs), including exosomes, exhibit robust therapeutic potential across diverse disease models, including neurological, renal, and musculoskeletal disorders [97]. The efficacy of exosomes often parallels that of their parent cells, as they recapitulate many therapeutic effects via their cargo.
Table: Comparative Therapeutic Efficacy by MSC Source
| MSC Source | Key Efficacy Findings | Noted Advantages |
|---|---|---|
| Bone Marrow (BMSCs) | Superior efficacy in attenuating inflammation and promoting cartilage protection in osteoarthritis models [95]. | Extensive research history; strong chondroprotective and anti-inflammatory effects [95] [1]. |
| Umbilical Cord (UMSCs) | Displayed superior anti-inflammatory efficacy and enhanced chondrocyte migration, comparable to BMSC-Exos [95]. | High proliferation capacity; strong immunomodulatory properties; young donor source [95] [17]. |
| Adipose Tissue (ADSCs) | Potent chondrogenic capabilities, but anti-inflammatory effects may be less pronounced than BMSC- or UMSC-Exos [95]. | Easily accessible source; promotes angiogenesis and wound healing [1]. |
In what ways do exosomes outperform synthetic nanoparticles as drug delivery vehicles? Exosomes offer distinct advantages as naturally derived delivery systems. Their lipid bilayer membrane provides high biocompatibility and lower toxicity compared to synthetic nanoparticles [32]. Crucially, their innate surface proteins facilitate membrane fusion and receptor-mediated uptake by recipient cells, enhancing delivery efficiency [96] [32]. Furthermore, their ability to cross biological barriers and inherent homing capabilities to injury sites are significant benefits over synthetic counterparts, which are often rapidly cleared and require complex engineering for targeting [97] [32].
Experimental Protocols for Efficacy Validation
What is a standard experimental workflow for comparing the anti-inflammatory efficacy of exosomes from different MSC sources? The following workflow, adapted from a comparative study on osteoarthritis models, outlines a robust methodology for this purpose [95].
Detailed Methodology:
Exosome Isolation & Characterization:
- Isolation: Use differential ultracentrifugation or aqueous two-phase system (ATPS) to isolate exosomes from conditioned media of BMSCs, ADSCs, and UMSCs [95].
- Characterization:
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration (e.g., BMSC-Exos: ~6.9 × 10^7 particles/mL) [95].
- Transmission Electron Microscopy (TEM): Confirm classic cup-shaped morphology [95].
- Western Blot (WB): Verify the presence of exosomal markers (CD63, CD81, ALIX) and absence of negative markers (e.g., Calnexin) [95].
In Vitro Model & Intervention:
Efficacy Assessment (Key Assays):
- Cell Viability (CCK-8 Assay): Confirm exosomes show low cytotoxicity at working concentrations [95].
- Western Blot Analysis of Signaling Pathways:
- NF-κB pathway: Analyze phosphorylation levels of p65 (pp65).
- MAPK pathway: Analyze phosphorylation levels of p38 (pp38), JNK (pJNK), and ERK (pERK).
- BMSC-Exos and UMSC-Exos typically show stronger reduction in these phosphorylated proteins than ADSC-Exos [95].
- Migration Assay (Scratch Assay): Quantify enhanced chondrocyte migration, a critical component of cartilage repair, after exosome treatment [95].
How do I investigate the molecular mechanisms behind exosome-mediated therapeutic effects? A critical pathway involves analyzing the modulation of the NF-κB and MAPK signaling pathways, which are central to inflammation.
Troubleshooting Common Experimental Challenges
How can I address the heterogeneity of MSC exosomes in my experiments? Heterogeneity is a major challenge arising from donor, tissue source, and culture conditions [1] [10]. Mitigation strategies include:
- Source Standardization: Clearly report the MSC source (e.g., bone marrow, umbilical cord), donor age, and health status [95] [10]. Consider pooling exosomes from multiple donors to minimize inter-individual variation [10].
- Process Control: Standardize culture conditions (e.g., oxygen levels, medium composition, passage number) and exosome isolation methods (e.g., ultracentrifugation vs. size-exclusion chromatography) across all experiment batches [1].
- Rigorous Characterization: Always characterize exosome preparations for size, concentration, and standard markers (CD63, CD81, ALIX) before functional experiments. This provides critical quality control and allows for result normalization [95] [98].
What are common pitfalls in exosome isolation and how can I avoid them? Isolation pitfalls can compromise exosome integrity and functionality.
- Pitfall: Co-isolation of Contaminants. Ultracentrifugation can co-precipitate protein aggregates and lipoproteins [98].
- Solution: Consider a purity-enhancing step like density gradient centrifugation or size-exclusion chromatography (SEC) [98].
- Pitfall: Low Yield or Damaged Exosomes. High centrifugal forces can damage exosomes and reduce functional yield [98].
- Solution: Optimize g-forces and duration. For sensitive applications, gentler methods like SEC or ultrafiltration may be preferable [98].
- Pitfall: Inconsistent Starting Material.
- Solution: Use a consistent volume of conditioned media or normalize to the total cell number or protein content during MSC culture to ensure reproducible exosome production [1].
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Materials and Reagents for MSC Exosome Research
| Reagent / Kit | Function / Application | Key Considerations |
|---|---|---|
| Mesenchymal Stem Cells | Source of exosomes. | Choose source (e.g., BM, UC, AD) based on desired efficacy profile [95] [1]. Monitor differentiation potential and surface markers (CD105+, CD73+, CD90+, CD45-) [10]. |
| Exosome Isolation Kits | Isolation from conditioned media or biofluids. | Polymer-based precipitation (high yield, potential polymer contamination); Size-Exclusion Chromatography (SEC) (high purity, preserves integrity) [98]. |
| Characterization Instruments | Confirm exosome identity and quantity. | NTA (size/concentration), TEM (morphology), Western Blot (marker validation). Use a multi-method approach [95]. |
| Cell-Based Assay Kits | Assess functional efficacy. | CCK-8 (cytotoxicity), ELISA (cytokine quantification), Apoptosis Assay (e.g., Annexin V), Migration Assay (e.g., scratch assay) [95]. |
| Antibodies | Characterize exosomes and analyze signaling. | Anti-tetraspanins (CD63, CD81, CD9), Anti-ALIX/TSG101; Phospho-specific antibodies (pp65, pp38, pJNK, pERK) for pathway analysis [95]. |
Frequently Asked Questions (FAQs)
Q: Are MSC exosomes FDA-approved for clinical use? A: As of now, the FDA has not officially approved any exosome products. However, numerous clinical trials are ongoing to evaluate their safety and efficacy for specific conditions, which is a necessary pathway toward future approvals [17].
Q: Can MSC exosomes be used as a drug delivery system? A: Yes, this is a major area of research. Exosomes can be loaded with therapeutic cargo (e.g., small molecule drugs, miRNAs, proteins) using techniques like electroporation, sonication, or transfection. Their natural targeting capabilities make them promising engineered delivery vehicles [96] [32].
Q: What is the primary advantage of using a cell-free exosome therapy over whole MSCs? A: Exosomes offer a cell-free therapy that circumvents risks associated with whole cells, such as uncontrolled differentiation, potential vascular occlusion, and immune rejection. They are also more stable, have a longer shelf life, and can be engineered for targeted delivery [1] [97] [98].
Q: How should I store my exosome preparations to maintain stability? A: Exosomes are sensitive to repeated freeze-thaw cycles and storage at -20°C. For long-term stability, aliquot exosome preparations and store them at -80°C. Avoid mechanical stress and document storage duration, as it can impact cargo integrity over time [98].
Mesenchymal stromal/stem cell-derived exosomes (MSC-Exos) represent a promising new class of therapeutic agents, shifting the paradigm from cell-based therapies to cell-free treatments. These nanoparticles (30-150 nm in diameter) mediate the therapeutic effects of MSCs through their cargo of proteins, lipids, and nucleic acids, influencing processes including immunomodulation, tissue repair, and angiogenesis [99]. Unlike traditional small molecule drugs with defined chemical structures, MSC-Exos are inherently heterogeneous products. This heterogeneity presents unique challenges for regulatory approval, as it complicates the establishment of consistent identity, purity, potency, and quality across manufacturing batches [1] [60].
The transition from MSC therapies to MSC-Exos offers clinical advantages, including reduced challenges with cell viability during storage, easier administration, and potentially improved pharmacological predictability. However, this transition introduces significant regulatory complexities. Manufacturing MSC-sEV (small extracellular vesicle) products faces challenges in defining critical quality attributes (CQAs) necessary for ensuring consistent product identity and potency [60]. This technical support guide addresses the specific regulatory and technical challenges researchers face when developing MSC-Exos therapeutics, with particular emphasis on managing heterogeneity throughout the product lifecycle.
Frequently Asked Questions (FAQs) on Regulatory Pathways
Q1: What are the current FDA regulatory considerations for MSC-derived exosome therapies?
The FDA's Center for Biologics Evaluation and Research (CBER) oversees the regulation of MSC-derived exosome products. While specific formal guidance documents for exosomes are still under development, the FDA's 2025 Guidance Agenda indicates several relevant topics that will inform exosome therapeutic development [100]. These include:
- Potency Assurance for Cellular and Gene Therapy Products: This guidance will be critical for establishing potency assays for exosome products, which is particularly challenging for heterogeneous products with multiple mechanisms of action [100].
- Post-Approval Methods to Capture Safety and Efficacy Data for Cell and Gene Therapy Products: This directly applies to exosome therapies for post-market safety monitoring [100].
- Considerations for the Use of Human- and Animal-Derived Materials and Components in Manufacture: This is relevant given the sensitivity of exosome cargo to culture conditions [100].
Q2: How does product heterogeneity impact the regulatory pathway for MSC-Exos?
Heterogeneity introduces complexity at multiple regulatory levels. Regulators require demonstration that your manufacturing process consistently produces a product with predictable characteristics and biological effects, despite inherent heterogeneity. Key considerations include:
- Defining Acceptable Ranges: Establishing quantitative ranges for critical quality attributes (CQAs) rather than single-point measurements.
- Multiple Potency Assays: Developing multiple potency assays that reflect the product's known mechanisms of action, as a single assay may not capture the full therapeutic profile [60].
- Process-as-Product Definition: Embracing that "the process defines the product" is particularly relevant for exosomes, where manufacturing conditions significantly influence the final product profile [60].
Q3: What are the key differences between the FDA and EMA in their approach to heterogeneous biological products?
While both agencies require demonstration of safety, quality, and efficacy, some philosophical differences exist:
Table: Comparison of Regulatory Approaches to Heterogeneous Products
| Aspect | FDA (U.S.) | EMA (Europe) |
|---|---|---|
| Guidance Specificity | Prefers product-specific guidance; 2025 agenda includes cell/gene therapy topics relevant to exosomes [100] | Often employs more overarching guidelines for advanced therapy medicinal products (ATMPs) |
| Risk-Based Approach | Flexible framework with opportunities for accelerated pathways for rare diseases | Similar risk-based approach but may emphasize different risk categories |
| Potency Requirements | Requires potency assays linked to biological activity; particularly challenging for heterogeneous products [60] | Similarly requires comprehensive potency testing with multiple assays |
| Manufacturing Controls | Focus on process validation and control strategies to manage heterogeneity | Similar focus with possible differences in required validation data |
Q4: What strategies can be employed to address heterogeneity in MSC-Exos during product development?
- Source Control: Carefully select and characterize your MSC source (bone marrow, adipose tissue, umbilical cord), as this significantly influences exosome composition and function [1] [99].
- Process Control: Implement tightly controlled culture conditions (media composition, 3D vs. 2D culture, oxygen tension) as these parameters dramatically affect exosome cargo and yield [1].
- Advanced Characterization: Employ multiple orthogonal methods for exosome characterization (NTA, SEC, AF4, electron microscopy) to fully understand the profile of your product [101].
- Function-Based Potency Assays: Develop potency assays based on biological function rather than solely on physical characteristics [60].
Troubleshooting Guides for Common Experimental Challenges
Challenge 1: Inconsistent Therapeutic Effects Between Batches
Potential Causes and Solutions:
- Cause: Uncontrolled variability in MSC culture conditions affecting exosome cargo.
- Solution: Implement standardized culture protocols with defined media components, consistent cell passage numbers, and monitored cell confluence at harvest.
- Protocol: Use serum-free, chemically-defined media to eliminate variability introduced by serum-derived exosomes [101]. Document population doubling levels and establish a maximum passage number for production cells.
Cause: Inadequate characterization of starting MSC population.
- Solution: Implement rigorous MSC characterization including surface marker profiling (CD73+, CD90+, CD105+, CD34-, CD45-), differentiation potential assays, and functional immunomodulation assays before exosome production.
Cause: Inconsistent exosome isolation methods.
- Solution: Standardize isolation protocols with quality control checkpoints. Consider moving from differential ultracentrifugation to more reproducible methods like size exclusion chromatography or tangential flow filtration for clinical-scale production [101].
Challenge 2: Inadequate Potency for Desired Therapeutic Effect
Potential Causes and Solutions:
- Cause: Suboptimal MSC priming or preconditioning.
- Solution: Implement MSC preconditioning strategies to enhance exosome potency:
- Inflammatory Priming: Pre-treat MSCs with IFN-γ or TNF-α to enhance immunomodulatory exosome cargo.
- Hypoxic Conditioning: Culture MSCs under mild hypoxia (1-5% O₂) to enhance pro-angiogenic and regenerative factors [1].
- 3D Culture Systems: Utilize spheroid culture or bioreactors to create more physiologically relevant cell interactions [1].
- Solution: Implement MSC preconditioning strategies to enhance exosome potency:
- Cause: Incorrect dosing rationale.
- Solution: Establish dose-response relationships in relevant preclinical models. Note that the most effective dose is not necessarily the highest dose [99].
Challenge 3: Failure to Meet Regulatory Purity Specifications
Potential Causes and Solutions:
- Cause: Co-isolation of non-exosomal contaminants.
- Solution: Implement orthogonal purification methods such as combining ultracentrifugation with size exclusion chromatography [101].
- Protocol: For blood samples, include density-based separation methods to remove lipoproteins, which are major contaminants in plasma-derived exosome preparations [101].
- Cause: Incomplete characterization of product composition.
- Solution: Implement comprehensive characterization using multiple complementary techniques:
Table: Essential Characterization Methods for MSC-Exos
| Method | Parameter Measured | Regulatory Purpose | Target Specification |
|---|---|---|---|
| Nanoparticle Tracking Analysis (NTA) | Size distribution and concentration | Identity, quality | Majority particles 30-150 nm; report PDI |
| Transmission Electron Microscopy | Morphology and ultrastructure | Identity | Cup-shaped morphology, intact membrane |
| Western Blot | Surface and cargo markers (CD63, CD81, CD9, TSG101) | Identity, purity | Positive for tetraspanins, negative for calnexin |
| MicroRNA/RNA Profiling | Nucleic acid cargo | Quality, potential potency | Batch-to-batch consistency in profile |
| Proteomic Analysis | Protein composition | Identity, potency | Consistent profile, presence of key functional proteins |
Experimental Protocols for Critical Characterization assays
Protocol 1: Comprehensive MSC-Exos Characterization Workflow
This standardized protocol ensures consistent characterization of MSC-Exos for regulatory submissions:
Step 1: Sample Preparation
- Isolate exosomes using your validated method (SEC recommended for reproducibility) [101].
- Resuspend exosome pellet in PBS and perform protein quantification using BCA assay.
- Aliquot samples for different characterization assays and store at -80°C if not used immediately.
Step 2: Size and Concentration Analysis (NTA)
- Dilute exosome preparation 1:100 - 1:1000 in filtered PBS to achieve 20-100 particles per frame.
- Acquire five 30-second videos using Nanosight NS300 system.
- Analyze with detection threshold set to 5 and screen gain at 10.
- Report mean size, mode size, D10, D50, D90, and concentration (particles/mL).
Step 3: Surface Marker Characterization (Flow Cytometry)
- Bind exosomes to 4μm aldehyde/sulfate latex beads (Invitrogen) by incubating 10μg exosome protein with 5μL beads in 100μL PBS overnight at 4°C.
- Block with 100mM glycine for 30 minutes.
- Incubate with fluorochrome-conjugated antibodies against CD63, CD81, CD9, and appropriate isotype controls.
- Analyze on flow cytometer, gating on bead population.
Step 4: Functional Potency Assay (Lymphocyte Proliferation)
- Isolate PBMCs from healthy donor blood by density gradient centrifugation.
- Label with CFSE cell division tracker dye.
- Activate with CD3/CD28 activation beads.
- Add MSC-Exos at multiple concentrations (e.g., 10, 50, 100 μg/mL).
- After 96 hours, analyze T cell proliferation by flow cytometry using CFSE dilution.
- Calculate percentage inhibition compared to activated control without exosomes.
Protocol 2: MSC Preconditioning to Enhance Exosome Potency
Hypoxic Preconditioning Protocol:
- Culture MSCs to 70-80% confluence in standard culture flasks.
- Place cells in hypoxia chamber with 1% O₂, 5% CO₂, and balance N₂ for 48 hours.
- Collect conditioned media and isolate exosomes using standard protocol.
- Validate enhanced potency using angiogenesis assay (HUVEC tube formation) or immunomodulation assay.
Inflammatory Priming Protocol:
- Culture MSCs to 70-80% confluence.
- Treat with 50 ng/mL IFN-γ for 48 hours.
- Wash cells thoroughly with PBS and replace with fresh media for exosome collection.
- Isolate exosomes and validate enhanced immunomodulatory potency using T cell proliferation assay.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table: Essential Materials for MSC-Exos Research and Development
| Category | Specific Items | Function/Purpose | Considerations for Regulatory Compliance |
|---|---|---|---|
| Cell Culture | Serum-free, xeno-free media (e.g., StemMACS MSC XF) | MSC expansion without introducing animal-derived contaminants | Essential for regulatory approval; eliminates variability from serum batches [1] |
| Characterization | CD63, CD81, CD9 antibodies; NTA system; TEM | Identity confirmation and quality assessment | Use GMP-grade antibodies for clinical lot testing; validate methods [101] |
| Isolation | Size exclusion columns (qEVoriginal); Ultracentrifugation equipment | Reproducible exosome isolation with minimal contaminants | SEC provides higher purity than UC alone; more suitable for scalable production [101] |
| Functional Assays | HUVEC cells for angiogenesis; PBMCs for immunomodulation | Potency assessment | Establish assay acceptance criteria; document control responses [99] |
| Storage | Cryopreservation solutions (e.g., Trehalose) | Maintain exosome stability during storage | Validate storage conditions and expiration dating [60] |
Regulatory Strategy Diagram
Navigating regulatory pathways for heterogeneous MSC-derived exosome products requires a strategic approach that acknowledges both their complexity and therapeutic potential. By implementing robust characterization methods, controlling manufacturing processes, developing relevant potency assays, and engaging early with regulatory agencies, developers can successfully advance these promising therapies through the regulatory landscape. The frameworks presented in this technical support guide provide a foundation for addressing the unique challenges presented by heterogeneous biological products, with emphasis on the current regulatory expectations and scientific best practices.
Conclusion
The heterogeneity of MSC-derived exosomes is not merely a challenge to be overcome but a fundamental property that, when understood and controlled, unlocks their immense therapeutic potential. Success in this field hinges on a multi-faceted approach: standardizing upstream processes and isolation methods, implementing robust characterization frameworks based on critical quality attributes, and advancing bioengineering strategies to direct cargo loading and targeting specificity. Future progress will depend on interdisciplinary collaboration to establish universally accepted manufacturing standards, comprehensive biodistribution studies, and clinical trials designed to correlate specific exosome subpopulations with therapeutic outcomes. By systematically addressing heterogeneity, the scientific community can transform MSC exosomes from a biologically complex mixture into a new class of programmable, cell-free nanomedicines capable of precise intervention in a wide range of diseases.
References
- 1. Heterogeneity of mesenchymal stem cell-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9063275/]
- 2. Mesenchymal stem cell-derived extracellular vesicles for ... [https://www.nature.com/articles/s41419-022-05034-x]
- 3. Mesenchymal Stem Cell-Derived Exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC8393426/]
- 4. Efficient methods of isolation and purification of extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12463815/]
- 5. Advances of extracellular vesicles isolation and detection ... [https://jnanobiotechnology.biomedcentral.com/articles/10.1186/s12951-025-03768-2]
- 6. Secretome and extracellular vesicle signatures in bone ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12367461/]
- 7. A comprehensive review on recent advances in exosome ... [https://www.sciencedirect.com/science/article/pii/S1936523324002481]
- 8. Advances in Mesenchymal Stem Cell-Derived Exosomes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8854766/]
- 9. Regulation of cargo selection in exosome biogenesis and ... [https://www.nature.com/articles/s12276-024-01209-y]
- 10. The heterogeneity of mesenchymal stem cells: an important ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-023-03587-y]
- 11. Extracellular Vesicle Heterogeneity and Its Impact for ... [https://www.sciencedirect.com/science/article/pii/S0031699724007907]
- 12. Diverse Long RNAs Are Differentially Sorted into ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6248336/]
- 13. Where Are Mesenchymal Stem Cells (MSCs) Found in the ... [https://celltexbank.com/blog-msc-tissue-sources/]
- 14. The Pros and Cons of Mesenchymal Stem Cell-Based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6719501/]
- 15. The Opportunities and Challenges of Mesenchymal Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11640604/]
- 16. Preconditioning and Engineering Strategies for Improving the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9124131/]
- 17. MSC Exosome Therapy FAQ | Safety and Compliance [https://mscexosometherapy.com/msc-exosome-therapy-faq]
- 18. Applications, challenges and prospects of mesenchymal stem ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-021-02596-z]
- 19. The issue of heterogeneity of MSC-based advanced ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2024.1400347/full]
- 20. The Influence of Circulating Exosomes Derived From ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11526583/]
- 21. The Comparative Effect of Plasma Exosomes of Young and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11512957/]
- 22. Exosome Source Matters: A Comprehensive Review from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11858990/]
- 23. Age-Related Changes in Plasma Extracellular Vesicle ... [https://www.nature.com/articles/s41598-017-01386-z]
- 24. Origin and Composition of Exosomes as Crucial Factors in ... [https://www.mdpi.com/2076-3417/12/23/12259]
- 25. Extracellular Vesicle Heterogeneity: Subpopulations ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2018.00738/full]
- 26. Cells release subpopulations of exosomes with distinct ... [https://www.nature.com/articles/srep22519]
- 27. The role of exosomal molecular cargo in ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1417758/full]
- 28. Exosomes Frequently Asked Questions (FAQs) [https://www.thermofisher.com/us/en/home/life-science/cell-analysis/exosomes/exosomes-webinar-q-and-a.html]
- 29. Challenges and opportunities in exosome research ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6481742/]
- 30. Current Strategies for Exosome Cargo Loading and Targeting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10216928/]
- 31. Mesenchymal stem cell-derived exosome subpopulations ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1550447/full]
- 32. Recent advances in extracellular vesicles for therapeutic ... [https://www.nature.com/articles/s12276-024-01201-6]
- 33. upstream bioprocessing, cell culture, biopharmaceuticals ... [https://www.leadventgrp.com/blog/upstream-bioprocessing-challenges-and-advances-in-cell-culture-systems]
- 34. Understanding Upstream Bioprocessing: Key Processes & ... [https://www.bioprocessonline.com/topic/understanding-upstream-bioprocessing-key-processes-and-trends]
- 35. A 3D culture system improves the yield of MSCs-derived ... [https://www.sciencedirect.com/science/article/pii/S0753332223003451]
- 36. Current Advances in 3D Dynamic Cell Culture Systems - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9778081/]
- 37. Advances in 3D Culture Models to Study Exosomes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10931050/]
- 38. The tiny giants of regeneration: MSC-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12310703/]
- 39. preconditioning strategies for MSC-derived extracellular ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1509418/full]
- 40. Exosomes from preconditioned mesenchymal stem cells [https://www.sciencedirect.com/science/article/pii/S2352320424000099]
- 41. Exosomes from preconditioned mesenchymal stem cells [https://pmc.ncbi.nlm.nih.gov/articles/PMC10875222/]
- 42. Advances in application of hypoxia-preconditioned ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11375678/]
- 43. Hypoxia preconditioned MSC exosomes attenuate high- ... [https://www.sciencedirect.com/science/article/pii/S2452199X25002464]
- 44. Harnessing engineered mesenchymal stem cell-derived ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04708-5]
- 45. engineering mesenchymal stem cell-derived small ... [https://cancerci.biomedcentral.com/articles/10.1186/s12935-025-03900-0]
- 46. Recent advances in CAR-MSCs: the new engine of cellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12557984/]
- 47. Advancing gene editing: the role of lipid nanoparticles in ... [https://www.drugtargetreview.com/article/188056/advancing-gene-editing-the-role-of-lipid-nanoparticles-in-crispr-delivery/]
- 48. CRISPR Delivery Methods: Cargo, Vehicles, and Challenges [https://www.synthego.com/blog/delivery-crispr-cas9/]
- 49. Mesenchymal stem cells-derived exosomes for drug delivery [https://pmc.ncbi.nlm.nih.gov/articles/PMC8557580/]
- 50. Mesenchymal stem cell-derived extracellular vesicles [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1626996/full]
- 51. Engineering strategies to enhance the research progress ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12220656/]
- 52. Exosomes vs Stem Cells: Differences & Utility (2025) [https://www.dvcstem.com/post/exosomes-vs-stem-cells]
- 53. Trends in mesenchymal stem cell-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488731/]
- 54. Functionalized exosomes for targeted therapy in cancer and ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-025-02268-y]
- 55. Extracellular vesicle-based drug overview: research ... [https://www.nature.com/articles/s41392-025-02312-w]
- 56. Multifunctional Engineering of Exosomes for Precision ... [https://link.springer.com/article/10.1007/s11095-025-03961-w]
- 57. Latest developments in advanced exosome engineering ... [https://link.springer.com/article/10.1007/s44395-025-00023-3]
- 58. Advancing exosome Isolation, Engineering, and imaging [https://www.sciencedirect.com/science/article/abs/pii/S0169409X25000079]
- 59. Extracellular vesicles for targeted drug delivery [https://pmc.ncbi.nlm.nih.gov/articles/PMC12486671/]
- 60. Overcoming challenges in MSC-sEV therapeutics [https://www.sciencedirect.com/science/article/pii/S1465324925005912]
- 61. Exosome-Based Drug Delivery: A Next-Generation Platform ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12567338/]
- 62. Current challenges and future directions of ATMPs in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12274769/]
- 63. What are CQAs?: Understanding the FDA's Critical Quality ... [https://mkainsights.com/what-are-cqas-understanding-the-fdas-critical-quality-attributes/]
- 64. Critical Quality Attributes (CQAs): Know Their Importance & ... [https://www.roosterbio.com/blog/critical-quality-attributes-cqas-know-their-importance-limitations-in-product-process-development/]
- 65. Identifying and Measuring Critical Quality Attributes - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK475684/]
- 66. Comparison of the efficiency of ultrafiltration, precipitation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10885727/]
- 67. Comparison of extracellular vesicle isolation processes for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10214056/]
- 68. Efficient methods of isolation and purification of extracellular ... [https://nanoconvergencejournal.springeropen.com/articles/10.1186/s40580-025-00509-x]
- 69. Exosomes - The Good, Bad, Ugly and Current State [https://www.americanpharmaceuticalreview.com/Featured-Articles/575432-Exosomes-The-Good-Bad-Ugly-and-Current-State/]
- 70. A comparison of methods for the isolation and separation ... [https://www.nature.com/articles/s41598-020-57497-7]
- 71. Limitations and challenges in the characterization of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12011935/]
- 72. Strategies for Scalable and GMP-Compliant Drug ... [https://www.bioanalysis-zone.com/from-lab-to-launch-strategies-for-scalable-and-compliant-drug-development-with-lenora-dieyi/]
- 73. CSV Strategies for a Growing Biotech Facility [https://assureallc.com/scaling-up-gmp-manufacturing-csv-strategies-for-a-growing-biotech-facility/]
- 74. Scaling Strategies in Drug Product Process Development [https://www.gbibio.com/scaling-strategies-in-drug-product-process-development-from-laboratory-to-commercial-production/]
- 75. Quantifying extracellular vesicle heterogeneity: the effect of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12049792/]
- 76. the effect of process conditions on protein cargo for skin therapy [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04279-5]
- 77. Navigating miRNA Sequencing Data Analysis Challenges [https://www.revvity.com/blog/navigating-complexities-mirna-sequencing-data-analysis]
- 78. Quantitative Proteomic Approach for MicroRNA Target ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5147440/]
- 79. Protein-level batch-effect correction enhances robustness ... [https://www.nature.com/articles/s41467-025-64718-y]
- 80. miRNA and pri-miRNA Support—Troubleshooting [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/real-time-digital-PCR-applications-support-center/mirna-and-noncoding-rna-support/mirna-and-noncoding-rna-support-troubleshooting.html]
- 81. Why You Must Correct Batch Effects in Transcriptomics Data? [https://www.metwarebio.com/transcriptomics-batch-effect-correction/]
- 82. Different storage and freezing protocols for extracellular vesicles [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-04005-7]
- 83. 8.2. Methods for the Characterization of Exosomes [https://bio-protocol.org/exchange/minidetail?id=10538017&type=30]
- 84. Labeling, isolation, and characterization of cell type-specific ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12505222/]
- 85. of Heterogeneity -derived extracellular... mesenchymal stem cell [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-022-00786-7]
- 86. Western Blot Troubleshooting [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-gel-electrophoresis-information/western-blot-troubleshooting.html]
- 87. Western Blot Troubleshooting Guide [https://www.rndsystems.com/resources/technical/troubleshooting-guide-western-blot]
- 88. vs In : Modern Vaccine Batch Release Vitro In Vivo Potency Assays [https://www.quantics.co.uk/blog/in-vitro-vs-in-vivo-potency-assays-modern-vaccine-batch-release/]
- 89. Establishing correlation between in ... vitro potency and in vivo [https://pmc.ncbi.nlm.nih.gov/articles/PMC12159153/]
- 90. Isolation and characterization of exosomes from adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8053584/]
- 91. Bio fluid exosomes: promises, challenges, and future ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06886-5]
- 92. Potential Signals of Serious Risks/New Safety Information ... [https://www.fda.gov/drugs/fdas-adverse-event-reporting-system-faers/potential-signals-serious-risksnew-safety-information-identified-fda-adverse-event-reporting-system]
- 93. 2025 FDAAA 801 Final Rule Changes Impacting Clinical ... [https://prorelixresearch.com/2025-fdaaa-801-clinical-trial-rule-impact/]
- 94. Exosome Research Market Size to Reach USD 961.41 ... [https://finance.yahoo.com/news/exosome-research-market-size-reach-084600173.html]
- 95. Comparative Efficacy of Exosomes Derived from Different ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12193399/]
- 96. Mesenchymal Stem Cell-Derived Exosomes as Drug Delivery ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11277139/]
- 97. Umbrella review of mesenchymal stem cell-derived ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1655623/full]
- 98. Applications, challenges and prospects of mesenchymal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8478268/]
- 99. Mesenchymal stromal/stem cell (MSC)-derived exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10079493/]
- 100. UPDATE: REGULATORY Releases FDA Agenda 2025 Guidance [https://www.aabb.org/news-resources/news/article/2025/01/03/regulatory-update--fda-releases-2025-guidance-agenda]
- 101. A comprehensive review on recent advances in exosome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11418158/]