Optimizing Reprogramming: A Comprehensive Comparison of Yamanaka Factor Combinations for iPSC Generation
This article provides a systematic analysis of the efficiency, safety, and application-specific suitability of various Yamanaka factor combinations for induced pluripotent stem cell (iPSC) reprogramming.
Optimizing Reprogramming: A Comprehensive Comparison of Yamanaka Factor Combinations for iPSC Generation
Abstract
This article provides a systematic analysis of the efficiency, safety, and application-specific suitability of various Yamanaka factor combinations for induced pluripotent stem cell (iPSC) reprogramming. Tailored for researchers and drug development professionals, it covers foundational mechanisms, methodological delivery systems, strategies for troubleshooting and optimizing reprogramming protocols, and comparative validation of established and emerging factor cocktails. The review synthesizes current evidence to guide the selection of optimal reprogramming strategies for disease modeling, drug screening, and clinical applications, addressing key challenges such as tumorigenicity and low efficiency.
The OSKM Blueprint and Beyond: Core Principles of Reprogramming Factor Combinations
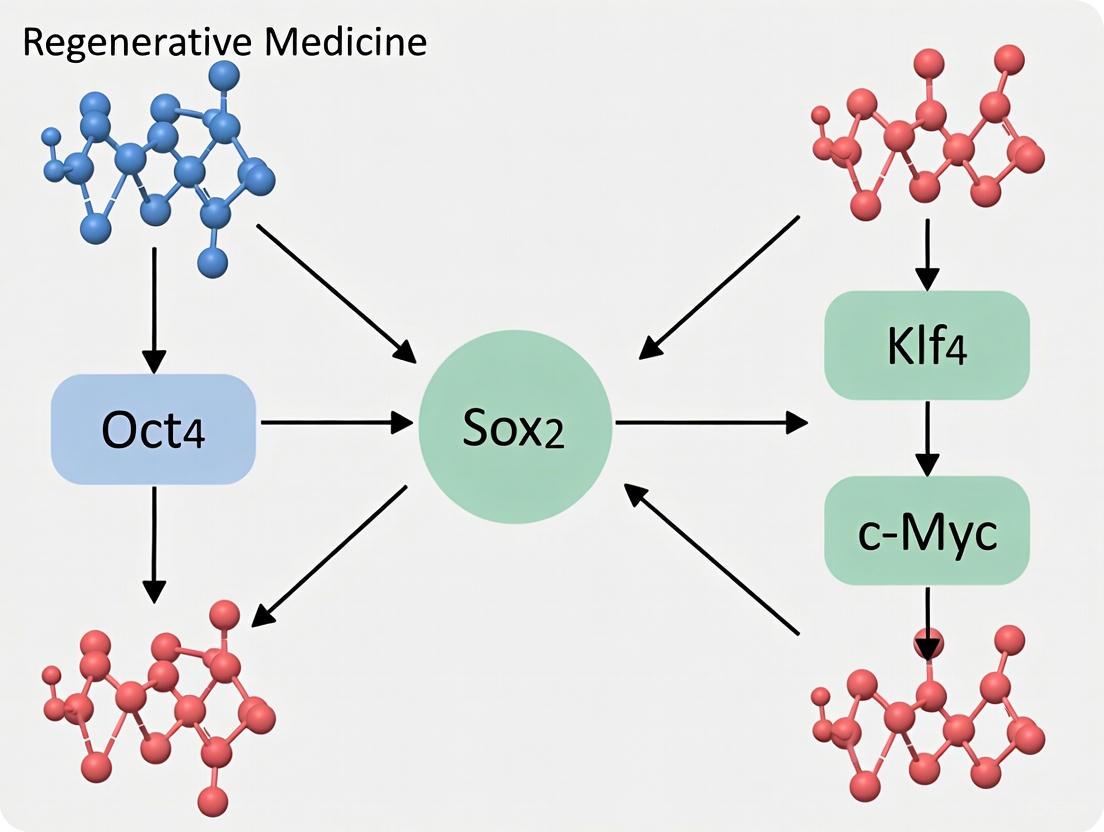
Since their groundbreaking discovery in 2006, the OSKM transcription factors—OCT4, SOX2, KLF4, and c-MYC—have represented the gold standard for reprogramming somatic cells into induced pluripotent stem cells (iPSCs) [1] [2]. This revolutionary combination demonstrated that mature, differentiated cells could be returned to an embryonic-like state through the forced expression of just four defined factors, fundamentally changing the landscape of developmental biology and regenerative medicine [2] [3]. While numerous alternative reprogramming approaches have emerged over the past two decades, the original Yamanaka factors continue to serve as the essential benchmark against which all new methodologies are measured. This review examines the enduring role of OSKM as the reference standard in reprogramming research, comparing its performance metrics against subsequently developed factor combinations and delivery systems, with particular focus on efficiency, safety, and practical application in research and therapeutic development.
Core Mechanisms and Molecular Pathways
The reprogramming process initiated by OSKM factors involves profound remodeling of the chromatin structure and epigenome, transitioning through distinct phases [2]. During the early phase, somatic genes are silenced while early pluripotency-associated genes are activated—a process characterized by stochastic binding events, particularly to closed chromatin sites [4] [2]. The late phase involves more deterministic activation of late pluripotency-associated genes, establishing a self-reinforcing pluripotency network [1].
OSKM Reprogramming Pathway
c-MYC initiates reprogramming by associating with histone acetyltransferase complexes to induce global histone acetylation, enabling exogenous OCT4 and SOX2 to access their target loci [1]. OCT4 and SOX2 then cooperate as key transcription factors that inhibit differentiation-associated genes while activating the pluripotency network [1]. KLF4 plays a dual role, simultaneously suppressing somatic gene expression and activating pluripotency-associated genes [1]. This coordinated action ultimately leads to the establishment of a self-sustaining pluripotent state maintained by endogenous factor expression.
Comparative Performance Analysis of Reprogramming Factor Combinations
Efficiency Metrics Across Factor Combinations
Table 1: Comparative Efficiency of Reprogramming Factor Combinations
| Factor Combination | Reprogramming Efficiency | Time to iPSC Formation | Key Advantages | Notable Limitations |
|---|---|---|---|---|
| OSKM (Original) | <0.1%-1% [5] | 3-4 weeks [5] | Gold standard, reliable, extensively validated | Low efficiency, tumorigenic risk with c-MYC |
| OSK (c-MYC omitted) | Significantly lower than OSKM [6] | Similar to OSKM | Reduced tumorigenic risk | Greatly reduced efficiency |
| OSNL (OCT4, SOX2, NANOG, LIN28) | Comparable to OSKM [1] | Similar to OSKM | Avoids c-MYC oncogene | LIN28 may affect proliferation similarly to c-MYC |
| OSKMNL (Six factors) | ~10x higher than OSNL [1] | Potentially reduced | Enables reprogramming of cells from aged donors | Increased genetic manipulation burden |
| OKS (OCT4, KLF4, SOX2) | Lower than OSKM but functional [7] | Not specified | Reduced oncogenic load, effective for partial reprogramming | Not sufficient for full reprogramming in all contexts |
| AI-Enhanced Variants | >50x higher than wild-type OSKM [5] | Markers appear days sooner [5] | Dramatically improved efficiency, enhanced DNA damage repair | Novel technology requiring further validation |
The original OSKM combination establishes the fundamental benchmark for reprogramming efficiency, typically achieving successful conversion in fewer than 1% of treated cells over approximately three weeks [5]. As illustrated in Table 1, efforts to improve upon this baseline have followed several strategic approaches: reducing oncogenic risk through factor omission (OSK), substituting alternative factors (OSNL), expanding the factor repertoire (OSKMNL), or recently, using artificial intelligence to engineer enhanced variants [6] [1] [5].
The critical role of c-MYC is evidenced by the significant efficiency reduction observed with its omission, though it is not strictly essential for reprogramming [6]. Similarly, the OSNL combination developed by Thomson's group demonstrates that completely different factor combinations can achieve similar outcomes, with LIN28 potentially functioning as a functional analog to c-MYC by accelerating cell proliferation [1]. The most dramatic efficiency improvements have recently emerged through AI-guided protein engineering, with OpenAI and Retro Biosciences reporting redesigned SOX2 and KLF4 variants that achieve over 50-fold higher expression of pluripotency markers compared to wild-type OSKM [5].
Species-Specific and Cell-Type Specific Variations
Table 2: Mouse vs. Human Reprogramming Characteristics with OSKM
| Characteristic | Mouse System | Human System |
|---|---|---|
| Reprogramming Timeline | 1-2 weeks [4] | 3-4 weeks [4] |
| c-MYC Dependency | Less critical; OSK often sufficient [4] | More critical for efficient reprogramming [4] |
| OSKM Binding Distribution | c-MYC binds proximally to TSS [4] | c-MYC binds distally to TSS [4] |
| Conserved Binding Events | Limited conservation in syntenic regions [4] | Limited conservation in syntenic regions [4] |
| Endpoint Pluripotency | Naïve state [4] | Primed state [4] |
Comparative analyses reveal significant differences in OSKM-mediated reprogramming between mouse and human systems, as summarized in Table 2. Mouse cells reprogram approximately twice as fast as human cells, with different dependencies on individual factors—notably, c-MYC plays a more essential role in human reprogramming [4]. Chromatin interaction studies demonstrate that while the general features of OSKM binding are largely conserved between species, including target genes and binding motifs, the specific genomic binding locations show limited conservation in syntenic regions [4]. These differences highlight the importance of considering species-specific effects when evaluating reprogramming efficiency across experimental systems.
Experimental Protocols and Methodologies
Standard OSKM Reprogramming Workflow
Fibroblast Reprogramming Protocol (Based on Original Yamanaka Method [2]):
- Factor Delivery: Introduce OSKM factors via integrating retroviral or lentiviral vectors into mouse embryonic fibroblasts (MEFs) or human dermal fibroblasts
- Culture Conditions: Maintain cells in ESC culture conditions with serum-containing media or defined formulations
- Timeline: Continue culture for 2-3 weeks (mouse) or 3-4 weeks (human) with regular medium changes
- Colony Identification: Monitor for emergence of ESC-like morphology with tight, dome-shaped colonies
- Validation: Confirm pluripotency through marker expression (SSEA-4, TRA-1-60, NANOG), teratoma formation, or differentiation potential [5]
Experimental Workflow for Reprogramming Efficiency Comparison
Advanced Methodology: Sequential Factor Delivery for Enhanced Efficiency
Recent optimization efforts have identified sequential delivery of reprogramming factors as a key strategy for improving knock-in efficiency in iPSCs. The following protocol has demonstrated success in achieving knock-in efficiencies exceeding 30%:
- Day 0: Preparative Culture - Switch to enriched culture medium two days prior to nucleofection to enhance cell fitness [8]
- Day 1: Donor Plasmid Delivery - Perform first nucleofection with donor plasmid DNA only, using 3×10ⶠcells per cuvette in P4 Nucleofection Buffer (Lonza 4D Nucleofector, program CA-167) [8]
- Post-Nucleofection Recovery - Incubate cells in RPMI medium for 10 minutes immediately after nucleofection to significantly improve cell survival [8]
- Day 2: RNP Complex Delivery - Perform second nucleofection with ribonucleoprotein (RNP) complexes containing Cas9/Cas12a nucleases and guide RNA [8]
- Cold Shock Incubation - Maintain cells at 32°C for enhanced homology-directed repair [8]
- Screening and Validation - Employ limiting dilution cloning and flow cytometry-based screening for modified clones [8]
This sequential approach demonstrates the continued optimization potential of reprogramming methodologies while maintaining compatibility with Good Manufacturing Practice (GMP) requirements for therapeutic applications [8].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for OSKM Reprogramming Studies
| Reagent Category | Specific Examples | Research Applications |
|---|---|---|
| Delivery Systems | Retroviral/lentiviral vectors, Sendai virus, mRNA transfection, recombinant protein [6] | Factor delivery with varying integration risks and efficiency |
| Reprogramming Enhancers | Valproic acid, 8-Br-cAMP, Sodium butyrate, RepSox [6] | Small molecules that improve reprogramming efficiency |
| Pluripotency Markers | SSEA-4, TRA-1-60, NANOG, alkaline phosphatase [5] | Validation of successful reprogramming |
| Selection Systems | Antibiotic resistance, FACS sorting markers [8] | Enrichment for successfully reprogrammed cells |
| Culture Media | DMEM/F12, defined media formulations, serum-free conditions [6] | Maintenance of pluripotent state |
| 4-Chloro-2-pyridin-3-ylquinazoline | 4-Chloro-2-pyridin-3-ylquinazoline|CAS 98296-25-4 | 4-Chloro-2-pyridin-3-ylquinazoline (CAS 98296-25-4) is a quinazoline-based chemical building block for anticancer research. This product is for research use only and not for human use. |
| 4-Chloro-2,6-bis(hydroxymethyl)phenol | 4-Chloro-2,6-bis(hydroxymethyl)phenol|CAS 17026-49-2 |
Applications and Therapeutic Translation
The original OSKM factors continue to enable diverse applications across regenerative medicine, disease modeling, and drug discovery. In disease modeling, iPSCs generated with OSKM have been particularly valuable for investigating neurodegenerative diseases like amyotrophic lateral sclerosis (ALS), where patient-specific iPSCs can be differentiated into affected cell types like motor neurons to recapitulate disease pathology [6]. In the therapeutic realm, partial reprogramming approaches using reduced factor combinations (often OKS without c-MYC) have shown promise for restoring youthful epigenetic patterns in aged tissues without completely reversing cell identity [7]. This approach has demonstrated efficacy in mitigating intervertebral disc degeneration by reducing senescence markers and restoring extracellular matrix homeostasis [7].
The emergence of HLA-matched iPSC banks, such as the initiative at Kyoto University led by Yamanaka, aims to provide allogeneic iPSC lines that could cover most of the Japanese population, potentially overcoming the limitations of autologous iPSC generation [1]. These developments, built upon the foundational OSKM technology, highlight the continuing translational impact of the original Yamanaka factors.
Nearly two decades after their discovery, the original Yamanaka factors (OSKM) maintain their status as the gold standard in cellular reprogramming. While numerous alternative factor combinations and methodologies have emerged—from reduced cocktails like OKS to expanded sets like OSKMNL—all are evaluated against the benchmark established by OSKM [6] [1] [7]. The recent development of AI-enhanced factor variants achieving unprecedented efficiency gains represents the next evolutionary step in this field, yet still relies on the original OSKM factors as both a conceptual framework and experimental control [5]. As reprogramming research progresses toward increasingly refined therapeutic applications, the original Yamanaka factors continue to provide the fundamental reference point for evaluating efficiency, safety, and practical utility across diverse biological contexts and applications.
The seminal discovery of induced pluripotent stem cells (iPSCs) using the OSKM (OCT4, SOX2, KLF4, c-MYC) transcription factors revolutionized regenerative medicine, offering a potential workaround to the ethical concerns of embryonic stem cell research [6] [9]. However, the clinical translation of this technology is significantly hampered by safety concerns, primarily the tumorigenic risk associated with the use of the oncogene c-MYC [6] [9]. This safety profile is a critical determinant for researchers and drug development professionals selecting a reprogramming system. Consequently, the quest for safer alternative factor combinations has become a central focus in the field. Among the most prominent alternatives is the OSNL combination, which replaces KLF4 and c-MYC with NANOG and LIN28 [6] [10]. This guide provides a objective comparison of these core combinations, detailing their performance, underlying mechanisms, and practical application in a research setting.
Core Combination Comparison
The choice of reprogramming factors profoundly impacts the efficiency, safety, and applicability of the resulting iPSCs. The table below provides a detailed comparison of the established OSKM and the alternative OSNL combination.
Table 1: Comparative Analysis of OSKM and OSNL Reprogramming Factor Combinations
| Feature | OSKM (OCT4, SOX2, KLF4, c-MYC) | OSNL (OCT4, SOX2, NANOG, LIN28) |
|---|---|---|
| Core Components | OCT4, SOX2, KLF4, c-MYC [6] | OCT4, SOX2, NANOG, LIN28 [6] [10] |
| Key Replacement | N/A | Replaces KLF4 and c-MYC with NANOG and LIN28 [6] |
| Primary Rationale | Original reprogramming "gold standard" [6] | To address tumorigenic risks associated with c-MYC [6] |
| Reprogramming Efficiency | High [6] | Sufficient, but reported to have lower success rates than OSKM in some comparative studies [10] |
| Tumorigenic Risk Profile | High; c-MYC is a potent oncogene, and its overexpression contributes to tumorigenesis [6] | Lower; avoids the use of the c-MYC oncogene, thereby reducing inherent oncogenic risk [6] [9] |
| Noted Advantages | High efficiency, well-characterized [6] | Reduced risk of tumor formation, successful reprogramming of human somatic cells demonstrated [6] [6] |
| Reported Limitations | Use of oncogene c-MYC poses significant safety risks for clinical applications [6] | May exhibit lower reprogramming efficiency compared to the OSKM combination [6] [10] |
Experimental Insights and Performance Data
Empirical data is crucial for evaluating the practical performance of these factor combinations. A comparative study offers direct insight into how OSNL and OSKM (delivered via the STEMCCA system) stack up against each other.
Table 2: Experimental Data from a Comparative Study of hiPSC Derivation
| Experimental Parameter | OSNL System | STEMCCA (OSKM) System |
|---|---|---|
| Reprogramming Efficiency | Significantly lower [10] | Significantly higher [10] |
| Success Rate of Generation | Lower [10] | Higher [10] |
| Pluripotency of Generated hiPSCs | Confirmed; all analysed hiPSCs were pluripotent and showed characteristics similar to human embryonic stem cells [10] | Confirmed; all analysed hiPSCs were pluripotent and showed characteristics similar to human embryonic stem cells [10] |
| Somatic Cell Source Impact | Successfully generated hiPSCs from hair keratinocytes, bone marrow cells (MSCs), and skin fibroblasts, though with varying efficiencies [10] | Successfully generated hiPSCs from hair keratinocytes, bone marrow cells (MSCs), and skin fibroblasts, with higher overall efficiency [10] |
| Downstream Differentiation (Cardiomyocytes) | All hiPSC lines could differentiate into functional cardiomyocytes [10] | All hiPSC lines could differentiate into functional cardiomyocytes [10] |
The same study also highlighted that the source of somatic cells significantly influences reprogramming outcomes. For instance, bone marrow-derived mesenchymal stem cells (MSCs) were more easily reprogrammed than keratinocytes and subsequently exhibited a significantly higher efficiency in spontaneously differentiating into beating cardiomyocytes, regardless of the reprogramming system used [10]. This underscores the importance of considering cell source in experimental design.
Detailed Experimental Workflow
To ensure reproducibility, this section outlines a generalized protocol for deriving and characterizing iPSCs using these factor combinations, based on methodologies cited in the literature [10] [11].
Somatic Cell Preparation and Reprogramming
- Cell Source Isolation and Culture: Obtain and expand the chosen somatic cell population (e.g., dermal fibroblasts, bone marrow MSCs, or hair keratinocytes) in their respective standard culture media [10].
- Factor Delivery: Introduce the transcription factors (OSKM or OSNL) into the somatic cells. This is typically achieved using viral delivery systems.
- Lentiviral Vectors: The STEMCCA system, a lentiviral vector expressing OSKM in a single polycistronic cassette, is noted for its high reprogramming efficiency [10].
- Retroviral Vectors: Traditional for expressing the original Yamanaka factors [6].
- Non-Integrating Methods: For higher clinical safety, consider Sendai virus (an RNA virus that does not integrate into the genome) or episomal plasmids [6] [11].
- iPSC Colony Formation: After transduction, culture the cells on a feeder layer or in feeder-free conditions using media that support pluripotent stem cells (e.g., mTeSR1). Monitor for the emergence of compact, ESC-like colonies over 3-4 weeks.
iPSC Characterization and Validation
- Molecular Analysis of Pluripotency:
- In Vitro Differentiation: Subject iPSCs to spontaneous differentiation via embryoid body (EB) formation. Assess the resulting cells for markers of all three germ layers (e.g., α-fetoprotein for endoderm, α-actinin for mesoderm, βIII-tubulin for ectoderm) [10] [9].
- Karyotype Analysis: Perform G-banding karyotyping to ensure genomic integrity and the absence of major chromosomal abnormalities.
- Teratoma Assay: For a gold-standard test of pluripotency, inject iPSCs into immunodeficient mice. After 8-12 weeks, a teratoma should form containing differentiated tissues from all three germ layers [9].
Molecular Mechanisms and Signaling Pathways
The OSNL and OSKM combinations work by reactivating the core pluripotency network, but they engage this network through partially distinct mechanisms. The following diagram illustrates the key pathways and logical relationships involved in OSNL-mediated reprogramming.
The OSNL combination operates by directly activating the core pluripotency circuitry while leveraging alternative pathways to enhance reprogramming. OCT4 and SOX2 form a heterodimer that binds to and activates the expression of genes essential for pluripotency, including itself and NANOG [6]. NANOG acts as a key reinforcing factor, stabilizing the pluripotent state by suppressing alternative differentiation pathways [6]. A distinctive mechanism of the OSNL combination is the action of LIN28. LIN28 promotes reprogramming primarily by inhibiting the let-7 family of microRNAs [6]. Since let-7 miRNAs repress several pro-reprogramming factors, including c-MYC, LIN28 inhibition effectively derepresses the c-MYC pathway without the need for the potentially dangerous c-MYC oncogene itself [6]. This results in a safer profile while still leveraging a critical proliferative and metabolic network.
The Scientist's Toolkit: Essential Research Reagents
The following table catalogs key reagents and their functions essential for conducting reprogramming experiments with the OSNL and OSKM factor combinations.
Table 3: Key Research Reagents for iPSC Reprogramming Experiments
| Reagent / Solution | Function in Reprogramming | Specific Examples & Notes |
|---|---|---|
| Reprogramming Factor Vectors | Deliver genes encoding transcription factors into somatic cells. | Lentiviral (e.g., STEMCCA for OSKM [10]), Retroviral, or non-integrating Sendai virus systems. |
| Pluripotency Support Media | Culture medium providing essential nutrients and signaling molecules to maintain pluripotency. | TeSR-E8, mTeSR1; used after transduction to support emerging iPSC colonies. |
| Feeder Cells / Substrate | Provides a physical and biochemical support layer for iPSC growth. | Mitotically-inactivated mouse embryonic fibroblasts (MEFs) or defined, feeder-free substrates like Matrigel. |
| Pluripotency Marker Antibodies | Detect expression of key proteins to validate pluripotent state via immunocytochemistry. | Antibodies against OCT4, SOX2, NANOG, SSEA-4, TRA-1-60, TRA-1-81 [10] [9]. |
| Differentiation Induction Media | Directs iPSC differentiation into specific lineages to confirm functional pluripotency. | Media formulations for generating cardiomyocytes, neurons, hepatocytes, etc. [10] |
| Small Molecule Enhancers | Chemicals that improve reprogramming efficiency or replace transcription factors. | Valproic acid (VPA), RepSox, Tranylcypromine; can be used to enhance efficiency or in novel chemical cocktails [6] [12] [13]. |
| 3-Amino-4-(phenylamino)benzonitrile | 3-Amino-4-(phenylamino)benzonitrile, CAS:68765-52-6, MF:C13H11N3, MW:209.25 g/mol | Chemical Reagent |
| 5'-Phosphopyridoxyl-7-azatryptophan | 5'-Phosphopyridoxyl-7-azatryptophan, CAS:157117-38-9, MF:C18H21N4O7P, MW:436.4 g/mol | Chemical Reagent |
The comparative data clearly illustrates the fundamental trade-off between the high efficiency of the OSKM system and the superior safety profile of the OSNL combination. For therapeutic applications where minimizing oncogenic risk is paramount, OSNL presents a compelling alternative, despite its potential efficiency drawbacks [6] [10] [9]. The field continues to evolve beyond these initial combinations, exploring factors like GLIS1 and ESRRB as substitutes, and moving towards non-genetic methods such as chemically-induced reprogramming to further enhance safety [6] [12] [14]. Innovations like the 2c small molecule cocktail (RepSox and Tranylcypromine) and the discovery of novel single-gene targets like SB000 promise a new generation of rejuvenation therapies that avoid the risks of pluripotency induction entirely [15] [13]. For researchers, the choice between OSKM, OSNL, or newer approaches must be strategically aligned with the specific goals of their work, whether for high-throughput basic research or the development of clinically translatable therapies.
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) through the ectopic expression of specific transcription factors revolutionized regenerative medicine and developmental biology [3]. The classic "Yamanaka factors" — OCT4, SOX2, KLF4, and c-MYC (OSKM) — provide a powerful combination for resetting cellular identity [16]. However, each factor contributes distinct mechanistic functions to the reprogramming process, with specific structural features, molecular interactions, and epigenetic activities that collectively orchestrate the transition from somatic to pluripotent state. This review systematically compares the functional mechanisms of these four core factors, examining how their individual properties influence reprogramming efficiency, trajectory, and the quality of resulting iPSCs.
Factor-by-Factor Functional Analysis
OCT4: The Master Pluripotency Regulator
Structural and Functional Domains: OCT4 (POU5F1) contains a bipartite POU DNA-binding domain composed of POU-specific (POUS) and POU-homeodomain (POUHD) regions connected by a linker, flanked by N-terminal (NTD) and C-terminal (CTD) effector domains [17]. While the DNA-binding domain is essential for both reprogramming and pluripotency maintenance, recent research has identified Short Linear Peptides Essential for Reprogramming (SLiPERs) within the intrinsically disordered regions (IDRs) of OCT4 that are critical for reprogramming but dispensable for embryonic stem cell self-renewal [17].
Key Functional Mechanisms:
- DNA Binding and Nucleosome Engagement: The structured POU domain enables OCT4 to bind specific DNA sequences and interact with nucleosomes, which is essential for accessing chromatin during reprogramming [17].
- Context-Specific Functionality: SLiPER domains within IDRs adopt quasi-ordered states and recruit unique protein complexes to closed chromatin specifically during reprogramming, but not during pluripotency maintenance [17].
- Developmental Essentiality: Removing SLiPERs prevents embryos from developing beyond late gastrulation and derails the transition of ESCs out of pluripotency [17].
- Dosage Sensitivity: OCT4 expression levels are critical for successful reprogramming, with both insufficient and excessive expression impairing the process [1].
Table 1: Functional Domains of OCT4 and Their Roles in Reprogramming
| Domain | Location | Key Functions | Essential for Reprogramming | Essential for ESC Self-Renewal |
|---|---|---|---|---|
| POU-Specific Domain (POUS) | a.a. 47-70 | DNA binding, nucleosome interaction | Yes | Yes |
| Linker Region | a.a. 71-78 | Connects POUS and POUHD domains | Partially (unstructured portions) | Yes (structured portions) |
| POU-Homeodomain (POUHD) | a.a. 79-95 | DNA binding, nucleosome interaction | Yes | Yes |
| N-Terminal Domain (NTD) | a.a. 1-47 | Contains SLiPER sequences | Yes (specific peptides) | No |
| C-Terminal Domain (CTD) | a.a. 271-352 | Contains SLiPER sequences | Yes (specific peptides) | No |
SOX2: The Chromatin Accessibility Factor
Structural and Functional Domains: SOX2 contains a high-mobility group (HMG) DNA-binding domain that enables sequence-specific DNA recognition and bending, facilitating chromatin remodeling [16].
Key Functional Mechanisms:
- Cooperative DNA Binding: SOX2 frequently binds DNA in partnership with OCT4, with the two factors recognizing adjacent binding sites to form enhanceosome complexes that activate pluripotency genes [16].
- Chromatin Architecture Remodeling: The HMG domain of SOX2 bends DNA, which helps remodel chromatin structure and make regulatory regions accessible during reprogramming [16].
- Retroviral Silencing Trigger: When co-expressed with c-MYC, SOX2 can trigger immediate retroviral silencing, which explains why previous attempts to generate iPSCs without OCT4 using retroviral vectors failed [18].
- Lineage Specifier Counterbalance: In the "seesaw model" of pluripotency, SOX2 counteracts OCT4's effects by promoting neuroectoderm specification while OCT4 promotes meso-endoderm specification [18].
KLF4: The Dual-Function Regulator
Structural and Functional Domains: KLF4 contains three carboxyl-terminal zinc fingers that mediate binding to GC-rich sequences (CACCC) in gene regulatory regions, plus distinct domains for gene activation and repression [19].
Key Functional Mechanisms:
- Chromatin Organization: KLF4 can form liquid-like biomolecular condensates with DNA that recruit OCT4 and SOX2, facilitating the assembly of pluripotency transcription complexes [19].
- Dual Regulatory Capacity: KLF4 can function as both an activator and repressor, suppressing somatic cell identity genes while simultaneously activating pluripotency genes [1].
- Epigenetic Flexibility: KLF4 uniquely binds both unmethylated and CpG-methylated DNA, allowing it to initiate stem-cell gene expression profiles during reprogramming despite repressive epigenetic marks [19].
- Self-Renewal Promotion: In embryonic stem cells, KLF4 works in synchrony with KLF2 and KLF5 to regulate self-renewal and expression of pluripotency genes like Nanog [19].
c-MYC: The Chromatin Priming Factor
Structural and Functional Domains: c-MYC belongs to the basic helix-loop-helix leucine zipper (bHLHLZ) family and requires heterodimerization with MAX for transcriptional activity [20].
Key Functional Mechanisms:
- Chromatin Opening: c-MYC associates with histone acetyltransferase complexes to induce global histone acetylation, creating a more open chromatin environment that allows other factors access to their targets [1].
- Metabolic Reprogramming: c-MYC promotes a transition to energy metabolism typical of cancer cells, enhancing cell proliferation during the early stages of reprogramming [1].
- Transcriptional Amplification: With binding sites far exceeding those of OCT4 and SOX2, c-MYC acts as a broad amplifier of transcriptional changes during reprogramming [1].
- Dispensability with Trade-offs: While c-MYC significantly enhances reprogramming efficiency, it is dispensable for iPSC generation, and its exclusion reduces transformation risk but slows the process [16].
Table 2: Comparative Functions of Yamanaka Factors in Reprogramming
| Factor | Main Function | Essentiality | Binding Specificity | Key Partners |
|---|---|---|---|---|
| OCT4 | Master pluripotency regulator | Conditionally essential | DNA sequence-specific | SOX2, KLF4, nucleosome remodeling complexes |
| SOX2 | Chromatin accessibility factor | Conditionally essential | DNA sequence-specific with OCT4 | OCT4, c-MYC |
| KLF4 | Dual activator/repressor | Replaceable | GC-rich sequences | OCT4, SOX2, KLF2/5 |
| c-MYC | Chromatin priming factor | Dispensable | E-box sequences | MAX, histone acetyltransferases |
Experimental Approaches and Methodologies
Reprogramming Efficiency Assays
Colony Formation Assays: The gold standard for quantifying reprogramming efficiency involves counting alkaline phosphatase-positive or pluripotency marker-positive (e.g., Nanog-GFP) colonies following factor expression [17] [18]. Typical protocols involve:
- Transducing somatic cells (usually mouse embryonic fibroblasts) with reprogramming factors
- Inducing factor expression for defined periods (typically 5-21 days)
- Fixing and staining for alkaline phosphatase or quantifying GFP-positive colonies
- Normalizing colony counts to initial cell numbers
Tetraploid Complementation Assays: This stringent test for developmental potential involves injecting iPSCs into tetraploid blastocysts and assessing their ability to generate entire mice [18]. iPSCs generated without OCT4 overexpression show significantly improved developmental potential in this assay compared to traditional OSKM-iPSCs [18].
Molecular Mechanism Studies
Domain Mapping Approaches: Systematic deletion mapping studies using overlapping 5-amino-acid deletions across OCT4 have identified functionally distinct regions [17]. This approach revealed SLiPER domains specifically required for reprogramming but not pluripotency maintenance.
Chromatin Immunoprecipitation (ChIP): Genome-wide binding analyses show that the core pluripotency factors (OCT4, SOX2, NANOG) co-occupy target genes at levels well beyond chance expectation, suggesting cooperative binding and function [16].
Biomolecular Condensate Imaging: Advanced microscopy techniques have demonstrated that KLF4 can form liquid-like biomolecular condensates with DNA that recruit OCT4 and SOX2, providing a mechanism for transcription hub formation [19].
Figure 1: Temporal Phases of Cellular Reprogramming Showing Distinct Factor Activities
Alternative Factor Combinations and Replacements
Research has identified several alternative factor combinations that can generate iPSCs, providing insights into the core requirements for reprogramming:
OCT4-Independent Reprogramming: The combination of SOX2, KLF4, and c-MYC (SKM) can reprogram mouse somatic cells to pluripotency, though with delayed kinetics and reduced efficiency compared to OSKM [18]. This process requires high cell proliferation rates and is facilitated by the observation that SOX2 and c-MYC co-expression triggers retroviral silencing [18].
Alternative Pluripotency Factors: The combination of OCT4, SOX2, NANOG, and LIN28 (OSNL) can also reprogram human somatic cells, with NANOG functioning as an essential pluripotency maintainer and LIN28 accelerating cell proliferation similarly to c-MYC [1].
GATA Factor Substitution: Endoderm lineage specifiers GATA3, GATA4, and GATA6 can replace OCT4 in reprogramming cocktails, supporting the "seesaw model" where pluripotency represents a balance between opposing developmental lineages [18].
Table 3: Experimentally Validated Alternative Reprogramming Factor Combinations
| Factor Combination | Efficiency vs. OSKM | Key Advantages | Limitations |
|---|---|---|---|
| SKM (SOX2, KLF4, c-MYC) | ~30% efficiency, 2-day delay | Improved developmental potential, fewer epigenetic aberrations | Requires high proliferation, cell type dependent |
| OSKM + Enhancers (e.g., Glis1) | Enhanced efficiency | Reduces partially reprogrammed colonies | Additional factors required |
| OSNL (OCT4, SOX2, NANOG, LIN28) | Comparable efficiency | Avoids oncogenic KLF4 and c-MYC | LIN28 has oncogenic potential |
| GATA + SKM (GATA3/4/6, SOX2, KLF4, c-MYC) | Higher than OSKM in some contexts | Provides alternative lineage balancing | May bias differentiation potential |
Research Reagents and Experimental Tools
Table 4: Essential Research Reagents for Reprogramming Studies
| Reagent/Tool | Function | Example Applications | Key Considerations |
|---|---|---|---|
| dox-Inducible Lentiviral Vectors | Controlled factor expression | Sequential reprogramming studies, kinetics analysis | Integration risks, potential silencing |
| Oct4-GFP Reporter MEFs | Visualization of reprogramming | Efficiency quantification, colony isolation | GFP activation indicates endogenous OCT4 expression |
| Episomal Vectors | Non-integrating factor delivery | Clinical-grade iPSC generation, safety studies | Lower efficiency, transient expression |
| TetO-Polycistronic Cassettes | Coordinated factor expression | OSKM, SKM, OSN combinations | Fixed factor ratios, compact design |
| Small Molecule Enhancers | Replace specific factors | Chemical reprogramming, efficiency boosting | Defined mechanisms, concentration critical |
The functional comparison of OCT4, SOX2, KLF4, and c-MYC reveals a sophisticated division of labor in cellular reprogramming. OCT4 serves as the master regulator with context-specific functionalities embedded in its disordered regions, while SOX2 provides chromatin accessibility and partnership. KLF4 offers dual regulatory capacity and biomolecular condensate formation, and c-MYC primes chromatin for broad transcriptional changes. The discovery that OCT4 can be omitted under specific conditions challenges long-standing dogmas and highlights the plasticity of reprogramming pathways.
These findings have substantial implications for regenerative medicine applications, particularly in optimizing factor combinations for specific cell types and clinical applications. The improved developmental potential of SKM-reprogrammed iPSCs suggests that factor selection significantly impacts the epigenetic fidelity and functionality of resulting stem cells. Future research directions should focus on further elucidating the mechanistic basis of SLiPER domain functions, biomolecular condensate dynamics, and developing small molecule approaches to mimic or enhance the specific functional contributions of each factor while minimizing oncogenic risk.
The discovery that somatic cells could be reprogrammed into induced pluripotent stem cells (iPSCs) using the transcription factors OCT4, SOX2, KLF4, and c-MYC (OSKM) revolutionized regenerative medicine and disease modeling [6] [21]. Subsequent research has focused on optimizing this reprogramming cocktail to enhance efficiency and safety, particularly by addressing the oncogenic potential of factors like c-MYC [6] [22]. A critical advancement in this optimization has been the identification of substitute factors capable of replacing their original counterparts in the reprogramming process while modulating functional outcomes.
Factor substitutability refers to the capability of certain transcription factors from the same family or with similar functional domains to replace the core Yamanaka factors in somatic cell reprogramming [6] [23]. Research has demonstrated that KLF2 and KLF5 can substitute for KLF4; SOX1 and SOX3 can replace SOX2; and L-MYC and N-MYC can stand in for c-MYC [6] [23]. Understanding the relative performance, efficiency, and safety profiles of these alternative factors is essential for designing optimized reprogramming protocols for specific research and therapeutic applications. This guide provides a systematic comparison of these substitute factors, presenting experimental data and methodologies to inform their use in scientific investigations.
Comparative Performance Data of Substitute Factors
Table 1: Functional Classification and Key Characteristics of Substitute Factors
| Original Factor | Substitute Factor | Family/Type | Key Reported Functions in Reprogramming | Oncogenic Risk Profile |
|---|---|---|---|---|
| KLF4 | KLF2 | Krüppel-like factor (KLF) | Activates pluripotency genes, suppresses somatic gene expression [6] | Lower than c-MYC [6] |
| KLF4 | KLF5 | Krüppel-like factor (KLF) | Similar transcriptional activation profile to KLF4 [6] | Lower than c-MYC [6] |
| SOX2 | SOX1 | SRY-related HMG-box (SOX) | Maintains pluripotency network, can bind similar targets [6] [23] | Not typically associated with high oncogenic risk |
| SOX2 | SOX3 | SRY-related HMG-box (SOX) | Functional redundancy with SOX2 in establishing pluripotency [6] [23] | Not typically associated with high oncogenic risk |
| c-MYC | L-MYC | MYC proto-oncogene family | Enhances proliferation with reduced tumorigenic potential compared to c-MYC [6] | Lower than c-MYC [6] |
| c-MYC | N-MYC | MYC proto-oncogene family | Promotes cell cycle progression and metabolic changes [23] | Lower than c-MYC, but caution is still advised [23] |
Table 2: Reported Reprogramming Efficiency and Key Experimental Findings
| Substitute Factor | Reprogramming Efficiency Relative to Original Factor | Key Experimental Observations and Context |
|---|---|---|
| KLF2 | Significantly lower efficiency than KLF4 [6] | Can establish pluripotency but with slower kinetics [6] |
| KLF5 | Significantly lower efficiency than KLF4 [6] | Shows overlapping target gene activation with KLF4 [6] |
| SOX1 | Significantly lower efficiency than SOX2 [6] | Effective in neural differentiation contexts [23] |
| SOX3 | Significantly lower efficiency than SOX2 [6] | Demonstrated functional redundancy in reprogramming circuitry [6] [23] |
| L-MYC | Comparable efficiency to c-MYC, with improved safety profile [6] | Associated with reduced teratoma formation in derived iPSCs [6] |
| N-MYC | Lower efficiency than c-MYC, but safer profile [23] | Used in alternative factor combinations (e.g., with Oct4, Sox2, Klf4) [23] |
Experimental Protocols for Evaluating Substitute Factors
Protocol for Fibroblast Reprogramming with Substitute Factors
This standard protocol is used to assess the efficacy of substitute factors in generating iPSCs from mouse or human fibroblasts [6] [21].
Cell Culture Preparation:
- Isolate and culture mouse embryonic fibroblasts (MEFs) or human dermal fibroblasts in standard fibroblast medium (e.g., DMEM with 10% FBS).
- Ensure cells are actively dividing and are at a low passage number for optimal reprogramming.
Factor Delivery:
- Vector Preparation: Clone the cDNA of the substitute factors (e.g., KLF2, SOX1, L-MYC) into appropriate delivery vectors. For comparative studies, use the same vector system (e.g., lentivirus, Sendai virus, or episomal plasmids) for all factors to ensure consistency [6] [21].
- Transduction/Transfection: Infect or transfect fibroblasts with viruses or plasmids carrying the reprogramming cassette. A common experimental setup includes:
- Use a defined multiplicity of infection (MOI) for viral delivery or optimized concentrations for non-viral methods to ensure consistent expression levels across groups.
Pluripotency Induction and Colony Culture:
- 24-48 hours post-transduction, replace the medium with specialized iPSC/ESC culture medium (e.g., containing KnockOut Serum Replacement, bFGF) [23].
- Culture cells on a feeder layer of mouse embryonic fibroblasts (MEFs) or on defined substrates like Matrigel in feeder-free conditions [23].
- Change the medium daily. Colonies with embryonic stem cell-like morphology will typically appear in 2-3 weeks.
Colony Picking and Expansion:
- Manually pick individual iPSC colonies based on morphology and transfer them to new culture plates for expansion.
- Expand clonal lines and characterize them for pluripotency markers.
Protocol for Pluripotency Validation
Rigorous validation is required to confirm the successful reprogramming of colonies generated with substitute factors [23] [21].
Immunofluorescence Staining:
- Fix iPSC colonies using 4% paraformaldehyde.
- Permeabilize cells with Triton X-100 and block with serum.
- Incubate with primary antibodies against core pluripotency transcription factors (e.g., Anti-Oct4, Anti-Nanog) and surface markers (e.g., Anti-SSEA-4, Anti-TRA-1-60).
- Incubate with fluorophore-conjugated secondary antibodies and visualize using a fluorescence microscope.
In Vitro Differentiation (Embryoid Body Formation):
- Harvest iPSCs and culture them in low-attachment plates in medium without bFGF to promote the formation of embryoid bodies (EBs).
- After 7-10 days, plate EBs on gelatin-coated dishes and allow for spontaneous differentiation for another 7-14 days.
- Analyze the resulting cells for markers of the three germ layers (ectoderm, mesoderm, endoderm) via immunofluorescence or RT-qPCR.
In Vivo Teratoma Formation Assay:
- Inject a suspension of iPSCs subcutaneously into immunodeficient mice (e.g., NSG mice).
- After 6-12 weeks, dissect the resulting tumors and fix them for histological analysis.
- Section and stain tumors with H&E. The presence of tissues derived from all three germ layers (e.g., neural rosettes for ectoderm, cartilage for mesoderm, gut-like epithelium for endoderm) confirms pluripotency [23]. This assay also provides critical safety data on the tumorigenic potential of the iPSCs.
Signaling Pathways and Molecular Mechanisms
The reprogramming process involves a complex network of interactions where the Yamanaka factors and their substitutes collaboratively suppress the somatic cell program and activate the pluripotency network. The diagram below illustrates the core functional relationships and the position of substitute factors in this process.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Reprogramming and Characterization Experiments
| Reagent / Material | Function in Experiment | Specific Examples / Notes |
|---|---|---|
| Delivery Vectors | Introducing reprogramming genes into somatic cells [6] | Retrovirus, Lentivirus (integrating); Sendai virus, Episomal plasmids (non-integrating) [6] [21]. Choice affects biosafety and efficiency. |
| Culture Matrix | Provides a scaffold for cell attachment and growth, supporting self-renewal [23] | Matrigel, recombinant Laminin, Vitronectin, or defined synthetic hydrogels [23]. |
| Defined Culture Medium | Maintains iPSCs in a pluripotent state; supports reprogramming [23] | TeSR1, mTeSR1, or similar formulations containing essential nutrients and growth factors like bFGF [23]. |
| Small Molecule Enhancers | Improve reprogramming efficiency by modulating epigenetic or signaling pathways [6] [23] | Valproic acid (VPA, HDAC inhibitor), 5-Azacytidine (DNA methyltransferase inhibitor), SB431542 (TGF-β inhibitor), PD0325901 (MEK inhibitor) [6] [23]. |
| Pluripotency Antibodies | Validation of successful reprogramming via immunostaining or flow cytometry [23] | Primary antibodies against Oct4, Nanog, Sox2, SSEA-4, TRA-1-60. |
| In Vivo Model | Testing pluripotency through teratoma formation [23] | Immunodeficient mice (e.g., NOD-scid gamma (NSG) mice). |
| 3,5-Dibromo-4-pyridinol | 3,5-Dibromo-4-pyridinol, CAS:141375-47-5, MF:C5H3Br2NO, MW:252.89 g/mol | Chemical Reagent |
| 3,4-Dimethyl-5-propyl-2-furannonanoic Acid | 3,4-Dimethyl-5-propyl-2-furannonanoic Acid|CAS 57818-38-9 | 3,4-Dimethyl-5-propyl-2-furannonanoic acid is a high-purity furan fatty acid (9D3) for lipid oxidation research. This product is For Research Use Only and not for human or veterinary diagnostics or therapeutic applications. |
The systematic comparison of KLF2, KLF5, SOX1, SOX3, L-MYC, and N-MYC reveals a critical trade-off in factor substitutability: while these alternatives generally offer improved safety profiles by reducing oncogenic risk, they often do so at the cost of significantly lower reprogramming efficiency compared to the original Yamanaka factors [6] [23]. This efficiency-safety balance is a central consideration for research applications.
Future work must focus on optimizing culture conditions and leveraging small molecule enhancers to close the efficiency gap [6] [21]. Furthermore, the choice between autologous iPSCs derived from a patient's own cells and the use of HLA-matched allogeneic iPSCs from biobanks will significantly influence which factor combinations are most appropriate for therapeutic development, with safety being paramount for clinical translation [21] [1]. Continued research into the mechanisms of these substitutes will further refine their application, pushing the field toward more efficient and safer reprogramming protocols.
In the field of regenerative medicine, induced pluripotent stem cell (iPSC) technology has revolutionized biological research and therapeutic development since its discovery by Shinya Yamanaka. The original Yamanaka factors—OCT4, SOX2, KLF4, and c-MYC (OSKM)—enable reprogramming of somatic cells into pluripotent stem cells, but with notoriously low efficiency, typically converting less than 0.1% of treated cells [5]. Defining and quantifying reprogramming efficiency is therefore critical for comparing different reprogramming factor combinations and advancing both basic research and clinical applications. This guide examines the key metrics, benchmarks, and experimental frameworks that researchers employ to objectively measure reprogramming efficiency, with particular emphasis on emerging technologies that are reshaping the landscape of iPSC generation.
Key Metrics for Assessing Reprogramming Efficiency
To systematically compare the performance of different Yamanaka factor combinations, researchers track multiple quantitative metrics throughout the reprogramming process. These parameters collectively provide a comprehensive picture of how effectively somatic cells are being converted to a pluripotent state.
Table 1: Core Metrics for Evaluating Reprogramming Efficiency
| Metric Category | Specific Metrics | Measurement Technique | Interpretation |
|---|---|---|---|
| Early Pluripotency Markers | SSEA-4 expression [5] | Flow cytometry, immunostaining | Indicates initiation of reprogramming |
| Late Pluripotency Markers | TRA-1-60, NANOG expression [5] | Immunostaining, RNA analysis | Confirms establishment of pluripotency |
| Functional Pluripotency | Alkaline phosphatase (AP) activity [5] | AP staining | Demonstrates functional pluripotency |
| Temporal Efficiency | Time to colony appearance [5] | Microscopy observation | Measures acceleration of process |
| Throughput Efficiency | Percentage of cells converting [5] | Colony counting, flow cytometry | Quantifies overall success rate |
| Characterization | Germ layer differentiation [5] | In vitro differentiation assays | Validates trilineage potential |
| Safety | Genomic stability [5] | Karyotyping, DNA sequencing | Ensures clinical suitability |
Benchmarking Yamanaka Factor Combinations
The original OSKM factors remain the benchmark against which novel reprogramming combinations are evaluated. However, recent advances have identified more efficient alternatives, including both modified versions of the original factors and completely new factor combinations.
Table 2: Performance Comparison of Reprogramming Factor Combinations
| Factor Combination | Reprogramming Efficiency | Key Advantages | Limitations | Experimental Validation |
|---|---|---|---|---|
| Traditional OSKM | <0.1% cell conversion [5] | Established benchmark, widely validated | Low efficiency, tumorigenic risk (c-MYC) [6] | Fibroblast reprogramming in 3+ weeks [5] |
| OSK (without c-MYC) | Lower than OSKM [6] | Reduced tumorigenic risk [6] | Significantly decreased efficiency [6] | Demonstrated in mouse and human fibroblasts [6] |
| OSNL (OCT4, SOX2, NANOG, LIN28) | Comparable to OSKM [6] | Avoids c-MYC oncogene [6] | May require additional enhancers | Human somatic cell reprogramming [6] |
| L-Myc substitution | Similar to c-MYC [6] | Reduced tumorigenic potential [6] | Potentially tissue-dependent effects | Mouse and human cell studies [6] |
| RetroSOX/RetroKLF + OK | >30% cell conversion (SSEA-4, TRA-1-60) [5] | 50x higher marker expression, faster onset [5] | AI-engineered, requires further validation | Multiple donors, cell types, and delivery methods [5] |
| Chemical Reprogramming | Varies with cocktail composition [6] | Non-integrating, enhanced safety profile [6] | Often lower than genetic methods | Human fibroblast reprogramming [6] |
Experimental Protocols for Efficiency Assessment
Standardized experimental approaches are essential for generating comparable data across different studies of reprogramming factors. The following protocols represent current best practices in the field.
Protocol 1: Quantitative Assessment of Reprogramming Efficiency
Cell Culture and Transduction
- Isolate human fibroblasts from tissue samples or use established lines (e.g., HDF, BJ fibroblasts)
- Culture in standard fibroblast medium (DMEM + 10% FBS)
- Transduce with lentiviral or sendai viral vectors encoding reprogramming factors at MOI 5-10
- Include control groups: wild-type OSKM, experimental factors, and empty vector
- For non-integrating methods, use mRNA transfection or episomal vectors
Pluripotency Marker Analysis
- At days 7, 14, and 21 post-transduction, dissociate cells for analysis
- For flow cytometry: stain with anti-SSEA-4 and anti-TRA-1-60 antibodies
- Fix parallel samples for immunocytochemistry (NANOG, SOX2, OCT4)
- Perform alkaline phosphatase staining using commercial kits according to manufacturer protocols
- Quantify positive cells across multiple fields (minimum n=5) and normalize to total cell count
Molecular Validation
- Extract RNA from emerging iPSC colonies for qRT-PCR analysis of endogenous pluripotency genes (OCT4, SOX2, NANOG)
- Perform bisulfite sequencing of OCT4 and NANOG promoters to assess epigenetic reprogramming
- Verify trilineage differentiation potential through embryoid body formation and specific staining for ectoderm (βIII-tubulin), mesoderm (α-smooth muscle actin), and endoderm (SOX17)
Protocol 2: Functional Characterization of Reprogrammed Cells
Long-Term Culture and Stability Assessment
- Passage emerging iPSC colonies onto feeder layers or defined matrices
- Monitor morphological stability over 10+ passages
- Perform G-banding karyotype analysis at passages 5, 10, and 15
- Use whole-genome sequencing to identify potential genetic abnormalities
Advanced Pluripotency Validation
- Perform in vitro differentiation toward specific lineages (neuronal, cardiac, hepatic)
- Conduct teratoma formation assays in immunodeficient mice (for research purposes)
- Validate global gene expression profiles by RNA sequencing compared to reference hESC lines
Visualization of Reprogramming Pathways and Workflows
Reprogramming Pathway Progression
Reprogramming Efficiency Workflow
The Scientist's Toolkit: Essential Research Reagents
Successful reprogramming experiments require carefully selected reagents and systems. The following table outlines critical components for iPSC generation studies.
Table 3: Essential Research Reagents for Reprogramming Studies
| Reagent Category | Specific Examples | Function | Considerations |
|---|---|---|---|
| Reprogramming Factors | Wild-type OSKM, OSNL, RetroSOX/RetroKLF [5] | Induce pluripotency | Delivery method affects efficiency and safety |
| Delivery Systems | Lentivirus, Sendai virus, mRNA, episomal plasmids [6] | Introduce factors into cells | Integrating vs. non-integrating approaches |
| Cell Culture Media | Fibroblast medium, iPSC maintenance medium | Support cell growth and reprogramming | Defined media improve reproducibility |
| Characterization Antibodies | Anti-SSEA-4, Anti-TRA-1-60, Anti-NANOG [5] | Detect pluripotency markers | Validate species cross-reactivity |
| Detection Kits | Alkaline phosphatase staining kits [5] | Identify pluripotent colonies | Some kits work on live cells for sorting |
| Culture Surfaces | Matrigel, laminin-521, feeder cells | Provide adhesion and signaling | Defined matrices reduce variability |
| Enhancer Molecules | Valproic acid, sodium butyrate, 8-Br-cAMP [6] | Improve reprogramming efficiency | Concentration optimization required |
| Demethylamino Ranitidine Acetamide Sodium | Demethylamino Ranitidine Acetamide Sodium|CAS 112251-56-6 | Demethylamino Ranitidine Acetamide Sodium is a Ranitidine impurity for research. This product is For Research Use Only and is not intended for diagnostic or personal use. | Bench Chemicals |
| GLYCOLURIL, 3a,6a-DIPHENYL- | GLYCOLURIL, 3a,6a-DIPHENYL-, CAS:5157-15-3, MF:C16H14N4O2, MW:294.31 g/mol | Chemical Reagent | Bench Chemicals |
Emerging Technologies and Future Directions
The field of cellular reprogramming is rapidly evolving with several emerging technologies poised to transform efficiency benchmarking.
AI-Guided Protein Engineering
Recent breakthroughs in artificial intelligence are enabling the design of novel reprogramming factors with significantly enhanced performance. The collaboration between OpenAI and Retro Biosciences has demonstrated that AI-generated variants of SOX2 and KLF4 (RetroSOX and RetroKLF) can achieve remarkable improvements in reprogramming efficiency—up to 50-fold higher expression of pluripotency markers compared to wild-type factors [5]. These AI-engineered factors also demonstrate enhanced DNA damage repair capabilities, suggesting improved rejuvenation potential alongside more efficient reprogramming [5].
Chemical Reprogramming
Small molecule approaches represent a promising alternative to genetic reprogramming methods. Chemical cocktails can replace some or all reprogramming factors, potentially offering enhanced safety profiles by avoiding genomic integration [6]. These approaches activate distinct molecular pathways compared to traditional OSKM-based reprogramming and may access different intermediate cell states [6].
Partial Reprogramming
For therapeutic applications targeting aging and cellular rejuvenation, partial reprogramming approaches are gaining traction. These methods involve transient expression of Yamanaka factors sufficient to restore youthful epigenetic patterns and cellular functions without completely reversing cell identity [24]. While promising, this approach requires precise control to avoid teratoma formation or loss of cellular identity [24].
The systematic assessment of reprogramming efficiency through standardized metrics and benchmarks is fundamental to advancing iPSC technology. As the field progresses beyond the original Yamanaka factors, rigorous comparison of novel factor combinations—including AI-engineered variants and chemical approaches—will be essential for identifying optimal strategies for both research and clinical applications. The experimental frameworks and benchmarks outlined in this guide provide a foundation for objective evaluation of reprogramming efficiency across different platforms and conditions. Future advances will likely focus not only on increasing efficiency but also on enhancing safety, reproducibility, and accessibility of iPSC generation across diverse cell types and donor backgrounds.
From Theory to Practice: Delivery Systems and Application-Specific Cocktails
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) has revolutionized regenerative medicine and disease modeling [25] [21]. A critical determinant of reprogramming success is the method used to deliver these factors into target cells. The choice between viral and non-viral delivery systems involves significant trade-offs between efficiency, safety, and clinical applicability. This guide provides a objective comparison of the most commonly used delivery methods—retrovirus, lentivirus, Sendai virus, and episomal vectors—focusing on their performance in reprogramming experiments, to inform researchers and drug development professionals.
Vector Comparison Tables
The following tables summarize the core characteristics and performance metrics of the four primary delivery vectors used for iPSC generation.
Table 1: Fundamental Characteristics of Delivery Vectors
| Feature | Retrovirus | Lentivirus | Sendai Virus (SeV) | Episomal Vectors |
|---|---|---|---|---|
| Vector Type | Viral, Integrating | Viral, Integrating | Viral, Non-integrating | Non-viral, Non-integrating |
| Genetic Material | RNA | RNA | RNA | DNA |
| Genomic Integration | Yes | Yes | No | No (Episomal) |
| Target Cell Requirement | Dividing cells only | Dividing & non-dividing cells | Dividing & non-dividing cells | Dividing & non-dividing cells |
| Primary Safety Concern | Insertional mutagenesis | Insertional mutagenesis | Cytopla smic persistence | Low transgene expression |
| Residual Transgene Expression | Yes, unless silenced | Yes, unless silenced | Diluted upon cell division | Diluted upon cell division |
Table 2: Experimental Performance Metrics in iPSC Generation
| Performance Metric | Retrovirus | Lentivirus | Sendai Virus (SeV) | Episomal Vectors |
|---|---|---|---|---|
| Reprogramming Efficiency | High [21] | High [26] | Highest among non-integrating methods [27] | Lower than SeV [27] |
| Typical Time to iPSC Colony | ~2-3 weeks | ~2-3 weeks | ~3-4 weeks | ~3-4 weeks |
| Aberrant Methylation Sites | Higher [28] | Not specified in results | Lowest number [28] | Intermediate [28] |
| Stability of Reprogramming | Stable | Stable | Stable, but virus must be lost | Can be unstable |
| Key Advantage | Robust, stable integration | Broad tropism, integrates non-dividing cells | High efficiency, non-integrating | Non-integrating, non-viral |
| Key Disadvantage | Silencing in pluripotent cells; safety risk | Safety risk of insertional mutagenesis | Requires protocol to clear virus | Low efficiency; requires optimization |
Detailed Experimental Protocols
To ensure reproducibility, this section outlines standardized protocols for implementing the two most prevalent non-integrating reprogramming methods, as derived from biobanking practices [27].
Sendai Virus (SeV) Reprogramming Protocol
- Source Cells: Fibroblasts or Peripheral Blood Mononuclear Cells (PBMCs).
- Transduction: Cells are transduced using a CytoTune Sendai Reprogramming Kit, which typically includes four separate SeV vectors, each expressing one of the factors hOCT4, hSOX2, hKLF4, and hC-MYC, alongside a reporter gene like EmGFP [27].
- Post-Transduction Culture: Twenty-four hours after transduction, the medium is refreshed. Cells are cultured for approximately 6 additional days, with medium exchanged every other day.
- Assessment: Transduction efficiency is estimated by examining GFP-positive cells using fluorescence microscopy.
- Rep plating: Approximately 7 days post-transduction for fibroblasts (or 3 days for PBMCs), the cells are harvested and replated onto feeder cells or a suitable substrate like Matrigel.
- Colony Picking: After an additional 2–3 weeks, emergent iPSC colonies reach the appropriate size for transfer. At this stage, at least 24 colonies are manually picked for further expansion and characterization [27].
Episomal Vector Reprogramming Protocol
- Source Cells: Lymphoblastoid Cell Lines (LCLs) or fibroblasts.
- Nucleofection: Cells are nucleofected (electroporation) using an Amaxa Nucleofector II device with specific programs (e.g., U-015 for LCLs, U-023 for fibroblasts). The vectors used are OriP/EBNA1 episomal plasmids expressing hOCT3/4, hSOX2, hKLF4, hL-MYC, LIN28, and a marker like EGFP [27].
- Post-Nucleofection Culture: Post-nucleofection, cells are maintained in a 5% O2 incubator and fed every other day.
- Assessment: Nucleofection efficiency is monitored by tracking GFP-positive cells.
- Rep plating: On days 6–7 after nucleofection, the transfected cells are replated.
- Colony Picking: After 1–2 additional weeks, at least 24 clones are manually selected for expansion, leading to master and distribution bank freezes [27].
Research Reagent Solutions
Critical quality control (QC) measures are paramount for ensuring the reliability and reproducibility of generated iPSC lines. The following table details key reagents and tests used in this process [27].
Table 3: Essential Reagents for iPSC Generation and Quality Control
| Reagent / Test | Function / Purpose | Example Details |
|---|---|---|
| CytoTune Sendai Kit | Delivers OSKM factors via non-integrating SeV | Contains separate viruses for hOCT4, hSOX2, hKLF4, hC-MYC [27] |
| Episomal Plasmids | Delivers reprogramming factors without viral components | OriP/EBNA1 vectors with hOCT3/4, hSOX2, hKLF4, hL-MYC, LIN28 [27] |
| mTeSR1 Medium | Defined, feeder-free culture medium for pluripotent stem cells | Used for maintaining established iPSCs; sometimes with "double feeding" protocol [27] |
| Y-27632 (ROCK inhibitor) | Improves survival of single pluripotent stem cells | Added to medium for 20-24 hours after thawing or passaging cells [27] |
| Alkaline Phosphatase (AP) Staining | Marker of pluripotency; undifferentiated cells stain positively | Standard QC test performed on iPSC colonies [27] |
| Karyotyping | Detects gross chromosomal abnormalities | Standard QC test (e.g., G-banding) to ensure genomic integrity [27] |
| Short Tandem Repeat (STR) Analysis | Confirms cell line identity and rules for cross-contamination | Standard QC test comparing the iPSC line to the source material [27] |
| Mycoplasma Testing | Ensures cell cultures are free from mycoplasma contamination | Essential routine test using dedicated kits on spent medium/cell suspension [27] |
Visualizing Reprogramming Workflows
The diagrams below illustrate the logical workflow for the two primary non-integrating reprogramming methods and the subsequent quality control pipeline.
Sendai Virus vs. Episomal Reprogramming Workflow
iPSC Line Quality Control Pipeline
The choice of delivery vector for iPSC generation is a critical decision that balances efficiency against safety. Retroviruses and lentiviruses offer high reprogramming efficiency and stable integration but carry the inherent risk of insertional mutagenesis, making them less suitable for clinical applications [26] [21]. Among non-integrating methods, the Sendai virus consistently demonstrates superior reprogramming efficiency and a lower number of aberrant methylation sites compared to episomal vectors [28] [27]. However, episomal vectors, while less efficient, provide a completely non-viral alternative, eliminating concerns related to viral components entirely [27]. The optimal vector is thus highly dependent on the research or therapeutic objective: SeV is ideal for robust iPSC generation where transient viral presence is acceptable, episomal vectors are preferable for applications demanding the highest safety profile, and lentiviral vectors remain powerful tools for research requiring stable genetic modification.
The generation of human induced pluripotent stem cells (hiPSCs) has revolutionized biomedical research, offering unprecedented opportunities in disease modeling, drug screening, and regenerative therapies [27]. Since the groundbreaking discovery of cellular reprogramming, the field has progressively shifted from integrating viral vectors to non-integrating reprogramming methodologies to minimize the risk of genomic alterations and enhance the clinical safety profile of resulting hiPSCs [27] [29]. Among the various non-integrating methods available, Sendai virus (SeV) and episomal (Epi) reprogramming have emerged as two of the most prevalent approaches due to their relative ease of manipulation and efficiency [27] [29].
Despite their widespread use, a clear consensus regarding their comparative success rates has been lacking, with different studies reporting apparently conflicting findings. This analysis directly addresses this uncertainty by synthesizing current scientific evidence to provide a definitive comparison of reprogramming success rates between Sendai viral and episomal methods. Framed within a broader investigation into the efficiency of different Yamanaka factor delivery strategies, this guide provides researchers, scientists, and drug development professionals with the experimental data and methodological details necessary to inform their reprogramming platform selection.
Defining and Quantifying Reprogramming Success
In hiPSC generation, "success" is typically evaluated through two primary metrics: reprogramming efficiency and reprogramming success rate.
- Reprogramming Efficiency refers to the percentage of starting somatic cells that give rise to iPSC colonies [29]. This metric provides a precise measure of how productive a specific reprogramming method is with a particular cell sample.
- Reprogramming Success Rate describes the percentage of independent somatic cell samples for which a method can generate at least three bona fide iPSC colonies [29]. This metric is particularly relevant for biobanking and clinical applications where generating lines from diverse donor samples is essential.
The following sections present a direct, data-driven comparison of these metrics for Sendai virus and episomal reprogramming methods.
Direct Comparative Analysis of Success Rates
Comprehensive Success Rate and Efficiency Data
A systematic comparison of non-integrating reprogramming methods provides the most direct evidence for their relative performance. Key findings from a landmark study are summarized in Table 1 [29].
Table 1: Comparative Performance of Non-Integrating Reprogramming Methods
| Reprogramming Method | Reprogramming Efficiency (Mean %) | Overall Success Rate (% of Samples) | Time Until Colony Picking (Days) | Relative Hands-on Time (Hours) |
|---|---|---|---|---|
| Sendai Virus (SeV) | 0.077% | 94% | ~26 | 3.5 |
| Episomal (Epi) | 0.013% | 93% | ~20 | 4.0 |
| mRNA | 2.1% | 27% (improves to 73% with miRNA) | ~14 | 8.0 |
| Lentiviral (Lenti) | 0.27% | 100% | Not Specified | Not Specified |
The data reveals a critical distinction: while the Sendai virus method offers a significantly higher reprogramming efficiency (approximately 6-fold higher) than the episomal method, both techniques demonstrate an equally high and reliable success rate exceeding 90% [29]. This means that for the vast majority of somatic cell samples, both methods are very likely to yield iPSC colonies, but the Sendai virus protocol typically generates a greater number of colonies from the same number of starting cells.
A more recent 2025 study corroborates the superior efficiency of the Sendai virus system, confirming that it "yields significantly higher success rates relative to the episomal reprogramming method" [27] [30]. However, an earlier study reported a contrasting finding, observing a higher colony count with episomal transfection [31]. This discrepancy highlights that success can be influenced by specific experimental protocols, vector designs, and source cell types.
Methodological Impact on Success
The experimental workflow for reprogramming involves several key stages where methodological choices can influence the final outcome. The diagram below illustrates the general workflow and critical decision points for Sendai virus and episomal methods.
Figure 1: A generalized workflow for hiPSC generation comparing the initial steps of the Sendai virus and episomal reprogramming paths. Key methodological differences in the delivery and expression of reprogramming factors significantly influence outcomes.
Detailed Experimental Protocols
Sendai Virus Reprogramming Protocol
The Sendai virus protocol utilizes a non-integrating, cytoplasmic RNA virus to deliver and express reprogramming factors.
Key Reagents:
- CytoTune-iPS Sendai Reprogramming Kit (Thermo Fisher Scientific): Contains four SeV-based vectors, each encoding one of the Yamanaka factors (Oct4, Sox2, Klf4, c-Myc) [32].
- Somatic Cells: Fibroblasts, Peripheral Blood Mononuclear Cells (PBMCs), or Lymphoblastoid Cell Lines (LCLs) [27].
- Culture Medium: Appropriate medium for the somatic cell type, followed by transition to human iPSC medium post-transduction.
Detailed Workflow:
- Day 0: Plate somatic cells at an optimal density (e.g., 5 x 10^5 cells per well of a 6-well plate) and culture overnight [32].
- Day 1: Transduce cells with a combination of the four CytoTune Sendai virus vectors. The Multiplicity of Infection (MOI) should be optimized; a total MOI of 3-5 is often used [27] [32].
- Day 2: Refresh the culture medium 24 hours after transduction to remove viral particles.
- Days 3-7: Continue culturing, exchanging the medium every other day. Transduction efficiency can be estimated if using a vector expressing a fluorescent reporter like EmGFP [27].
- Approximately Day 7: Harvest and re-plate the transduced cells onto feeder layers or Matrigel-coated plates.
- Days 10-28: Continue daily feeding with hiPSC medium. Colonies typically become large enough for manual picking around day 26 post-transduction [27] [29].
- Colony Expansion: Pick at least 24 individual colonies for expansion and screening. The SeV genome is gradually diluted out over 10-15 passages [29] [33].
Episomal Reprogramming Protocol
The episomal method uses OriP/EBNA1-based plasmids that replicate extra-chromosomally in dividing cells but are gradually lost in the absence of selection.
Key Reagents:
- Episomal Plasmids: Typically a combination of plasmids expressing OCT4, SOX2, KLF4, L-MYC, LIN28, and an shRNA against p53 [27] [34].
- Somatic Cells: Fibroblasts or LCLs.
- Nucleofection Device: Such as the Amaxa Nucleofector II (Lonza) [27].
Detailed Workflow:
- Day 0: Prepare somatic cells for nucleofection.
- Day 1: Co-nucleofect cells with the episomal plasmid combination using a cell-type-specific nucleofection program (e.g., U-023 for fibroblasts) [27]. Culture transfected cells in a low-oxygen (5% O2) incubator to enhance efficiency [27].
- Days 2-6: Feed cells every other day. Transfection efficiency can be monitored if plasmids contain a reporter like EGFP [27].
- Days 6-7: Re-plate the transfected cells onto feeder layers or Matrigel.
- Days 14-21: Feed daily. Colonies are often ready for picking around day 20, slightly earlier than with SeV [29].
- Colony Expansion: Manually pick and expand at least 24 clones. Passaging cells continuously is required to dilute out the episomal plasmids. A significant fraction of lines (over 30%) may retain plasmids even at passage 10-11, necessitating PCR validation [29].
The Scientist's Toolkit: Essential Reagents and Materials
Successful reprogramming relies on a suite of specialized reagents and tools. Table 2 lists the core components of a reprogramming toolkit for the methods discussed.
Table 2: Essential Research Reagent Solutions for hiPSC Reprogramming
| Reagent/Material | Function/Purpose | Example Product/Catalog Number |
|---|---|---|
| Sendai Virus Vectors | Delivery of OSKM factors; non-integrating | CytoTune-iPS Sendai Reprogramming Kit (Thermo Fisher, A1378001) [32] |
| Episomal Vectors | Delivery of OSKM, L-MYC, LIN28, sh-p53; non-integrating | OriP/EBNA1-based plasmids (e.g., Addgene #41855-41858) [27] |
| Nucleofector Device | High-efficiency plasmid delivery for episomal method | Amaxa Nucleofector II Device (Lonza) [27] |
| Y-27632 (ROCK inhibitor) | Improves survival of single pluripotent cells; used during passaging and thawing | Y-27632 (e.g., Thermo Fisher, PHZ1234) [27] |
| mTeSR1 Medium | Defined, feeder-free culture medium for hiPSCs | mTeSR1 (StemCell Technologies, 85850) [27] |
| Matrigel | Extracellular matrix for feeder-free culture of hiPSCs | Matrigel (Corning, 354230) [27] |
| Alkaline Phosphatase Staining Kit | Identification of pluripotent colonies | e.g., Millipore Sigma, SCR004 |
| Pluripotency Antibodies | Immunostaining for markers (TRA-1-60, SSEA4, NANOG) | Anti-Tra-1-60 (Thermo Fisher, 41-1000), Anti-SSEA4 (Thermo Fisher, 41-4000) [29] [32] |
| Tetrabutylammonium bibenzoate | Tetrabutylammonium bibenzoate, CAS:116263-39-9, MF:C30H47NO4, MW:485.7 g/mol | Chemical Reagent |
| Decapreno-|A-carotene | Decapreno-|A-carotene, CAS:5940-03-4, MF:C50H68, MW:669.1 g/mol | Chemical Reagent |
Critical Considerations Beyond Success Rate
While success rate is a primary concern, other factors are crucial for method selection, particularly for clinical applications. The diagram below illustrates the multi-faceted decision-making process.
Figure 2: Key decision factors beyond initial success rate for selecting a reprogramming method. Colors indicate relative advantage (green), caution (red), or neutral (yellow).
Summary of Critical Factors:
Genomic Safety and Transgene Clearance: Both methods are "footprint-free" in principle. However, the Sendai virus is an RNA virus that remains in the cytoplasm and is reliably diluted out over passages [32]. In contrast, episomal plasmids can be retained in a significant subset of hiPSC lines (over 30% at passage 10), requiring rigorous quality control to confirm loss [29]. A noted concern with Sendai virus is the potential for persistence, particularly when hiPSCs are cultured in naive media conditions, which may select for cells retaining the virus and its exogenous factors [35].
Genetic Stability: Karyotypic analyses reveal that Sendai virus-derived hiPSCs have a lower rate of aneuploidy (4.6%) compared to episomal-derived lines (11.5%) [29]. This makes SeV a potentially safer choice from a genetic integrity standpoint.
Workload and Practicality: The Sendai virus method demands the least hands-on time (approximately 3.5 hours until colony picking) and is relatively straightforward, involving a single transduction step [29]. The episomal method requires nucleofection expertise and slightly more hands-on time (4 hours). However, the need to screen a larger number of SeV-derived clones for viral clearance and Epi-derived clones for plasmid retention adds to the downstream workload for both [29].
The direct comparative analysis of Sendai virus and episomal reprogramming methods reveals a nuanced picture. The Sendai virus method demonstrates a statistically significant advantage in reprogramming efficiency, generating more iPSC colonies per input cell, while both methods show equally high success rates in terms of being able to reprogram most somatic cell samples [27] [29].
For research applications where the highest efficiency and lowest aneuploidy rate are prioritized, and where rigorous screening for viral clearance is feasible, the Sendai virus method is often the superior choice. Conversely, for projects or facilities where viral vectors are prohibited or heavily restricted, the episomal method provides a robust, non-viral alternative, albeit with a requirement for careful monitoring of plasmid retention.
Future advancements in reprogramming will likely focus on refining these protocols further, perhaps through the use of optimized small-molecule cocktails that can modulate signaling pathways like TGF-β to enhance efficiency, particularly for challenging senescent or pathologic somatic cells [34] [14]. The ultimate goal remains the consistent generation of clinically safe, high-quality hiPSCs, a pursuit for which this direct comparison provides essential guidance.
The efficiency of generating induced pluripotent stem cells (iPSCs) is highly dependent on the synergistic combination of reprogramming factors and the specific somatic cell source used. This guide compares established and emerging reprogramming strategies for three common cell sources—fibroblasts, peripheral blood mononuclear cells (PBMCs), and lymphoblastoid cell lines (LCLs)—by presenting consolidated experimental data, detailed protocols, and key reagents to inform research and development.
Comparative Efficiency of Reprogramming Strategies
The table below summarizes reprogramming efficiencies for fibroblasts, PBMCs, and LCLs using different factor delivery methods and factor combinations, as reported in recent studies.
| Cell Source | Reprogramming Method | Key Factors | Reported Efficiency / Outcome | Key Findings |
|---|---|---|---|---|
| Fibroblasts | Sendai Virus (SeV) | OCT4, SOX2, KLF4, C-MYC (OSKM) | Significantly higher success rates vs. episomal method [27] | A comparative analysis found SeV yield superior for fibroblasts [27]. |
| Fibroblasts | Episomal Vectors | OCT4, SOX2, KLF4, L-MYC, LIN28, sh-p53 | Lower success rates vs. SeV method [27] | Non-integrating method with lower efficiency [27]. |
| Fibroblasts | mRNA & Viral Vectors | AI-enhanced SOX2 (RetroSOX) & KLF4 (RetroKLF) | >50x higher reprogramming marker expression vs. wild-type [5] | Novel AI-designed factor variants dramatically boost efficiency [5]. |
| PBMCs | Sendai Virus (SeV) | OCT4, SOX2, KLF4, C-MYC (OSKM) | Successful reprogramming achieved [27] | A viable and minimally invasive cell source [27]. |
| PBMCs | Chemical Reprogramming | Small Molecule Cocktail | Higher efficiency vs. OSKM + p53 knockdown in fresh/cryopreserved samples [36] | Efficient, non-genetic method; works with finger-prick samples [36]. |
| LCLs | Episomal Vectors | OCT4, SOX2, KLF4, L-MYC, LIN28, sh-p53 | Successful reprogramming achieved [27] | Nucleofection used for delivery; no significant impact on overall success rates [27]. |
| T Cells (from Blood) | Sendai Virus (SeV) | NEUROD1 + OSKM | ~22.4% of surviving cells became βIII-Tubulin+ neurons [37] | Direct conversion to functional neurons, bypassing pluripotency [37]. |
Detailed Experimental Protocols
Sendai Virus (SeV) Reprogramming for Fibroblasts and PBMCs
This non-integrating, highly efficient method is widely used for fibroblasts and PBMCs [27].
- Cell Culture Pre-Processing:
- Fibroblasts: Culture in standard fibroblast growth medium until ~80% confluent.
- PBMCs: Isolate from whole blood via density gradient centrifugation (e.g., Ficoll). Stimulate with anti-CD3 antibody and IL-2 for T-cell expansion if needed [37].
- Transduction:
- Use the CytoTune Sendai Reprogramming Kit or similar.
- Transduce cells with SeV vectors expressing OSKM and EmGFP at an appropriate multiplicity of infection (MOI).
- Incubation: 24 hours post-transduction, refresh the medium. Culture for approximately 6 more days, changing the medium every other day.
- Monitoring & Picking:
- Estimate transduction efficiency by examining GFP-positive cells.
- Around day 7 (fibroblasts) or day 3 (PBMCs), harvest and replate cells onto feeder layers or Matrigel-coated plates.
- After 2-3 weeks, manually pick at least 24 distinct colonies for expansion [27].
Episomal Reprogramming for LCLs and Fibroblasts
This non-integrating method uses OriP/EBNA1 plasmids, suitable for LCLs and fibroblasts [27].
- Cell Preparation:
- Culture LCLs or fibroblasts to a density of ~1x10^6 cells per nucleofection sample.
- Nucleofection:
- Use episomal vectors expressing OCT4, SOX2, KLF4, L-MYC, LIN28, and EGFP, often with sh-p53 to enhance efficiency.
- Use the Amaxa Nucleofector II device with cell-type-specific programs: U-015 for LCLs and U-023 for fibroblasts [27].
- Post-Nucleofection Culture:
- Maintain transfected cells in a 5% O2 incubator, feeding every other day.
- Monitor nucleofection efficiency via GFP-positive cells.
- On days 6-7, replate the transfected cells. After 1-2 additional weeks, manually pick at least 24 clones for expansion [27].
Direct Neuronal Conversion from Peripheral Blood T Cells
This protocol directly converts T cells into neurons using a 5-factor (5F) combination, bypassing the iPSC stage [37].
- Cell Source & Activation: Isolate T cells from human peripheral blood and stimulate them using an anti-CD3 antibody for expansion.
- Viral Transduction:
- Cotransduce activated T cells using Sendai virus vectors expressing NEUROD1, OCT3/4, SOX2, KLF4, and c-MYC.
- Use a defined MOI (e.g., 10) and remove the virus by medium replacement after 24 hours.
- Culture & Differentiation:
- Plate transduced cells on Matrigel-coated plates.
- Maintain in optimized neuronal differentiation medium (e.g., a combination of MHM and StemFit basal media), supplemented with 10% FBS to enhance cell survival.
- Timeline: Adherent cells appear within a week. Neuron-like processes develop in the second week, and complex dendritic morphology is observed by the third week [37].
Experimental Workflow Visualization
The following diagram illustrates the key decision points and pathways for selecting a reprogramming strategy based on cell source and desired outcome.
The Scientist's Toolkit: Key Research Reagent Solutions
The table below lists essential reagents and kits used in the featured studies, providing a practical resource for protocol design.
| Reagent / Kit | Function / Application | Cell Source Compatibility |
|---|---|---|
| CytoTune Sendai Reprogramming Kit (Thermo Fisher) | Delivery of OSKM factors via non-integrating RNA virus. | Fibroblasts, PBMCs [27] |
| OriP/EBNA1 Episomal Vectors | Non-integrating plasmid system for factor delivery. | LCLs, Fibroblasts [27] |
| Amaxa Nucleofector II Device (Lonza) | High-efficiency delivery of episomal vectors into hard-to-transfect cells. | LCLs (Program U-015), Fibroblasts (Program U-023) [27] |
| mTeSR1 / mTeSR Plus Medium (STEMCELL Tech.) | Defined, feeder-free culture medium for hiPSC maintenance. | All cell sources post-reprogramming [27] [38] |
| iMatrix-511 / Laminin E8 Fragment | Recombinant laminin substrate for feeder-free cell adhesion. | Improving plating efficiency for neuronal conversion & iPSC culture [37] |
| Y-27632 (ROCK Inhibitor) | Small molecule that enhances survival of dissociated single cells. | Used during passaging and thawing of iPSCs [27] |
| CRISPR-Cas9 System | Gene editing for generating hypoimmunogenic universal iPSCs. | iPSCs (e.g., knockout of HLA genes) [38] |
| 2-(2-Methoxyethyl)phenol | 2-(2-Methoxyethyl)phenol, CAS:330976-39-1, MF:C9H12O2, MW:152.19 g/mol | Chemical Reagent |
| 2-Phenylpropanenitrile | 2-Phenylpropanenitrile|CAS 1823-91-2|RUO |
Emerging research focuses on partial reprogramming, which aims to reverse age-related epigenetic marks without fully erasing cell identity, and the use of small molecule cocktails to replace transcription factors, offering a more controlled and potentially safer approach for clinical applications [25] [39]. Furthermore, AI-assisted protein engineering is now creating novel, highly efficient variants of core reprogramming factors like SOX2 and KLF4, pushing the boundaries of reprogramming efficiency [5].
In conclusion, the choice of cell source and reprogramming method is not trivial and should be guided by the specific research or therapeutic goals. While Sendai virus offers high efficiency for fibroblasts and PBMCs, episomal vectors provide a viable non-viral alternative, particularly for LCLs. The ongoing development of factor combinations, delivery methods, and novel reagents continues to enhance the efficiency, safety, and applicability of cell reprogramming technologies.
Table 1: At-a-Glance Comparison of Reprogramming Modalities
| Approach | Key Components | Reported Efficiency/Success Rate | Primary Advantages | Key Safety/Toxicity Concerns |
|---|---|---|---|---|
| Genetic Factors (OSKM) | OCT4, SOX2, KLF4, c-MYC [25] | Typically <0.1% cell conversion [5] | Gold standard; proven pluripotency [25] | Tumorigenicity (especially c-MYC), insertional mutagenesis [27] [25] |
| Small Molecule Cocktail (7c) | CHIR99021, VPA, RepSox, Forskolin, TTNPB, DZNep, Tranylcypromine [13] [40] | Extends lifespan in C. elegans [41] | Non-genetic; can modulate concentration [40] | Higher toxicity noted in mice [13] |
| Small Molecule Cocktail (2c) | RepSox, Tranylcypromine [13] [41] | 42.1% lifespan extension in C. elegans [41] | Simplified formulation; reduces DNA damage & senescence [41] | In vivo toxicity concerns remain [13] |
| AI-Enhanced Proteins | RetroSOX, RetroKLF (AI-designed) [5] [42] | >50x pluripotency marker expression; >30% hit rate for improved variants [5] | Novel, highly active variants; deep sequence changes [5] | Early R&D stage; long-term safety not yet established [42] |
The discovery that somatic cell identity could be reprogrammed by the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC, or OSKM) represented a paradigm shift in regenerative biology [25]. However, the translational application of this genetic approach is limited by low efficiency, risks of insertional mutagenesis, and the oncogenic potential of factors like c-MYC [27] [22]. Consequently, the field has pursued non-integrating and non-genetic methods. Chemical reprogramming using small molecules presents a promising alternative, offering advantages such as non-immunogenicity, cost-efficiency, minimal genomic residual effects, and the ability for fine-tuned, reversible intervention [40]. This guide provides a comparative analysis of small molecule replacements for genetic reprogramming factors, detailing their experimental performance, protocols, and mechanisms for a research audience.
Comparative Performance Data of Reprogramming Modalities
Table 2: Quantitative Performance Metrics of Reprogramming Strategies
| Approach | Pluripotency Marker Expression | Epigenetic Clock Reversal | DNA Damage Reduction | In Vivo Lifespan/Healthspan Impact | Teratoma/Toxicity Risk |
|---|---|---|---|---|---|
| OSKM (Genetic) | Baseline | Yes [14] | Yes [14] | Extended in progeroid mice [14] | High (teratomas) [22] [25] |
| OSK (Genetic, safer) | Not specified | Yes [22] | Not specified | Improved vision in aged mice [22] | Lower (excludes c-MYC) [22] |
| 7c Cocktail | Indicates pluripotency [13] | Yes (multi-omic rejuvenation) [14] | Yes [41] | Lifespan extension in C. elegans [41] | Higher toxicity in mice [13] |
| 2c Cocktail | Indicates pluripotency [13] | Ameliorates epigenetic alterations [41] | Significant reduction [41] | Extends lifespan & healthspan in C. elegans [41] | Toxicity noted [13] |
| AI-Designed Factors | >50x vs. wild-type [5] | Confirmed genomic stability [5] | Enhanced repair, reduced γ-H2AX [5] | Not yet reported | No teratomas in derived iPSCs [5] |
Detailed Methodologies for Key Experiments
In Vitro Chemical Reprogramming of Human Cells (7c/2c Cocktails)
This protocol is adapted from research demonstrating the amelioration of cellular aging hallmarks and is a foundation for partial reprogramming studies [41].
1. Cell Culture:
2. Chemical Reprogramming Treatment:
- Cocktail Formulation:
- 7c Cocktail: Prepare a cocktail containing CHIR99021 (GSK3 inhibitor), DZNep (HMT inhibitor), Forskolin (cAMP activator), TTNPB (RAR agonist), Valproic acid (HDAC inhibitor), RepSox (Tranylcypromine) (TGF-β inhibitor/LSD1 inhibitor) [13] [40]. Concentrations are typically in the low micromolar range and must be optimized for specific cell types.
- 2c Cocktail: Prepare a simplified cocktail containing RepSox (TGF-β inhibitor) and Tranylcypromine (LSD1 inhibitor) [13] [41].
- Treatment Regimen: Replace the standard culture medium with medium containing the chemical cocktail. For partial reprogramming, treatment is typically short-term and cyclic (e.g., several days of treatment followed by a recovery period in normal medium) to avoid full dedifferentiation and maintain cell identity [14] [41].
- Cocktail Formulation:
3. Assessment of Rejuvenation Markers:
- Senescence-Associated β-galactosidase (SA-β-Gal) Staining: A core marker. A significant decrease in SA-β-Gal positive cells is expected post-treatment [41] [22].
- DNA Damage Assay: Immunofluorescence staining for γ-H2AX foci (a marker for DNA double-strand breaks). Effective treatment shows reduced γ-H2AX intensity [5] [41].
- Gene Expression Analysis: qRT-PCR to measure the expression of youth-associated genes (e.g., OCT4, NANOG) and senescence-associated genes (e.g., p16, p21, IL-6). Successful reprogramming upregulates the former and downregulates the latter [41] [22].
- Metabolic and Epigenetic Analysis: Assess mitochondrial function, reactive oxygen species (ROS) levels, and global changes in histone modifications (e.g., H3K9me3, H3K27me3) [14] [41].
In Vivo Partial Reprogramming in Mouse Models
This methodology, used to demonstrate organismal rejuvenation, often employs genetically engineered mice but is moving toward gene therapy delivery [14].
1. Animal Model:
- Transgenic Model: Use progeroid mice (e.g., LmnaG609G/G609G) or wild-type aged mice with a doxycycline (dox)-inducible polycistronic OSKM or OSK cassette [14].
- Gene Therapy Model: Intravenous injection of adeno-associated viruses (e.g., AAV9) encoding OSK factors and the rtTA protein for dox-controlled expression in wild-type aged mice [14].
2. Induction of Reprogramming:
- Cyclic Induction Protocol: Administer doxycycline cyclically to trigger factor expression. A common regimen is a 2-day pulse of dox in the drinking water, followed by a 5-day chase without dox. This cycle is repeated multiple times (e.g., over several months) to achieve partial, but not complete, reprogramming [14].
- Chemical Cocktail Delivery: For small molecules, in vivo delivery can be achieved via intraperitoneal injection or oral administration, though optimizing bioavailability and tissue penetration remains a challenge [41].
3. Outcome Assessment:
- Healthspan and Lifespan: Monitor survival and calculate a frailty index based on physiological parameters. Successful intervention extends median lifespan and reduces the frailty index [14] [41].
- Molecular and Cellular Analysis: Post-mortem, tissues are analyzed for transcriptomic, epigenomic, and metabolomic rejuvenation using RNA-seq, DNA methylation clocks, and mass spectrometry. Tissue regeneration capacity (e.g., in skin and pancreas) is also assessed [14].
- Safety Monitoring: Animals are meticulously monitored for teratoma formation, a key risk of reprogramming [14] [25].
Signaling Pathways in Chemical Reprogramming
The small molecules in reprogramming cocktails function by targeting specific signaling and epigenetic pathways to mimic the action of the Yamanaka factors. The following diagram illustrates the core mechanistic logic.
Diagram 1: Logical framework of small molecule (SM) actions in cell reprogramming. SMs target three core cellular processes: epigenetic remodeling to open chromatin, signaling pathway modulation to mimic key transcription factors, and metabolic switching to a glycolytic state. Converging actions on these pathways enable pluripotency induction or partial rejuvenation [40].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Chemical Reprogramming Research
| Reagent / Tool | Function / Mechanism | Example & Key Details |
|---|---|---|
| CHIR99021 | GSK-3β inhibitor; promotes metabolic switching to glycolysis and enhances self-renewal [40]. | Mandatory component in all major cocktails; critical for establishing a permissive metabolic state [40]. |
| RepSox | TGF-β receptor inhibitor; modulates signaling to replace Sox2 function [40]. | Found in 7 of 10 documented cocktails; high functional substitution for a core Yamanaka factor [40]. |
| Tranylcypromine (Parnate) | LSD1 inhibitor; epigenetic modifier that prevents demethylation of H3K4, promoting a open chromatin state [13] [40]. | Key component of the simplified 2c cocktail [13] [41]. |
| Valproic Acid (VPA) | Histone deacetylase (HDAC) inhibitor; promotes chromatin relaxation and gene accessibility [40]. | Common epigenetic modifier; used in 6 of 10 documented cocktails [40]. |
| Forskolin | Activator of adenylate cyclase, increasing cAMP levels; can replace Oct4 function [40]. | A signaling modifier used in multiple cocktail formulations [40]. |
| DZNep | Inhibitor of S-adenosylhomocysteine hydrolase, impacting histone methyltransferases (HMT) [40]. | An epigenetic modifier that targets EZH2, involved in 4 documented cocktails [40]. |
| Sendai Virus (SeV) Vectors | Non-integrating viral delivery method for genetic factors; episomal persistence [27]. | Gold-standard for non-integrating genetic reprogramming; higher success rates than episomal vectors [27]. |
| AAV9 Vectors | In vivo gene delivery capsid; broad tissue tropism for delivering OSK factors in animal models [14]. | Enables in vivo partial reprogramming studies in wild-type mice without transgenesis [14]. |
| 4-(N-Carboxymethyl-N-methylamino)-tempo | 4-(N-Carboxymethyl-N-methylamino)-TEMPO|CAS 139116-75-9 | |
| 3-(Methylphosphinico)propionic acid | 3-(Methylphosphinico)propionic acid, CAS:15090-23-0, MF:C4H9O4P, MW:152.09 g/mol | Chemical Reagent |
The discovery that somatic cells can be reprogrammed using transcription factors has revolutionized regenerative medicine, disease modeling, and drug discovery. The original Yamanaka factors—OCT4, SOX2, KLF4, and c-MYC (OSKM)—established the fundamental paradigm for cellular reprogramming [43]. However, researchers now recognize that different applications demand specialized factor combinations to optimize efficiency, safety, and functional outcomes. The selection of optimal transcription factor cocktails has emerged as a critical variable determining success across research and therapeutic domains.
This comparative analysis synthesizes current evidence and experimental data to establish application-driven guidelines for selecting reprogramming factor combinations. By evaluating performance metrics across disease modeling, drug discovery, and rejuvenation studies, we provide a structured framework for researchers to match specific experimental goals with the most efficacious factor combinations, thereby accelerating scientific progress and therapeutic development.
Fundamental Reprogramming Factor Combinations
Core Transcription Factors and Their Functions
The Yamanaka factors function as master regulators that reset epigenetic markers and redirect cellular fate. Each factor plays a distinct role in the reprogramming process:
- OCT4 (POU5F1): A POU-family transcription factor that maintains pluripotency and self-renewal in embryonic stem cells. It regulates expression of genes critical for early development and represses differentiation pathways [11].
- SOX2: Partners with OCT4 to co-regulate many pluripotency-associated genes. It maintains the epigenetic landscape necessary for self-renewal and prevents differentiation into trophectoderm [11].
- KLF4: Functions as both an activator and repressor of gene transcription. It promotes reprogramming by suppressing p53-mediated senescence pathways while activating pluripotency genes [4].
- c-MYC: A potent oncogene that enhances reprogramming efficiency by promoting global chromatin remodeling and increasing proliferation. However, its inclusion raises significant safety concerns for therapeutic applications [44].
Species-Specific Considerations in Factor Utilization
Research has revealed important species-specific differences in reprogramming factor function. While mouse cells can be reprogrammed efficiently with OSK alone (omitting c-MYC), human reprogramming demonstrates greater dependence on all four factors [4]. Additionally, the genomic binding patterns and target sites of these factors show significant divergence between species, with only a limited fraction of binding events conserved in syntenic regions between human and mouse fibroblasts during early reprogramming [4]. These differences necessitate careful consideration when translating findings between model systems.
Table 1: Core Yamanaka Factors and Their Functions
| Transcription Factor | Primary Function in Reprogramming | Key Molecular Interactions | Safety Considerations |
|---|---|---|---|
| OCT4 | Establishes and maintains pluripotent state | Binds with SOX2 to regulatory elements; activates NANOG | Ectopic expression promotes teratoma formation |
| SOX2 | Partners with OCT4; regulates chromatin accessibility | Forms heterodimers with OCT4; maintains pluripotency network | Dysregulation associated with neural tumors |
| KLF4 | Modulates p53 pathway; promotes cell cycle progression | Suppresses p53; activates TCF3 expression | Context-dependent oncogenic or tumor suppressor activity |
| c-MYC | Enhances proliferation; remodels chromatin | Binds broadly to transcriptional amplifiers; regulates metabolic genes | Potent oncogene; significantly increases cancer risk |
Application-Specific Factor Selection
Disease Modeling: Balancing Fidelity and Efficiency
For disease modeling, the primary objective involves creating accurate cellular representations of pathological states that faithfully recapitulate disease mechanisms. Research indicates that OCT4, SOX2, and KLF4 (OSK) frequently provide the optimal balance between reprogramming efficiency and maintenance of disease-relevant phenotypes, particularly when c-MYC might obscure pathological signatures [44].
Recent advances have enabled direct lineage reprogramming approaches that bypass the pluripotent state altogether. For neurological disease modeling, induced neural stem cells (iNSCs) can be generated using a modified Yamanaka factor approach followed by neuroinduction media, producing transcriptionally similar counterparts to native neural stem cells in a four-times faster protocol compared to iPSC-based methods [11]. This approach has demonstrated utility in modeling multiple sclerosis and spinal cord injury, with transplanted iNSCs reducing neuroinflammation by modulating proinflammatory metabolites in cerebrospinal fluid [11].
For corneal epithelial cells in limbal stem cell deficiency, researchers have successfully differentiated human iPSCs into functional corneal epithelial cell sheets using a specialized protocol, with clinical trials showing improved vision and reduced corneal cloudiness over two years [11]. Similarly, kidney organoids with PKD1 or PKD2 mutations have effectively modeled autosomal dominant polycystic kidney disease, displaying characteristic cyst formation for mechanistic studies and therapeutic screening [45].
Table 2: Optimal Cocktails for Disease Modeling Applications
| Disease Area | Recommended Factors | Differentiation Target | Key Performance Metrics | Notable Studies |
|---|---|---|---|---|
| Neurological Disorders | OSK-modified protocol | Induced neural stem cells (iNSCs) | 4x faster than iPSC route; reduces succinate-mediated neuroinflammation | Edenhofer et al. [11] |
| Ocular Diseases | OSKM (allogeneic) | Corneal epithelial cells | Improved vision over 24 months; reduced corneal cloudiness | Japanese clinical trial [11] |
| Kidney Diseases | OSKM | Kidney organoids | Recapitulates cyst formation in ADPKD; enables drug screening | Scarlat et al. [45] |
| Metabolic Disorders | OSKM | Pancreatic beta cells | Restored glycemic control in Type 1 diabetes (1 patient) | Chinese clinical study [11] |
Drug Discovery and Toxicity Testing
In pharmaceutical applications, the emphasis shifts toward standardization, predictability, and scalability. Complete OSKM cocktails remain the gold standard for generating consistent iPSC lines that can be differentiated into various cell types for high-throughput screening. The ability to create patient-specific iPSCs enables researchers to assess compound efficacy and toxicity across diverse genetic backgrounds [43].
The FDA Modernization Act 2.0, which permits cell-based assays as alternatives to animal testing for drug applications, has accelerated the development of iPSC-based testing platforms [43]. Companies like bit.bio offer optimized iPSC-derived cells with specific mutations for incorporation into standardized drug discovery workflows, improving translatability in early-stage development [43]. Similarly, BrainXell specializes in large-scale production of iPSC-derived neural cells for neuropharmacology applications, supplying 13 of the top 15 pharmaceutical firms [43].
For cardiotoxicity testing, iPSC-derived cardiomyocytes have become particularly valuable. Companies like Ncardia and REPROCELL (with its ReproCardio product) have commercialized human iPSC-derived cardiomyocytes that enable more accurate prediction of drug-induced cardiac effects compared to traditional animal models [43]. These systems help address the high attrition rates in drug development caused by species-specific differences in physiology and drug response [45].
Rejuvenation Studies: Partial Reprogramming Approaches
Partial reprogramming represents the most rapidly evolving area of factor optimization, with significant implications for treating age-related diseases. The dominant approach involves cyclic, transient expression of OSKM to reverse age-related epigenetic changes without completely altering cellular identity [24]. Research indicates that carefully calibrated exposure to these factors can restore youthful gene expression patterns, improve mitochondrial function, and enhance tissue regeneration capacity.
Groundbreaking recent research has demonstrated that AI-designed variants of SOX2 and KLF4 can dramatically enhance reprogramming efficiency. In collaboration with Retro Biosciences, OpenAI developed GPT-4b micro, a specialized AI model that generated novel SOX2 and KLF4 variants achieving a 50-fold increase in expression of stem cell reprogramming markers compared to wild-type controls [5]. These engineered variants also demonstrated enhanced DNA damage repair capabilities, indicating higher rejuvenation potential [5].
The optimal dosing strategy for rejuvenation appears to be short, cyclic induction of factors. In progeria mouse models, inducing Yamanaka factors for two days followed by five days of withdrawal in repeated cycles extended lifespan by 20-30% [44]. Similarly, a single one-week cycle of OSKM in aged mice (55 weeks old) elicited systemic rejuvenation across multiple organs, as evidenced by DNA methylation reprogramming in the pancreas, liver, spleen, and blood [24].
Small molecule alternatives to genetic reprogramming are also emerging. The 7c cocktail (CHIR99021, DZNep, Forskolin, TTNPB, Valproic acid, Repsox, and Tranylcypromine) and the simplified 2c cocktail (Repsox and Tranylcypromine) have shown promise in restoring multiple aging phenotypes, including genomic instability, epigenetic dysregulation, cellular senescence, and elevated ROS [13]. The 2c combination has demonstrated lifespan and healthspan extension in C. elegans models [13].
Table 3: Rejuvenation Approaches and Their Efficacy
| Reprogramming Approach | Factor Combination | Key Findings | Advantages | Limitations/Risks |
|---|---|---|---|---|
| Partial Reprogramming | Cyclic OSKM | 20-30% lifespan extension in progeria mice; reversed DNA methylation age | Avoids teratoma formation; maintains cellular identity | Precise timing critical; cancer risk with over-exposure [44] |
| AI-Optimized Factors | RetroSOX & RetroKLF | 50x higher expression of pluripotency markers; enhanced DNA damage repair | High hit rate (30-50%); works across cell types and delivery methods | Novel variants require extensive safety profiling [5] |
| Small Molecule Cocktails | 7c or 2c combinations | Rescued aging phenotypes in human cells; extended lifespan in C. elegans | Non-genetic approach; potentially easier to deliver | Toxicity concerns; in vivo efficacy limited [13] |
| Tissue-Specific Reprogramming | OSK (excluding c-MYC) | Rejuvenated Schwann cells enhanced peripheral nerve regeneration | Tissue-restricted effects; reduced oncogenic risk | Variable effects across tissue types [46] |
Experimental Protocols and Methodologies
In Vitro Partial Reprogramming Protocol
The following methodology details the cyclic reprogramming approach used to achieve rejuvenation without complete pluripotency induction:
Cell Preparation: Plate human fibroblasts (e.g., HDFs) at 60-70% confluence in standard culture medium 24 hours before transfection.
Factor Delivery:
- Viral Method: Transduce with lentiviral vectors carrying inducible OCT4, SOX2, KLF4, and c-MYC (OSKM) at MOI 5-20. Use polycistronic constructs for balanced expression.
- Non-Viral Method: Transfert with episomal plasmids containing OSKM using approved transfection reagents.
- Small Molecule Alternative: Treat with 2c cocktail (10µM RepSox + 5µM Tranylcypromine) for 48-96 hours [13].
Induction Cycling:
- Activate transgene expression with doxycycline (1-2µg/mL) for 48 hours.
- Withdraw doxycycline for 96-120 hours to allow phenotypic stabilization.
- Repeat for 4-8 cycles total, assessing rejuvenation markers after each cycle.
Assessment of Rejuvenation Markers:
- Epigenetic Clocks: Analyze DNA methylation patterns using Illumina EPIC arrays.
- Senescence Markers: Assess SA-β-galactosidase activity and p16INK4a expression.
- Functional Assays: Measure mitochondrial membrane potential (JC-1 staining), ROS levels (DCFDA), and DNA damage response (γ-H2AX foci) [5] [46].
AI-Guided Protein Engineering Workflow
The development of enhanced Yamanaka factors through artificial intelligence represents a cutting-edge methodology:
Model Training: Initialize GPT-4b micro from a scaled-down version of GPT-4o, then further train on a dataset composed of protein sequences, biological text, and tokenized 3D structure data [5].
Context Enrichment: Augment training data with additional contextual information about proteins in the form of textual descriptions, co-evolutionary homologous sequences, and groups of proteins with known interactions [5].
Sequence Generation: Prompt the model to propose diverse sets of "RetroSOX" and "RetroKLF" sequences with specific desired properties, leveraging the 64,000-token context window for enhanced controllability [5].
Wet Lab Validation:
- Screen AI-generated variants in human fibroblast cells using a validated platform.
- Assess early (SSEA-4) and late (TRA-1-60, NANOG) pluripotency markers via flow cytometry.
- Validate functional improvements through alkaline phosphatase staining at day 10 and DNA damage assays measuring γ-H2AX intensity [5].
Safety Considerations and Risk Mitigation
The tremendous potential of cellular reprogramming approaches is balanced by significant safety concerns that must be addressed through careful experimental design.
Tumorigenic Risk
The most significant safety consideration involves the potential for teratoma or tumor formation, particularly when using integrating viral vectors and oncogenic factors like c-MYC. Research demonstrates that incomplete reprogramming can cause pediatric-like cancers in mouse models, revealing that epigenetic changes alone can have the same effect as DNA damage in cancer development [44]. The difference between rejuvenation and carcinogenesis may depend on subtle variations in factor expression duration, with one study showing that inducing Yamanaka factors for just one week resulted in cancer development during the reprogramming process [44].
Risk mitigation strategies include:
- Precise temporal control using inducible systems with carefully calibrated expression windows
- Exclusion of c-MYC from factor combinations when possible
- Non-integrating delivery methods such as Sendai virus, episomal plasmids, or mRNA
- Small molecule alternatives that provide transient effects without genetic modification
- Comprehensive tumorigenicity screening in preclinical models, including long-term observation
Tissue-Specific Toxicity
Different cell types exhibit varying susceptibility to reprogramming factors, with potential for cell identity loss, tissue dysfunction, and organ failure. What benefits one cell type may harm others—for example, optimal reprogramming conditions for lung cells may cause liver toxicity [24].
Risk mitigation strategies include:
- Tissue-specific promoters to restrict factor expression to target cells
- Dose-response optimization for each cell type and application
- Comprehensive histopathological analysis across all major organ systems
- Functional assessment of tissue-specific performance post-reprogramming
Essential Research Reagent Solutions
The following toolkit represents critical reagents for implementing the reprogramming methodologies discussed in this review:
Table 4: Essential Research Reagents for Reprogramming Studies
| Reagent Category | Specific Products | Primary Function | Application Notes |
|---|---|---|---|
| Reprogramming Factors | Lentiviral OSKM, Sendai OSKM, Episomal plasmids | Deliver transcription factors to somatic cells | Sendai virus non-integrating; episomal for clinical applications |
| Small Molecule Cocktails | 7c cocktail, 2c (RepSox + Tranylcypromine) | Chemical induction of pluripotency | Lower efficiency but reduced tumorigenic risk |
| Cell Culture Media | mTeSR, Essential 8, ReproTeSR | Maintain pluripotent stem cells | Defined, xeno-free formulations recommended |
| Differentiation Kits | STEMdiff Cardiomyocyte, Neural Induction Medium | Direct iPSCs to specific lineages | Standardized protocols improve reproducibility |
| Characterization Antibodies | OCT4, SOX2, NANOG, SSEA-4, TRA-1-60 | Assess pluripotency markers | Essential for quality control during reprogramming |
| Epigenetic Analysis Kits | DNA methylation arrays, Chromatin immunoprecipitation | Evaluate epigenetic remodeling | Critical for assessing rejuvenation outcomes |
The optimization of Yamanaka factor combinations for specific applications represents a maturation of reprogramming technology from a blanket approach to a precision tool. The experimental evidence compiled in this review demonstrates that application-driven selection of factor cocktails significantly enhances outcomes across disease modeling, drug discovery, and rejuvenation studies.
Future progress will likely focus on several key areas: First, spatiotemporal control of factor expression through improved delivery systems and regulatory circuits will help maximize benefits while minimizing risks. Second, tissue-specific optimization will address the current challenge of variable responses across different cell types. Third, small molecule approaches may eventually provide the safety profile necessary for widespread therapeutic application, though current efficacy limitations must be overcome.
The emergence of AI-guided protein engineering has demonstrated remarkable potential to revolutionize the field, with GPT-4b micro generating enhanced factor variants achieving unprecedented 50x improvements in reprogramming efficiency [5]. This approach, combined with increasingly sophisticated safety measures, suggests that optimized reprogramming cocktails will continue to drive advances across biomedical research and clinical applications.
As Professor Yamada Yasuhiro of the University of Tokyo cautions, the field must balance enthusiasm with careful consideration of mechanisms and safety: "We shouldn't jump to the conclusion that we can simply prevent aging through partial reprogramming of cells. In fact, the research hasn't even reached the starting line yet" [44]. Nonetheless, the methodological frameworks and comparative data presented here provide researchers with evidence-based guidance for selecting optimal factor combinations specific to their experimental goals and applications.
Enhancing Efficiency and Safety: Strategies for Overcoming Reprogramming Hurdles
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using the Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) represents a landmark achievement in regenerative medicine. However, the clinical application of this technology faces significant challenges, primarily due to the tumorigenic risk associated with the c-MYC oncogene. c-MYC is a transcription factor constitutively and aberrantly expressed in over 70% of human cancers and functions as a powerful driver of tumorigenesis through multiple mechanisms including promoting cell cycle progression, altering metabolism, blocking DNA repair, and increasing cell stemness [47] [48]. While c-MYC enhances reprogramming efficiency, its integration into the host genome can lead to teratoma formation and other malignancies, creating a substantial barrier to therapeutic applications [47] [22].
This guide provides a comprehensive comparison of emerging strategies designed to mitigate tumorigenic risk by either replacing, omitting, or controlling c-MYC activity in reprogramming protocols. We objectively evaluate the efficiency and safety profiles of these approaches, presenting quantitative experimental data to inform researchers and drug development professionals in selecting optimal factor combinations for specific applications. The development of effective c-MYC-independent reprogramming strategies represents a critical advancement toward making iPSC-based therapies clinically viable, balancing the dual priorities of efficiency and safety.
Comparative Analysis of Reprogramming Strategies
Quantitative Comparison of Yamanaka Factor Combinations
Table 1: Efficiency and Safety Profiles of Different Reprogramming Factor Combinations
| Factor Combination | Reprogramming Efficiency | Tumorigenic Risk | Key Advantages | Documented Limitations |
|---|---|---|---|---|
| OSKM (Standard) | High (Baseline) | Very High | Well-characterized, reliable | High teratoma incidence, genomic instability |
| OSK (c-MYC omitted) | 0.1% or less (Significant reduction) | Substantially reduced | Eliminates oncogenic transgene | Greatly reduced efficiency, incomplete reprogramming |
| OSK + TERT | Significantly enhanced vs. OSK alone | Low | Restores aging markers, maintains safety profile | Requires additional genetic manipulation |
| AI-Enhanced Factors | >50x increase in pluripotency markers vs. wild-type | Potentially reduced (under evaluation) | Dramatically improved efficiency, novel variants | Limited long-term safety data |
The conventional OSKM approach demonstrates high reprogramming efficiency but carries unacceptable tumorigenic risk for clinical applications, as continuous c-MYC expression induces teratomas and significant mortality in model systems [22]. Simply omitting c-MYC to create OSK combinations substantially reduces tumorigenic risk but at the cost of drastically diminished reprogramming efficiency—typically falling to 0.001% or less with nonintegrating methods [47]. This efficiency reduction presents significant practical challenges for research and clinical applications.
Promising alternatives have emerged that balance efficiency and safety more effectively. The combination of OSK with TERT (telomerase reverse transcriptase) demonstrates significantly enhanced reprogramming efficiency compared to OSK alone while maintaining a favorable safety profile [22]. In human fibroblast models, OSK + TERT treatment markedly increased expression of youth-related genes (OCT4, SOX2, KLF4, NANOG) while reducing senescence-associated genes (p16, p21, ZSCAN4, ATF3, MMP13) and inflammatory markers (IL-6) [22]. Most notably, AI-guided protein engineering has yielded redesigned factors that achieve remarkable efficiency improvements—greater than 50-fold increases in stem cell reprogramming markers compared to wild-type controls while potentially mitigating tumorigenic risk through sequence optimization [5].
Molecular and Functional Outcomes of Alternative Approaches
Table 2: Experimental Outcomes of c-MYC-Independent Reprogramming Strategies
| Experimental Measure | OSKM (Control) | OSK Only | OSK + TERT | AI-Enhanced Factors |
|---|---|---|---|---|
| Pluripotency Marker Expression | Baseline | Significantly reduced | Enhanced vs. OSK | >50x increase vs. wild-type |
| Cell Viability | High | Moderate | Significantly improved | High |
| Senescence Reduction | Significant | Limited | Markedly improved (β-galactosidase reduction) | Enhanced DNA damage repair |
| Genomic Stability | Poor due to insertional mutagenesis | Good | Good | Healthy karyotypes maintained |
| In Vivo Tumor Formation | High incidence | Rare | Not reported | Significantly reduced |
The functional benefits of c-MYC-independent strategies extend beyond basic reprogramming metrics. In cellular senescence models, the OSK + TERT combination significantly improved cell viability, reduced β-galactosidase expression (a senescence marker), and promoted favorable cell cycle progression by decreasing the percentage of cells in G0/G1 phase [22]. Perhaps more importantly, cells reprogrammed with optimized factors demonstrate enhanced functionality, including improved DNA damage repair capacity—a key indicator of rejuvenation potential [5]. In DNA damage assays, cells treated with AI-enhanced factor cocktails showed visibly reduced γ-H2AX intensity (a marker of double-strand breaks) compared to those reprogrammed with standard factors, indicating more effective DNA repair [5].
Furthermore, iPSCs generated using these optimized approaches maintain genomic stability and full differentiation potential. AI-engineered factor variants have demonstrated robust performance across multiple donors, cell types, and delivery methods, with confirmation of full pluripotency and genomic stability in derived iPSC lines [5]. These functional advantages position c-MYC-independent strategies as viable alternatives for clinical-grade iPSC generation.
Experimental Protocols and Methodologies
OSK + TERT Reprogramming Protocol
The combination of OSK factors with TERT represents a well-characterized approach to enhance reprogramming efficiency while minimizing oncogenic risk. The following protocol has been validated in replicative senescence models using human MRC-5 fibroblasts:
Cell Culture and Transfection:
- Culture MRC-5 fibroblasts until they reach replicative senescence (approximately 50 population doublings)
- Transfect cells with OSK-expressing plasmid (pcDNA3.3 vector) with or without TERT-expressing plasmid at the onset of senescence
- Culture transfected cells for extended passages (60-70 generations) with regular medium changes
Efficiency Assessment:
- Measure mRNA expression levels of youth-related genes (OCT4, SOX2, KLF4, NANOG, TERT) using quantitative reverse transcription PCR
- Quantify senescence-associated genes (p16, p21, ZSCAN4, ATF3, MMP13) and inflammatory markers (IL-6)
- Perform western blotting to confirm protein level expression of key pluripotency factors
- Assess cell viability using Cell Counting Kit-8 (CCK-8) assays
- Evaluate cell cycle progression via propidium iodide staining and flow cytometry
- Detect senescence-associated β-galactosidase expression using specific staining protocols [22]
This approach demonstrates that OSK + TERT co-expression significantly enhances the expression of youth-related genes while reducing aging markers, effectively delaying cellular senescence by modulating aging-related gene expression and maintaining cell viability [22].
AI-Guided Factor Engineering Workflow
The development of AI-enhanced reprogramming factors represents a cutting-edge approach to improving the efficiency and safety of cellular reprogramming:
Model Training and Optimization:
- Initialize a specialized model (GPT-4b micro) from a scaled-down version of GPT-4o to leverage existing biological knowledge
- Further train the model on a dataset composed primarily of protein sequences, supplemented with biological text and tokenized 3D structure data
- Enrich training data with additional contextual information including textual descriptions, co-evolutionary homologous sequences, and protein interaction groups
- Utilize an extended context window (up to 64,000 tokens) during inference to enhance controllability and output quality
Experimental Validation:
- Establish a wet lab screening platform using human fibroblast cells
- Validate the platform with baseline OSKM and manually designed SOX2 variants in pilot screens
- Generate diverse "RetroSOX" and "RetroKLF" sequences through model prompting
- Screen AI-generated variants for pluripotency marker expression (SSEA-4, TRA-1-60, NANOG)
- Validate top-performing variants across multiple independent experiments, cell types (including mesenchymal stromal cells from middle-aged human donors), and delivery methods (viral vectors and mRNA)
- Confirm genomic stability through karyotype analysis and differentiation potential into all three germ layers [5]
This methodology has yielded remarkably high hit rates, with over 30% of AI-suggested SOX2 sequences outperforming wild-type SOX2 at expressing key pluripotency markers, and nearly 50% of KLF4 variants showing superior performance [5].
Signaling Pathways and Molecular Mechanisms
c-MYC Oncogenic Signaling Pathways
Figure 1: c-MYC Oncogenic Signaling Network
c-MYC drives tumorigenesis through multifaceted mechanisms encompassing tumor cell intrinsic effects, tumor microenvironment modulation, and metabolic reprogramming. The oncogene promotes cell cycle progression by upregulating cyclins and CDKs while suppressing CDK inhibitors, and enhances ribosomal biogenesis to support rapid proliferation [48]. Through metabolic reprogramming, c-MYC increases glycolysis, glutaminolysis, and lipogenesis while reducing oxidative phosphorylation, creating energy and building blocks for tumor growth [48] [49]. Additionally, c-MYC modulates the tumor microenvironment to promote immune evasion by causing T-cell efflux, increasing macrophage and myeloid-derived suppressor cell influx, suppressing MHC-I production, and enhancing PD-L1 expression [48].
Safer Reprogramming Signaling Pathways
Figure 2: Safer Reprogramming Strategies
c-MYC-independent reprogramming strategies activate alternative signaling pathways that maintain efficiency while reducing oncogenic risk. The OSK + TERT approach enhances youth-related gene expression while simultaneously suppressing senescence-associated genes, with TERT providing critical telomere maintenance functions that c-MYC typically supports [22]. AI-enhanced factors function through optimized protein sequences that improve DNA binding efficiency, protein stability, or interaction with co-factors, leading to more robust pluripotency establishment without activating oncogenic pathways [5]. These approaches collectively enable efficient reprogramming while maintaining genomic stability, preserving differentiation potential, and reducing tumorigenic risk—addressing the fundamental limitations of conventional OSKM approaches.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for c-MYC-Independent Reprogramming
| Reagent Category | Specific Products/Systems | Research Application | Safety Considerations |
|---|---|---|---|
| Non-Integrating Vectors | Sendai virus, Adenovirus, Episomal plasmids | Factor delivery without genomic integration | Reduced insertional mutagenesis risk |
| Gene Editing Systems | Cas9/Cas12a RNP complexes (IDT Alt-R systems) | Targeted transgene integration | GMP-compatible, virus-free workflow |
| Senescence Detection | β-galactosidase staining kits, CCK-8 assays | Cellular aging assessment | Quality control for reprogrammed cells |
| Pluripotency Validation | Antibodies for SSEA-4, TRA-1-60, NANOG | Stem cell characterization | Confirmation of complete reprogramming |
| Metabolic Assays | Seahorse Analyzer reagents | Metabolic profiling | Detection of aberrant metabolic reprogramming |
| GMP-Compliant Media | Commercial stem cell culture systems | Clinical-grade cell culture | Xeno-free, defined components |
| 1-(Benzyloxy)-3-(chloromethyl)benzene | 1-(Benzyloxy)-3-(chloromethyl)benzene, CAS:24033-03-2, MF:C14H13ClO, MW:232.7 g/mol | Chemical Reagent | Bench Chemicals |
| 1-(1,3-Benzodioxol-5-yl)pentan-1-ol | 1-(1,3-Benzodioxol-5-yl)pentan-1-ol|CAS 5422-01-5 | Bench Chemicals |
The implementation of c-MYC-independent reprogramming requires specialized reagents and systems that prioritize both efficiency and safety. Non-integrating vector systems, including Sendai virus, adenovirus, and episomal plasmids, enable factor delivery without genomic integration, substantially reducing the risk of insertional mutagenesis [47]. Advanced gene editing systems based on Cas9 or Cas12a ribonucleoprotein (RNP) complexes facilitate precise transgene integration while maintaining GMP compatibility through virus-free workflows [8]. Comprehensive validation reagents, including senescence detection kits and pluripotency marker antibodies, provide essential quality control measures to ensure complete reprogramming without aberrant cell states. Additionally, metabolic assay platforms enable researchers to monitor metabolic reprogramming—a known hallmark of both pluripotency and tumorigenesis—to identify potentially problematic cell populations early in the differentiation process.
The field of cellular reprogramming has made significant strides in addressing the fundamental challenge of tumorigenic risk associated with c-MYC. The strategies compared in this guide—from factor omission to AI-enhanced engineering—demonstrate that efficient reprogramming can be achieved while substantially improving safety profiles. Quantitative evidence indicates that optimized approaches like OSK + TERT combinations and AI-designed factors can overcome the efficiency limitations of simple c-MYC omission while maintaining favorable safety characteristics.
Future research directions should focus on further refining these approaches through improved factor engineering, optimized delivery methods, and enhanced safety switches. The integration of suicide genes or other safety mechanisms alongside reprogramming factors represents a promising avenue for clinical translation [8]. Additionally, continued advancement in AI-guided protein design will likely yield further improvements in both efficiency and specificity. As these technologies mature, the balance between efficiency and safety will continue to improve, ultimately enabling the widespread clinical application of iPSC-based therapies for regenerative medicine, disease modeling, and drug discovery.
The comparative data presented in this guide provides researchers and drug development professionals with evidence-based insights for selecting appropriate reprogramming strategies based on specific application requirements. While the optimal approach may vary depending on the specific research or clinical context, it is evident that c-MYC-independent reprogramming has evolved from a concept to a practical reality with tremendous potential for advancing regenerative medicine.
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using the Yamanaka factors (OCT4, SOX2, KLF4, and c-Myc) revolutionized regenerative medicine and disease modeling. However, a significant challenge remains the low efficiency of this reprogramming process. In the pursuit of optimizing iPSC generation, small molecules and epigenetic modulators have emerged as powerful tools to enhance reprogramming efficiency and safety. This guide provides a comparative analysis of key small molecules—Valproic Acid (VPA), RepSox, and 8-Bromoadenosine 3′,5′-cyclic monophosphate (8-Br-cAMP)—detailing their mechanisms, experimental data, and protocols to inform research and drug development.
Comparative Efficiency Data of Small Molecules
The table below summarizes the quantitative impact of these small molecules on reprogramming efficiency, based on experimental findings.
| Small Molecule | Primary Mechanism of Action | Impact on Reprogramming Efficiency | Key Signaling Pathways Affected |
|---|---|---|---|
| Valproic Acid (VPA) | Histone Deacetylase (HDAC) Inhibitor [6] | Synergistic effect: 6.5-fold increase (with 8-Br-cAMP) [6] [50] | p53 signaling, Cytokine & inflammatory pathways [50] |
| RepSox | TGF-β Receptor Inhibitor; replaces Sox2 [6] [51] | Enables reprogramming without exogenous Sox2 [6] | TGF-β/SMAD signaling, Mesenchymal-to-Epithelial Transition (MET) [51] [52] |
| 8-Br-cAMP | cAMP Pathway Activator [53] | 2-fold increase (alone); 6.5-fold increase (with VPA) [6] [50] | p53 signaling, Self-renewal genes (CCND2) [50] |
Detailed Experimental Protocols and Workflows
Protocol for Enhancing Reprogramming with 8-Br-cAMP and VPA
This protocol is adapted from a study demonstrating the synergistic effect of 8-Br-cAMP and VPA on reprogramming human fibroblast cells [50].
- Cell Line: Human neonatal foreskin fibroblast (HFF1) cells.
- Reprogramming Factors: Cells are transduced with the classic Yamanaka factors (OCT4, SOX2, KLF4, c-Myc) using a retroviral or lentiviral system.
- Small Molecule Treatment:
- 8-Br-cAMP: The cAMP analog is added to the culture medium at a concentration of 250 µM.
- VPA: The HDAC inhibitor is used at a concentration of 0.5 mM.
- Combination: For synergistic effect, both 8-Br-cAMP (250 µM) and VPA (0.5 mM) are added to the medium.
- Treatment Duration: The small molecules are introduced at the initiation of the reprogramming process and maintained in the culture medium for the first 7-10 days.
- Key Findings: The study found that this combined treatment led to a transient decrease in p53 protein levels during the early stages of reprogramming, alongside upregulation of self-renewal genes like CCND2. This resulted in a 6.5-fold synergistic increase in iPSC colony formation compared to the control with factors alone [50].
Protocol for RepSox in Factor Replacement
This methodology outlines the use of RepSox to replace the transcription factor Sox2 in the reprogramming cocktail [6] [51].
- Cell Line: Typically, mouse or human fibroblasts.
- Reprogramming Factors: Cells are transduced with a modified combination of factors, often OCT4, KLF4, and c-Myc (OKM), omitting SOX2.
- Small Molecule Treatment: RepSox is added to the culture medium at a concentration of 1-2 µM, starting from the initiation of reprogramming and continuing for 7-14 days [54].
- Mechanism: RepSox inhibits the TGF-β receptor, which suppresses the expression of genes specific to the somatic cell identity (e.g., fibroblast markers) and promotes the Mesenchymal-to-Epithelial Transition (MET), a critical early step in reprogramming [51] [52]. By modulating this pathway, it functionally replaces the need for exogenous SOX2 expression.
Signaling Pathways in Small Molecule-Augmented Reprogramming
The following diagram illustrates the core mechanisms through which VPA, 8-Br-cAMP, and RepSox enhance the reprogramming process.
The Scientist's Toolkit: Essential Research Reagents
The table below lists key reagents used in experiments with these small molecules, along with their primary functions in reprogramming.
| Reagent / Tool | Primary Function in Reprogramming |
|---|---|
| Valproic Acid (VPA) | Epigenetic modulator that inhibits histone deacetylases (HDACs), relaxing chromatin and facilitating gene expression [6] [51]. |
| 8-Br-cAMP | Cell-permeable cAMP pathway activator that enhances reprogramming, partly through transient p53 downregulation [50] [53]. |
| RepSox | TGF-β receptor inhibitor that promotes MET and can replace the transcription factor Sox2 [6] [51]. |
| CHIR99021 | GSK-3β inhibitor that activates Wnt signaling, a key pathway for sustaining pluripotency [54]. |
| BIX-01294 | G9a histone methyltransferase inhibitor; opens chromatin to improve factor access [51]. |
| Lentiviral / Retroviral Vectors | Common but integrating delivery systems for the OSKM transcription factors [6]. |
| Sendai Virus / Episomal Vectors | Non-integrating delivery systems for transient factor expression, enhancing clinical safety [6] [1]. |
| 2,3-Bis(hexadecyloxy)propan-1-ol | 2,3-Bis(hexadecyloxy)propan-1-ol, CAS:13071-60-8, MF:C35H72O3, MW:540.9 g/mol |
The integration of small molecules like VPA, RepSox, and 8-Br-cAMP into reprogramming protocols represents a significant advancement in iPSC technology. These compounds directly target the major bottlenecks of reprogramming—epigenetic barriers, specific signaling pathways, and cell identity transitions. Quantitative data confirms they can dramatically boost efficiency, sometimes synergistically, and even reduce reliance on exogenous transcription factors. As research progresses, the use of such chemical approaches is poised to improve the safety, efficiency, and clinical applicability of iPSC generation for disease modeling, drug discovery, and regenerative medicine.
The discovery of induced pluripotent stem cells (iPSCs) revolutionized regenerative medicine by demonstrating that somatic cells could be reprogrammed to a pluripotent state using defined transcription factors [1]. However, since the initial breakthrough, the field has consistently grappled with the challenge of low reprogramming efficiency, which often falls below 1% even under optimized conditions [6] [1]. This significant barrier has prompted extensive research into understanding the molecular roadblocks that inhibit reprogramming and developing strategies to overcome them.
Central to this efficiency challenge is the tumor suppressor protein p53, which acts as a potent barrier to reprogramming by initiating apoptosis or senescence in response to the cellular stress induced by reprogramming factors [55] [56]. Simultaneously, specific microRNA families, particularly miR-302/367 and miR-372, have emerged as powerful enhancers of reprogramming efficiency, in part through their ability to modulate the p53 pathway [55] [56]. This review systematically compares the performance of p53 suppression and microRNA supplementation strategies, providing experimental data and methodologies to guide researchers in optimizing reprogramming protocols for basic research and therapeutic applications.
Molecular Mechanisms of Reprogramming Barriers and Enhancers
p53: A Master Barrier to Reprogramming
The p53 pathway serves as a critical safeguard mechanism that protects cells from reprogramming-induced stress. During the early phases of iPSC generation, the expression of Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) triggers DNA damage responses and oncogenic stress that activate p53 [55]. Once activated, p53 transcriptionally upregulates numerous downstream targets that collectively inhibit reprogramming through multiple mechanisms: inducing apoptosis via Bax and Bak activation; promoting cell cycle arrest through p21 activation; and reinforcing senescence programs [56]. The fundamental role of p53 as a reprogramming barrier is evidenced by studies demonstrating that its suppression through genetic knockout, RNA interference, or protein inhibition can increase reprogramming efficiency by 3- to 10-fold [55] [56].
microRNAs as Potent Reprogramming Enhancers
MicroRNAs (miRNAs) have emerged as master regulators of reprogramming efficiency, with the miR-302/367 cluster and miR-372 representing particularly potent enhancers. The miR-302/367 cluster is highly expressed in embryonic stem cells and plays multifaceted roles in promoting pluripotency [55] [56]. Similarly, miR-372 facilitates reprogramming through parallel mechanisms [55]. These miRNAs function through several interconnected mechanisms: directly targeting and suppressing p53 expression; promoting cell cycle progression by overcoming G1-S phase barriers; facilitating epigenetic remodeling through targeting histone modifiers; and inhibiting epithelial-mesenchymal transition (EMT) by targeting ZEB1 and ZEB2 [55] [56].
The following diagram illustrates the molecular interplay between p53, microRNAs, and the reprogramming process:
Comparative Performance Analysis of Efficiency Enhancement Strategies
Quantitative Comparison of Reprogramming Enhancers
The table below summarizes key experimental findings comparing the efficiency improvements achieved through p53 suppression and microRNA supplementation:
Table 1: Efficiency Comparison of p53 Suppression vs. microRNA Strategies
| Strategy | Experimental System | Efficiency Enhancement | Key Molecular Targets | Reference |
|---|---|---|---|---|
| p53 suppression | Dgcr8-/- ESCs | Enables neural differentiation of previously incompetent cells | Direct p53 inhibition | [56] |
| p53 suppression | Mouse & human fibroblasts | 3-10 fold increase in iPSC generation | p53 and its downstream effectors | [55] |
| p53 suppression | Vitamin C treatment | Significant improvement in iPSC induction | p53 and p21 reduction | [6] |
| miR-302/367 cluster | Human & mouse somatic cells | Enables TF-free reprogramming | p53, TGFBR2, LATS2, cell cycle regulators | [55] |
| miR-372 | Fibroblast reprogramming | Enhances iPSC generation | Unknown targets in p53 pathway | [55] |
| miR-302 | Dgcr8-/- ESCs | Enables neural differentiation | Direct p53 suppression | [56] |
| Combined approaches | Various somatic cells | Highest efficiency reported | Multiple parallel pathways | [55] [56] |
Direct Performance Comparison in Experimental Models
When comparing the absolute and relative performance of these strategies across standardized experimental systems, distinct patterns emerge:
Table 2: Direct Performance Comparison in Reprogramming Models
| Strategy | Baseline Efficiency | Enhanced Efficiency | Fold Improvement | Differentiation Capacity | Safety Profile |
|---|---|---|---|---|---|
| Standard OSKM | 0.1-1% | Reference | 1x | Normal | Standard risk |
| p53 suppression | 0.1-1% | 3-10% | 3-10x | Preserved | Elevated oncogenic risk |
| miR-302/367 | 0.1-1% | 1-8% | 1-8x | Enhanced | Moderate risk |
| miR-372 | 0.1-1% | 2-5% | 2-5x | Preserved | Moderate risk |
| Combined approach | 0.1-1% | Up to 15% | Up to 15x | Variable | Highest risk |
Experimental Protocols for Efficiency Enhancement
p53 Suppression Methods
Genetic Ablation of p53:
- Utilize Trp53 knockout mice as cell donors or employ CRISPR/Cas9-mediated TP53 knockout in human somatic cells
- Transfer somatic cells with Yamanaka factors using lentiviral or Sendai virus systems
- Culture in standard iPSC media with serum replacement and bFGF
- Monitor emerging iPSC colonies 14-21 days post-transduction [56]
RNA Intermediation-Based p53 Suppression:
- Design shRNAs targeting human TP53 or mouse Trp53
- Clone into lentiviral vectors with U6 or H1 promoters
- Co-transduce with OSKM factors into somatic cells
- Assess p53 knockdown efficiency by Western blotting 72 hours post-transduction
- Evaluate reprogramming efficiency by pluripotency marker expression at day 21 [56]
Pharmacological Inhibition:
- Treat OSKM-transduced cells with p53 inhibitors such as Pifithrin-α
- Optimize concentration (typically 5-20μM) and timing (days 3-10 post-transduction)
- Replace inhibitors every 48 hours during the critical reprogramming window
- Monitor for reduced apoptosis and enhanced colony formation [55]
microRNA-Enhanced Reprogramming Protocols
miR-302/367 Cluster Expression:
- Clone miR-302/367 cluster into lentiviral or episomal vectors
- Transduce somatic cells with miRNA vectors alone or in combination with OSKM factors
- For TF-free reprogramming: use miRNA vectors alone with valproic acid (VPA) treatment
- Culture in hypoxic conditions (5% Oâ‚‚) to enhance efficiency
- Monitor for epithelial-like morphological changes days 7-14 [55]
miR-372 Supplementation:
- Express miR-372 using lentiviral delivery systems
- Co-transduce with OSKM factors at optimal ratios
- Alternatively, use synthetic miRNA mimics transfected days 3 and 7 post-OSKM transduction
- Culture in standard iPSC conditions with enhanced cell survival factors
- Assess alkaline phosphatase activity from day 14 onward [55]
Combined miRNA Approaches:
- Implement miR-302/367 with miR-372 co-expression
- Optimize vector ratios to maximize synergy while minimizing cellular stress
- Consider inducible systems for temporal control of miRNA expression
- Include small molecule enhancers (e.g., VPA, 8-Br-cAMP) for additive effects [55]
The following workflow diagram illustrates a typical experimental setup for comparing these efficiency enhancement strategies:
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Reprogramming Efficiency Research
| Reagent Category | Specific Examples | Function in Reprogramming | Considerations |
|---|---|---|---|
| p53 Modulators | shTP53 vectors, Pifithrin-α, Nutlin-3a (control) | Overcome primary reprogramming barrier | Monitor genomic instability; optimize timing |
| microRNA Expression Systems | miR-302/367 lentivectors, miR-372 mimics, inducible systems | Enhance efficiency through multiple pathways | Seed sequence fidelity critical for function |
| Reprogramming Factors | OSKM lentivirus, Sendai virus (non-integrating), mRNA kits | Core reprogramming machinery | Consider integration-free methods for therapy |
| Small Molecule Enhancers | Valproic acid, 8-Br-cAMP, sodium butyrate | Epigenetic modulation, signaling activation | Can reduce miRNA requirements |
| Cell Culture Supplements | bFGF, L-ascorbic acid, TGF-β | Support pluripotency and cell survival | Serum-free formulations improve consistency |
| Characterization Tools | Anti-TRA-1-60, anti-OCT4, alkaline phosphatase kits | iPSC colony identification and validation | Multiple markers recommended for confirmation |
The comparative analysis of p53 suppression and microRNA enhancement strategies reveals a complex efficiency landscape with complementary strengths and limitations. p53 suppression approaches provide more dramatic efficiency improvements but carry significant oncogenic risks that may limit their therapeutic application [55] [56]. In contrast, miR-302/367 and miR-372 supplementation offers more moderate efficiency enhancements but through more nuanced mechanisms that may preserve genomic integrity [55] [56].
Future research directions should focus on optimizing temporal control of p53 suppression to minimize oncogenic risk while preserving efficiency benefits, developing novel delivery systems for miRNA mimics that enable transient, dose-controlled expression, exploring synergistic combinations of moderate p53 modulation with miRNA enhancement, and establishing safety-profiling protocols specific to efficiency-enhanced iPSCs, particularly regarding tumorigenicity and genomic stability [6] [55] [56]. As reprogramming methodologies evolve toward clinical applications, the strategic integration of these efficiency enhancement approaches will be crucial for generating clinically usable iPSCs with optimal safety profiles.
Partial cellular reprogramming represents a groundbreaking strategy in regenerative medicine and aging research, aiming to reverse age-related cellular decline without altering a cell's fundamental identity. This approach harnesses the power of the Yamanaka factors—OCT4, SOX2, KLF4, and c-MYC (collectively known as OSKM)—but applies them transiently to avoid complete dedifferentiation into pluripotent stem cells [57] [58]. The fundamental premise is that brief exposure to these reprogramming factors can restore youthful epigenetic patterns, mitochondrial function, and gene expression while allowing cells to maintain their specialized functions [24] [57].
The central challenge lies in the delicate balance between achieving sufficient rejuvenation and preserving cellular identity. As one review notes, "What is good for lung cells is bad for liver cells. What is good for one type of cell in the liver is bad for its neighbor," highlighting the tissue-specific complexities of this approach [24]. The "bad" outcomes in this context can include cell death, tissue dysfunction, and cancer, underscoring the critical importance of precise methodological control [24]. This comparison guide examines the efficiency and safety profiles of different reprogramming factor combinations and protocols, providing researchers with evidence-based insights for experimental design.
Performance Comparison of Reprogramming Approaches
Efficiency and Safety Profiles of Factor Combinations
Table 1: Comparative Analysis of Reprogramming Factor Combinations
| Factor Combination | Reprogramming Efficiency | Key Advantages | Major Risks/Limitations | Primary Applications |
|---|---|---|---|---|
| OSKM (Full Set) | High efficiency in epigenetic rejuvenation | Most comprehensive rejuvenation; resets multiple aging hallmarks | High tumorigenic potential; significant identity loss | Basic research; in vitro studies with strict controls |
| OSK (Without c-MYC) | Moderate efficiency | Reduced cancer risk; better safety profile | Slower epigenetic remodeling; potentially less potent | In vivo applications; therapeutic development |
| Sequential OSKM | ~300% improvement over simultaneous delivery [59] | More natural transition through reprogramming stages | Complex protocol requiring precise timing | High-quality iPSC generation; mechanistic studies |
| Chemical Reprogramming | Variable (depends on cocktail) | Non-integrating; potentially safer delivery | Less established protocols; unknown long-term effects | Future clinical applications; sensitive cell types |
Quantitative Outcomes in Model Systems
Table 2: Experimental Outcomes Across Different Reprogramming Protocols
| Reprogramming Method | Lifespan Extension | Epigenetic Age Reduction | Tumor Formation Incidence | Functional Improvement |
|---|---|---|---|---|
| Cyclic OSKM (Progeria Mice) | 33% increase [58] [14] | Significant multi-omics rejuvenation [14] | Low with careful cycling [58] | Improved tissue function in multiple organs [58] |
| OSK in Wild-Type Aged Mice | 109% remaining lifespan increase [14] | Not specifically quantified | Minimal (c-MYC excluded) [14] | Reduced frailty index scores [14] |
| Short-Term OSKM (Aged Mice) | Not measured | DNA methylation reprogramming across tissues [24] | Not observed in short protocols | Enhanced wound healing, reduced fibrosis [24] |
| Chemical Partial Reprogramming | 42.1% (C. elegans) [14] | Epigenetic clock reversal [14] | Not reported | Reduced oxidative stress, decreased senescence [14] |
Molecular Mechanisms of Partial Reprogramming
Factor-Specific Contributions to Rejuvenation
Each Yamanaka factor plays a distinct role in the reprogramming process, with specific contributions to both rejuvenation and potential identity loss:
OCT4: Functions as the master regulator of epigenetic reprogramming, with studies suggesting it alone can induce pluripotency when other factors are endogenously expressed [57]. During reprogramming, OCT4 directly recruits the BAF chromatin remodeling complex to promote a euchromatic state, binds enhancers of Polycomb-repressed genes, and establishes autoregulatory pluripotency networks [57]. Optimal reprogramming requires a threefold excess of OCT4 relative to other factors [57].
SOX2: Acts as a pioneering factor that engages chromatin first and primes target sites for subsequent OCT4 binding [57]. SOX2 alone can open chromatin and bind target DNA sites before OCT4 arrival, with OCT4/SOX2-shared sites showing the most profound increase in accessibility during early reprogramming [57].
KLF4: Drives the first wave of transcriptional activation during reprogramming and exhibits dual functionality as both an activator and repressor depending on context [57]. OCT4-SOX2 binding increases KLF4 binding by several folds, particularly in chromatin regions that are closed in human fibroblasts [57].
c-MYC: Serves as a potent amplifier of reprogramming rather than a pioneering factor [57]. The presence of c-MYC increases OSK binding by twofold, but its strong pro-proliferative effects underlie its significant oncogenic potential [57]. Many recent protocols exclude c-MYC to improve safety profiles [14].
Diagram 1: Molecular Dynamics of Partial Reprogramming. The process involves sequential phases where SOX2 initially opens chromatin, followed by coordinated factor activity that can lead to either rejuvenation or identity loss depending on exposure duration and control.
Epigenetic Remodeling During Partial Reprogramming
The rejuvenating effects of partial reprogramming primarily operate through epigenetic remodeling, reversing age-related changes without altering DNA sequence. During aging, organisms experience progressive loss of epigenetic information that disrupts cellular homeostasis [14]. Partial reprogramming appears to reverse these changes by:
- Resetting DNA methylation patterns toward more youthful states [24] [14]
- Reducing repressive histone marks such as H3K9me3 and H3K27me3 [14]
- Restoring chromatin accessibility through recruitment of remodeling complexes [57]
- Reestablishing transcriptional networks characteristic of younger cells [24]
Notably, different tissues show varying susceptibility to epigenetic reprogramming. A review of current research notes that cyclic induction of OSKM in aged mice "restores youthful multi-omics signatures - including DNA methylation, transcriptomic, and lipidomic profiles - across multiple organs such as the spleen, liver, skin, kidney, lung, and skeletal muscle" [24]. This demonstrates the broad potential of the approach while highlighting the need for tissue-specific optimization.
Experimental Protocols for Controlled Partial Reprogramming
Standard Cyclic Induction Protocol
The most widely adopted method for in vivo partial reprogramming involves cyclic induction of Yamanaka factors, typically using doxycycline-inducible systems in transgenic mouse models [58] [14]. The standard protocol includes:
- Transgene Activation: Administer doxycycline (typically 0.1-2 mg/mL in drinking water) to activate OSKM expression
- Pulse Phase: Maintain induction for 24-48 hours to initiate reprogramming
- Chase Phase: Withdraw doxycycline for 5-7 days to allow stabilization and identity recovery
- Cycle Repetition: Repeat pulses weekly or biweekly for extended periods (up to 10 months in some studies) [14]
This approach has demonstrated significant benefits including lifespan extension, improved tissue regeneration, and multi-omics rejuvenation without widespread teratoma formation when carefully controlled [58] [14]. One study implementing this protocol in progeria mice reported a 33% lifespan extension with no weight loss or mortality effects from the treatment itself [14].
Sequential Factor Delivery Protocol
Research indicates that sequential addition of reprogramming factors, rather than simultaneous expression, can significantly improve reprogramming efficiency and potentially enhance safety. Pei and colleagues demonstrated that adding factors in a specific sequence—Oct4 and Klf4 first, followed by c-Myc, and finally Sox2—improves reprogramming efficiency by 300% compared to standard simultaneous OSKM delivery [59].
This protocol works by promoting a more natural transition through reprogramming stages, including a transient hyper-mesenchymal state before mesenchymal-epithelial transition (MET) [59]. The sequential approach appears to facilitate more extensive epigenetic reorganization before pluripotency gene activation, potentially allowing for better preservation of cellular identity during the rejuvenation process.
Chemical Reprogramming Methods
For translational applications, non-genetic approaches using small molecules offer potential safety advantages. Chemical reprogramming typically involves:
- Multi-stage protocols using different molecule combinations at each phase
- Extended treatment duration compared to factor-based reprogramming
- Induction of a plastic intermediate state rather than direct pluripotency
Notably, chemical reprogramming operates through distinct mechanisms from OSKM approaches. While OSKM-mediated reprogramming typically downregulates the p53 pathway, chemical reprogramming using the 7c cocktail upregulates p53—a key difference that may influence both safety and efficacy profiles [14].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Partial Reprogramming Studies
| Reagent/Category | Specific Examples | Function | Considerations |
|---|---|---|---|
| Inducible Systems | Tet-On polycistronic OSKM cassettes [60] [58] | Controlled temporal expression | Enables cycling protocols; reduces tumor risk |
| Delivery Vectors | Lentiviral, AAV9 (for in vivo) [14] | Factor delivery to target cells | AAV9 provides broad tissue distribution [14] |
| Small Molecules | Vitamin C, 7c chemical cocktail [14] [59] | Enhance efficiency or enable non-genetic reprogramming | Vitamin C increases efficiency by reducing p53 [24] |
| Cell Identity Markers | Cell-type specific antibodies, RNA-seq panels | Monitor differentiation status | Essential for detecting identity loss |
| Aging Biomarkers | DNA methylation clocks, senescence assays (SA-β-gal) [58] [14] | Quantify rejuvenation effects | Multi-omics approaches provide comprehensive assessment |
| Safety Assays | Teratoma formation assays, cancer markers [24] [25] | Assess tumorigenic risk | Critical for in vivo applications |
Safety Considerations and Identity Preservation Strategies
Managing Oncogenic Risk
The most significant safety concern in partial reprogramming is the potential for teratoma formation and other tumorigenic outcomes. Several strategies have emerged to mitigate this risk:
Excluding c-MYC: Many recent protocols omit this potent oncogene from the reprogramming cocktail, with one study demonstrating that OSK alone can extend remaining lifespan in wild-type mice by 109% without c-MYC [14].
Optimized Cycling Protocols: Short, intermittent pulses of factor expression followed by extended recovery periods prevent complete dedifferentiation while still achieving rejuvenation. Research shows that even 35 cycles of such treatment caused no weight loss or mortality effects in mouse models [14].
Tissue-Specific Approaches: Targeted delivery to specific tissues or systems reduces systemic exposure and potential off-target effects. This approach has shown promise for ocular, neural, and musculoskeletal applications [14].
Monitoring Identity Loss
Preserving cellular identity requires careful monitoring throughout the reprogramming process. Key indicators of identity compromise include:
- Downregulation of lineage-specific markers
- Activation of pluripotency genes such as Nanog
- Morphological changes toward stem-like characteristics
- Functional impairment in tissue-specific activities
Studies demonstrate that the timing of factor expression is critical for identity preservation. As one review notes, upon cessation of Yamanaka factor expression, aging symptoms "quickly begin to accumulate again, including the methylation age reverting to its original state" [24], suggesting that continuous or excessive expression may be necessary for permanent epigenetic reset—but this comes with increased identity loss risks.
Partial reprogramming represents a promising but complex approach for combating age-related degeneration, with different factor combinations offering distinct efficiency and safety profiles. The current evidence suggests that:
- OSK combinations without c-MYC provide the most favorable safety profile for in vivo applications
- Sequential factor delivery may significantly improve efficiency while potentially better preserving cellular identity
- Chemical reprogramming offers a promising non-genetic alternative but requires further development
- Tissue-specific optimization is essential given varying responses across different cell types
Future research directions include developing more precise temporal control systems, identifying factor combinations that maximize rejuvenation while minimizing identity loss, and establishing standardized safety monitoring protocols. As the field advances, balancing the remarkable rejuvenation potential of partial reprogramming with the fundamental need to preserve cellular identity remains the central challenge for researchers and therapeutic developers alike.
The groundbreaking discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using transcription factors has revolutionized regenerative medicine and aging research. However, the efficiency and kinetics of this process vary significantly across different cell types, presenting substantial challenges for research and therapeutic applications. The original Yamanaka factors—OCT4, SOX2, KLF4, and c-MYC (OSKM)—established the foundation for cellular reprogramming, but researchers have since pursued optimized factor combinations to overcome limitations in efficiency, safety, and applicability to challenging cell sources. This guide provides a comprehensive comparison of Yamanaka factor combinations, detailing their experimental performance across diverse cell types to inform protocol selection and optimization.
Established Yamanaka Factor Combinations: Mechanisms and Trade-offs
The core transcription factors function through coordinated mechanisms to erase somatic cell identity and establish pluripotency. OCT4 and SOX2 form a central partnership that activates the pluripotency network while suppressing differentiation genes. KLF4 contributes to chromatin remodeling and regulates the expression of both somatic and pluripotency genes. C-MYC acts as a global amplifier of transcription, accelerating cell cycle progression and facilitating epigenetic changes [25] [21].
While effective, the canonical OSKM combination presents significant challenges. The oncogenic potential of c-MYC raises safety concerns for therapeutic applications, and even the remaining three factors (OCT4, SOX2, KLF4) can induce teratoma formation if reprogramming is not carefully controlled [22] [44]. Additionally, primary cells from aged or diseased donors often demonstrate reduced reprogramming efficiency due to accumulated epigenetic barriers and cellular senescence [5] [25]. These limitations have motivated the development of safer, more efficient alternatives.
Table 1: Core Yamanaka Factors and Their Functions in Reprogramming
| Transcription Factor | Primary Function in Reprogramming | Key Molecular Interactions | Safety Considerations |
|---|---|---|---|
| OCT4 (POU5F1) | Master regulator of pluripotency; essential for establishing embryonic gene expression patterns | Forms heterodimers with SOX2; activates NANOG expression | Critical dosage sensitivity; aberrant expression promotes teratoma formation |
| SOX2 | Partners with OCT4 to activate pluripotency network; suppresses differentiation pathways | Binds DNA with OCT4 at composite SOX-OCT elements; regulates SOX2OT lncRNA | Required for maintenance of cellular identity; depletion leads to spontaneous differentiation |
| KLF4 | Dual role in gene activation/repression; promotes mesenchymal-to-epithelial transition (MET) | Induces p53; regulates cell cycle progression; modulates chromatin accessibility | Context-dependent oncogene or tumor suppressor; can induce cell cycle arrest in some contexts |
| c-MYC | Global chromatin modifier; accelerates cell proliferation; enhances reprogramming efficiency | Associates with histone acetyltransferases; alters metabolic programming | Potent oncogene; increases tumor risk in partially reprogrammed cells; not strictly essential |
Comparative Analysis of Reprogramming Factor Combinations
Researchers have developed numerous factor combinations to address the limitations of the original Yamanaka cocktail. These optimized protocols vary in their factor composition, efficiency, safety profiles, and suitability for different cell types. The table below provides a systematic comparison of established combinations and their performance characteristics.
Table 2: Comparative Performance of Yamanaka Factor Combinations Across Cell Types
| Factor Combination | Reported Efficiency Range | Key Applications & Cell Types | Advantages | Limitations & Risks |
|---|---|---|---|---|
| OSKM (Canonical) | 0.1%-1% (varies by cell source) | Mouse and human fibroblasts; established baseline | Comprehensive pluripotency induction; well-characterized | Low efficiency; teratoma risk; c-MYC oncogenicity |
| OSK (c-MYC excluded) | 0.01%-0.5% (generally lower than OSKM) | Safer in vivo reprogramming; aging intervention studies | Reduced oncogenic potential; improved safety profile | Slower kinetics; may require supplemental factors |
| OSK + TERT | Not quantified; significant functional improvement in aged cells | Replicatively senescent MRC-5 fibroblasts (50+ passages) | Enhanced rejuvenation markers; combats telomere attrition | Limited efficiency data; delivery optimization required |
| OSKNL (Oct4, Sox2, Klf4, Nanog, Lin28) | Up to 10-fold increase over OSKM in some studies | Fibroblasts from aged donors; refractory cell types | Improved reprogramming of aged cells; enhanced efficiency | Increased genetic payload; potential regulatory complexity |
| AI-Optimized Variants (RetroSOX/RetroKLF) | >30% hit rate (vs. <10% typical); 50x marker expression | Human fibroblasts from multiple aged donors; MSCs | Dramatically enhanced efficiency; deep sequence edits | Novel approach requiring specialized computational resources |
Recent breakthroughs in AI-guided protein engineering have demonstrated remarkable improvements in reprogramming efficiency. In collaboration with Retro Biosciences, OpenAI developed GPT-4b micro, a specialized model that designed novel variants of SOX2 and KLF4. These optimized factors achieved over 50-fold higher expression of stem cell reprogramming markers compared to wild-type controls when tested in human fibroblasts from multiple donors. The AI-generated variants also showed enhanced DNA damage repair capabilities, indicating higher rejuvenation potential [5].
For challenging cell types like those from aged donors, the exclusion of c-MYC (OSK combination) has emerged as a critical safety modification. Research on progeroid syndromes demonstrates that while OSKM can reverse aging markers, the oncogenic risk of c-MYC necessitates careful consideration [25]. Alternative approaches combining OSK with TERT gene therapy have shown promise in reversing senescence in aged MRC-5 fibroblasts, significantly reducing aging markers while improving cell viability and delaying aging phenotypes [22].
Experimental Protocols for Challenging Cell Types
AI-Optimized Factor Protocol for Aged Fibroblasts
The Retro Biosciences team established a screening platform using human fibroblast cells from aged donors. They initially validated the platform with baseline OSKM and manually designed SOX2 variants, then employed GPT-4b micro to generate diverse "RetroSOX" sequences. The AI-generated variants differed by more than 100 amino acids on average from wild-type SOX2, yet over 30% outperformed wild-type SOX2 in expressing pluripotency markers. For KLF4, the largest Yamanaka factor, nearly 50% of model-generated variants outperformed the best cocktails from the RetroSOX screen. The combination of top RetroSOX and RetroKLF variants produced dramatic gains, with fibroblasts showing accelerated marker onset and robust alkaline phosphatase activity indicative of pluripotency [5].
OSK + TERT Combination for Senescent Cells
To address cellular senescence, researchers utilized MRC-5 fibroblasts that had undergone 50 population doublings (replicative senescence). Cells were transfected with OSK-expressing plasmid with or without TERT-expressing plasmid using a pcDNA3.3 vector. Quantitative analysis at subsequent passages (60 and 70 generations) showed significantly increased expression of youth-related genes (OCT4, SOX2, KLF4, NANOG, c-MYC, TERT) and reduced expression of aging-related genes (p16, p21, ZSCAN4, ATF3, MMP13) and inflammatory cytokine IL-6. Functional assays confirmed improved cell viability, reduced G0/G1 phase arrest, and decreased β-galactosidase expression, demonstrating successful reversal of senescence markers [22].
Small Molecule Reprogramming Approaches
For researchers seeking non-genetic integration methods, small molecule cocktails offer an alternative pathway. The 7c combination (CHIR99021, DZNep, Forskolin, TTNPB, Valproic acid, Repsox, and Tranylcypromine) and the optimized 2c cocktail (Repsox and Tranylcypromine) can induce rejuvenation without genetic manipulation. These approaches partially restore youthful gene expression patterns and improve molecular hallmarks of aging, though in vivo applications may face challenges with toxicity and lipid accumulation [13].
Signaling Pathways and Experimental Workflows
The diagram above illustrates the core experimental workflow for reprogramming challenging cell types, highlighting multiple entry points through different factor combinations and the key molecular processes that lead to successful reprogramming outcomes.
The Scientist's Toolkit: Essential Research Reagents
Successful reprogramming of difficult cell types requires careful selection of reagents and delivery systems. The following table details essential materials and their functions in optimizing reprogramming protocols.
Table 3: Essential Research Reagents for Reprogramming Challenging Cell Types
| Reagent Category | Specific Examples | Function in Reprogramming | Application Notes |
|---|---|---|---|
| Factor Delivery Systems | Lentiviral vectors, Sendai virus, mRNA transfection, episomal plasmids | Introduction of reprogramming factors into target cells | Non-integrating systems (Sendai, mRNA) preferred for clinical applications; optimize for cell type-specific transduction efficiency |
| Culture Media Optimization | iCD1 defined medium, small molecule supplements (e.g., VPA, CHIR99021) | Support reprogramming efficiency and kinetics | Serum-free conditions improve reproducibility; specific small molecules can replace certain transcription factors |
| Small Molecule Enhancers | Vitamin C, RepSox, Tranylcypromine, Valproic acid, Forskolin | Modulate signaling pathways; reduce epigenetic barriers | Vitamin C reduces p53/p21 expression; TGF-β inhibitors replace SOX2 requirement; optimize concentration for specific cell types |
| Senescence Countermeasures | TERT expression, OSK factors, p53 inhibition | Overcome proliferation limits in aged cells | Critical for reprogramming cells from elderly donors; balance safety concerns with efficiency needs |
| Characterization Tools | Alkaline phosphatase staining, pluripotency marker antibodies, karyotyping | Validate successful reprogramming and genomic integrity | Essential quality control steps; combine multiple methods to confirm full pluripotency and exclude partially reprogrammed cells |
The optimization of Yamanaka factor combinations for challenging cell types represents a rapidly advancing frontier in reprogramming research. While the canonical OSKM factors established the field, modified approaches including c-MYC-free combinations, supplementation with TERT, and AI-optimized factors demonstrate significant improvements in both efficiency and safety. The selection of an appropriate factor combination must consider the specific cell source, desired application, and safety requirements. As research progresses, particularly in computational protein design and small molecule approaches, researchers now have an expanding toolkit to address even the most refractory cell types, accelerating both basic research and clinical translation in regenerative medicine and aging intervention.
Head-to-Head Comparisons and Validation: Evaluating Reprogramming Outcomes
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using defined transcription factors has revolutionized regenerative biology. The original combination of Oct4, Sox2, Klf4, and c-Myc (OSKM) established by Shinya Yamanaka demonstrated that cellular identity could be rewritten [44]. However, this protocol faces challenges in efficiency, kinetics, and safety concerns, particularly regarding the oncogenic potential of c-Myc. These limitations have spurred investigations into alternative factor combinations, including the omission of c-Myc (OSK) or its replacement with other factors such as Lin28 (OSNL). Understanding the relative performance of these combinations is crucial for advancing both basic research and therapeutic applications. This guide provides a systematic comparison of OSKM, OSNL, and OSK combinations, synthesizing experimental data on reprogramming efficiency, kinetics, and functional outcomes to inform research and development decisions.
Quantitative Efficiency Comparison
Table 1: Comparative Efficiency of Yamanaka Factor Combinations
| Factor Combination | Reprogramming Efficiency | Key Markers Enhanced | Time to Pluripotency Markers | Reported Teratoma/Tumor Formation Risk |
|---|---|---|---|---|
| OSKM (Oct4, Sox2, Klf4, c-Myc) | Typically <0.1% of cells convert; ~0.05% in human fibroblasts [61]. Can be dramatically improved with engineered factors (e.g., >50x marker expression with AI-designed RetroSOX/RetroKLF) [5]. | SSEA-4, TRA-1-60, NANOG, Alkaline Phosphatase [5]. Late markers appear several days sooner with engineered variants [5]. | Several days faster with engineered variants; human reprogramming can take up to a month [4] [5]. | Higher risk; incomplete reprogramming can cause cancer in mouse models [44]. |
| OSK (Oct4, Sox2, Klf4) | Lower efficiency than OSKM; mouse cells can be reprogrammed with OSK alone, but c-Myc is more critical in human systems [4] [61]. | Pluripotency markers achieved, but process is less efficient and slower [61]. | Takes considerably longer than OSKM, especially in human cells [4]. | Presumed lower risk due to absence of oncogenic c-Myc. |
| OSNL (Oct4, Sox2, Nanog, Lin28) | Information not available in search results - direct comparative quantitative data against OSKM/OSK was not found. | Information not available in search results. | Information not available in search results. | Information not available in search results. |
The data reveals a clear trade-off between efficiency and safety. The OSKM combination, particularly with enhanced factors, offers superior speed and efficiency but carries a significant risk of tumorigenesis [5] [44]. In contrast, the OSK combination is safer but less efficient, with a more pronounced efficiency drop in human cells compared to mouse cells [4]. A direct, quantitative comparison with the OSNL combination could not be established from the available data.
Detailed Experimental Protocols and Methodologies
OSKM Reprogramming Protocol
A standard protocol for OSKM reprogramming involves lentiviral transduction of human fibroblasts. Key steps include:
- Cell Source: Human fetal foreskin fibroblasts or other somatic cell types.
- Factor Delivery: Use of doxycycline (dox)-inducible lentiviral vectors to transduce OSKM factors [61]. This system allows for controlled temporal expression.
- Timeline: Cells are harvested for initial binding studies (e.g., ChIP-seq) at 48 hours post-transfection/induction. This early time point captures the initial chromatin engagement of the factors before major transcriptional changes occur [61]. The cells are then transferred to human ES cell culture conditions, with stable iPSC lines emerging over several weeks [4] [61].
- Efficiency Assessment: Reprogramming efficiency (~0.05%) is quantified by the number of stable iPSC colonies that express endogenous pluripotency markers (e.g., via immunostaining) and can form embryoid bodies capable of differentiating into all three germ layers [61].
In Vivo Reprogramming and Partial Reprogramming
Moving beyond in vitro iPSC generation, researchers also induce reprogramming in living organisms (in vivo) but for short durations to achieve partial reprogramming. This approach aims to rejuvenate cells by resetting epigenetic aging markers without fully converting them into pluripotent stem cells, thereby avoiding teratoma formation [24].
- Model System: Progeric mice or physiologically aged wild-type mice.
- Factor Delivery: Transgenic systems or viral vectors (e.g., AAV9, adenovectors) that allow for cyclic, transient induction of the factors (e.g., 2 days on/5 days off) [44] [24].
- Outcome Measures: Successful partial reprogramming is assessed by multi-omics analysis, including restoration of youthful DNA methylation patterns, transcriptomic profiles, and functional improvements in tissue regeneration (e.g., enhanced wound healing, improved muscle repair) [24]. Critically, this is achieved without a full loss of cellular identity.
The following diagram illustrates the logical decision pathway and key outcomes for selecting a reprogramming protocol.
Signaling Pathways and Molecular Mechanisms
The molecular journey from a somatic cell to a pluripotent state involves dramatic restructuring of the genomic regulatory network.
Initial Chromatin Engagement
During the early stages (e.g., 48 hours), OSK factors act as pioneer factors, binding to many closed chromatin sites and initiating the rewiring of the cellular program [61]. A key finding is the role of c-Myc in facilitating the binding of OSK to chromatin, providing a direct mechanistic explanation for its efficiency-boosting effect, beyond simply promoting proliferation [61]. In the first 48 hours, OSKM binding is highly collaborative, with a significant fraction of genes being co-targeted by all four factors, which is distinct from the binding pattern seen in established ES cells [61].
Overcoming Epigenetic Barriers
A major impediment to efficient reprogramming is the somatic cell's chromatin structure. Large megabase-scale domains marked by the repressive H3K9me3 modification act as barriers, preventing OSKM factors from accessing genes essential for pluripotency [61]. Knocking down the methyltransferases responsible for H3K9me3 allows OSKM binding to these previously inaccessible regions and enhances reprogramming efficiency [61]. This highlights a critical functional interaction between the reprogramming factors and the host cell's epigenetic landscape.
The following flowchart summarizes the key molecular events and major barriers in the early phase of OSKM-induced reprogramming.
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Reprogramming Research
| Reagent | Function and Application in Reprogramming |
|---|---|
| Lentiviral Vectors (Dox-inducible) | Allow stable integration and controlled, inducible expression of OSKM factors in target somatic cells (e.g., fibroblasts) [61]. |
| Adenoviral / AAV Vectors | Used for in vivo delivery of reprogramming factors. Particularly useful for transient expression in partial reprogramming protocols to reduce integration risk [62] [63]. |
| STEMCCA Polycistronic Cassette | A single vector system expressing OSKM from a single transcript via 2A "self-cleaving" peptides. Ensures co-expression of all four factors in a near-stoichiometric ratio in every infected cell, improving consistency [62] [63]. |
| ChIP-seq Kits | Critical for mapping the genomic binding sites (cistrome) of OSKM factors (e.g., at 48 hours) and comparing them to epigenetic marks like H3K9me3 [4] [61]. |
| Pluripotency Marker Antibodies | Essential for assessing reprogramming success. Key markers include: SSEA-4 (early marker), TRA-1-60 (late marker), NANOG (core pluripotency factor), and Alkaline Phosphatase (AP) activity [5]. |
| H3K9me3 Inhibitors (e.g., KDM4A) | Chemical inhibitors or genetic tools (siRNA/shRNA) targeting H3K9 methyltransferases. Used to relax heterochromatin barriers and enhance reprogramming efficiency [61]. |
The direct comparison between OSKM, OSNL, and OSK remains experimentally incomplete, with a pronounced lack of head-to-head studies quantifying OSNL performance against the established benchmarks of OSKM and OSK. The available data firmly establishes OSKM as the most potent combination for full reprogramming, albeit with significant safety trade-offs. The emergence of AI-engineered factor variants (e.g., RetroSOX, RetroKLF) represents a paradigm shift, moving beyond naturally occurring factors to create purpose-built molecules with dramatically enhanced activity and efficiency [5]. Furthermore, the concept of partial reprogramming has expanded the application of these factors from a binary cell-fate conversion tool to a potential modulator of cellular aging [24].
Future research must prioritize standardized, direct comparisons of all major factor combinations, including OSNL, under identical experimental conditions. Key challenges include optimizing delivery systems for spatiotemporal control in vivo and fully understanding the mechanistic link between partial reprogramming and functional tissue rejuvenation. The ultimate goal is to decouple the beneficial rejuvenating effects of these factors from the risks of tumorigenesis and loss of cellular identity, paving the way for their safe application in regenerative medicine and age-related diseases.
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC, collectively OSKM) has revolutionized regenerative medicine, disease modeling, and drug development [64]. However, the translational application of these cells depends heavily on rigorous quality control to ensure their safety and functionality. Evaluating genomic integrity, pluripotency, and differentiation potential is paramount, as the reprogramming process and subsequent culture can introduce genomic abnormalities and affect the cells' developmental potential [65] [66]. This guide provides a comparative analysis of quality control metrics essential for researchers evaluating different Yamanaka factor combinations and their resulting iPSCs.
Quality Control Pillars for iPSC Characterization
The quality assessment of iPSCs revolves around three fundamental pillars. A comprehensive understanding of the technical workflows and the logical relationship between these pillars is crucial for robust experimental design.
Experimental Workflow for iPSC Quality Control
The journey from somatic cell to characterized iPSC line involves a multi-stage process, each requiring specific quality checks. The following diagram outlines the key stages of reprogramming and the corresponding critical quality control checkpoints.
Logical Relationship of Core Regulatory Circuitry
The core transcriptional regulatory network governed by the Yamanaka factors is the foundation of pluripotency. Understanding this network is key to assessing a successful reprogramming outcome.
Assessing Genomic Integrity
The reprogramming process can induce various genomic abnormalities, making genomic integrity a primary safety concern. Different Yamanaka factor combinations influence the type and frequency of these mutations [65] [66].
Types and Origins of Genomic Aberrations in iPSCs
The following table summarizes the common genomic abnormalities, their detection methods, and their potential origins.
Table 1: Genomic Abnormalities in iPSCs and Detection Methods
| Abnormality Type | Description | Common Detection Techniques | Potential Origin |
|---|---|---|---|
| Karyotype Aberrations | Aneuploidies (e.g., trisomy 12, 17, X) or large chromosomal rearrangements [65]. | G-banding karyotyping | Often acquired during long-term culture [65]. |
| Copy Number Variations (CNVs) | Deletions or amplifications of DNA sections, frequently at fragile sites [65]. | SNP genotyping, CGH-arrays | Pre-existing in somatic population or acquired de novo during reprogramming [65]. |
| Single Point Mutations | Single-nucleotide changes in protein-coding regions [65]. | Whole exome sequencing | Preexisting in somatic cells (mosaicism) or acquired during reprogramming [65]. |
| Uniparental Disomy (UPD) | Inheritance of two chromosomes from one parent [65]. | SNP genotyping (for Loss of Heterozygosity) | Can occur during reprogramming, even with non-integrative methods [65]. |
Impact of Reprogramming Factors and Methods on Genomic Integrity
The choice of reprogramming factors and delivery methods significantly impacts the genomic stability of the resulting iPSCs.
- Factor Combinations: The omission of the oncogene c-Myc, using only OSK, is a common strategy to reduce the risk of tumorigenesis in derived cells, though it may lower reprogramming efficiency [66] [14].
- Reprogramming Methods: Non-integrating delivery methods (e.g., episomal vectors, Sendai virus, mRNA) are preferred over integrating retroviruses to avoid insertional mutagenesis [66] [64].
- Starting Cell Type: The initial somatic cell population matters. Somatic mosaicism means that pre-existing mutations in a subset of somatic cells can be selected for or amplified during reprogramming [65].
Evaluating Pluripotency
Pluripotency confirms that the reprogrammed cells have successfully acquired a stem cell identity. Assessment typically involves evaluating marker expression and functional potential.
Key Pluripotency Markers and Assays
A combination of molecular and functional tests is required to conclusively demonstrate pluripotency.
Table 2: Pluripotency Assessment Metrics
| Assay Category | Specific Marker/Test | Function and Significance |
|---|---|---|
| Surface Marker Expression | SSEA-4, TRA-1-60, TRA-1-81 [67] | Detected by flow cytometry or immunocytochemistry; indicates a characteristic pluripotent cell surface profile. |
| Transcription Factor Expression | OCT4, SOX2, NANOG [67] [25] | Detected by immunostaining or RT-qPCR; core regulators of the pluripotency network. |
| Enzyme Activity | Alkaline Phosphatase [64] | Histochemical staining; highly active in pluripotent stem cells. |
| Functional Assay In Vivo | Teratoma Formation [64] [25] | Injection of iPSCs into immunodeficient mice; a validated test for differentiation into tissues of all three germ layers (ectoderm, mesoderm, endoderm). |
Determining Differentiation Potential
The ultimate test of iPSC quality is their ability to efficiently and faithfully differentiate into functional somatic cell types, which is critical for disease modeling and cell therapy.
Directed Differentiation and Analysis of Derivatives
The differentiation potential is assessed by guiding iPSCs toward specific lineages and characterizing the resulting cells.
- Ectoderm Differentiation: iPSCs can be efficiently differentiated into neural cells, including dopaminergic neurons, using inhibitors of TGF-β/Smad signaling (e.g., Noggin, SB431542) [67].
- Mesoderm Differentiation: Protocols exist to generate functional cardiomyocytes expressing markers like Nkx2.5 and Troponin T, and adipocytes expressing C/EBPα and PPARγ [67].
- Endoderm Differentiation: iPSCs can be differentiated into pancreatic progenitor cells and further into insulin-producing cells expressing MafA and C-peptide [67].
Comparative Analysis of Yamanaka Factor Combinations
Different combinations of reprogramming factors yield iPSC lines with varying properties, necessitating tailored quality control approaches.
Table 3: Comparison of Yamanaka Factor Combinations and QC Focus
| Factor Combination | Reprogramming Efficiency | Key Characteristics | Primary QC Concerns | Ideal Application |
|---|---|---|---|---|
| OSKM (Canonical) | High [64] | Gold standard; rapid reprogramming [64]. | High tumorigenic risk (c-MYC); genomic instability [66]. | Basic research where efficiency is prioritized. |
| OSK | Moderate [14] | Improved safety profile; reduced tumorigenicity [14]. | Lower efficiency; requires rigorous check of pluripotency [14]. | Therapeutic applications and disease modeling. |
| OS (Oct4, Sox2) | Low [25] | Minimal factor setup; context-dependent function [68]. | Low efficiency; may require additional chemical supplements. | Studies on core pluripotency circuitry. |
| Chemical Cocktails (e.g., 7c/2c) | Variable [13] [12] | Non-genetic integration; potential for in vivo use [13] [12]. | Cytotoxicity (e.g., 7c); optimal dosing; molecular off-target effects [13]. | Rejuvenation studies and future in vivo therapies. |
The Scientist's Toolkit: Essential Research Reagents
A successful iPSC generation and characterization workflow relies on a suite of essential reagents and tools.
Table 4: Key Research Reagent Solutions for iPSC QC
| Reagent/Tool Category | Example Products | Function in QC Workflow |
|---|---|---|
| Reprogramming Vectors | Episomal vectors, Sendai virus, mRNA [66] [64] | Non-integrating delivery of Yamanaka factors to somatic cells. |
| Pluripotency Antibodies | Anti-OCT4, Anti-SOX2, Anti-NANOG, Anti-SSEA-4 [67] [25] | Immunostaining and flow cytometry for pluripotency marker verification. |
| Differentiation Kits | Neural Induction Medium, Cardiomyocyte Differentiation Kits [67] | Directed differentiation of iPSCs into specific somatic cell lineages. |
| Genomic Analysis Kits | SNP Genotyping Arrays, Whole Exome Sequencing Kits [65] | Detection of CNVs, point mutations, and UPD for genomic integrity assessment. |
| Senescence Assay Kits | SA-β-Galactosidase Staining Kits [12] | Detection of senescent cells in iPSC cultures, an indicator of cellular health. |
Emerging Trends: Partial Reprogramming and Rejuvenation
A rapidly advancing frontier is the application of Yamanaka factors for rejuvenation without full reprogramming to pluripotency. This approach, known as partial reprogramming, uses short, cyclic expression of OSKM or chemical cocktails (e.g., 2c: RepSox and tranylcypromine) to reverse age-associated epigenetic and transcriptomic changes in cells, restoring youthful function without erasing cellular identity [14] [12]. This has shown promise in ameliorating aging hallmarks in vivo and improving tissue function in models of aging and disease [13] [14]. Quality control in this context focuses on ensuring that youthful molecular signatures are restored without any loss of the cell's original identity or induction of pluripotency.
The ability to differentiate induced pluripotent stem cells (iPSCs) into specific somatic cell types represents the critical bridge between pluripotency and practical application in disease modeling, drug screening, and regenerative medicine. Functional validation of these differentiated cells confirms their morphological, molecular, and physiological similarity to their native counterparts, ensuring the reliability of data generated from iPSC-based platforms. The efficiency and success of differentiation are intrinsically linked to the initial reprogramming methodology, including the specific combination of Yamanaka factors used to create the iPSCs. This guide objectively compares the differentiation performance into three therapeutically crucial cell types—motor neurons, cardiomyocytes, and hepatocytes—and details the experimental protocols and reagent solutions that underpin robust functional validation.
Yamanaka Factor Combinations and Their Impact on Differentiation Potential
The foundational reprogramming factors used to create iPSCs can influence their subsequent differentiation behavior. While the original Yamanaka factors (OCT4, SOX2, KLF4, c-MYC, or OSKM) remain a gold standard, research has explored variations to optimize safety and efficacy.
- Standard OSKM Cocktail: The combination of OCT4, SOX2, KLF4, and c-MYC establishes a robust pluripotent state [1]. However, the oncogenic potential of c-MYC raises concerns for clinical applications, and its presence can sometimes skew differentiation potential or increase tumorigenicity risk in downstream derivatives [22] [44].
- Alternative Factor Combinations: The Thomson lab's combination (OCT4, SOX2, NANOG, LIN28) provides an effective alternative for reprogramming human somatic cells [1] [2]. Some studies suggest that excluding c-MYC (using OSK alone) can yield iPSCs with a improved safety profile, though potentially at the cost of lower reprogramming efficiency [22]. Research indicates that the specific ratio and expression levels of these factors, particularly OCT4 and SOX2, can significantly affect both reprogramming efficiency and the quality of the resulting iPSC colonies, which in turn influences their differentiation capacity [1].
Comparative Differentiation Efficiency and Functional Validation
The following section provides a comparative analysis of differentiation protocols for motor neurons, cardiomyocytes, and hepatocytes, summarizing key efficiency metrics and functional outcomes.
Table 1: Comparative Differentiation Efficiency for Key Cell Types
| Target Cell | Reported Efficiency | Key Markers for Validation | Critical Functional Assays |
|---|---|---|---|
| Hepatocytes | ~39% FOXA2+ endodermal cells with optimized protocols [69] | SOX17, GATA4 (DE); HNF4α, FOXA2 (Hepatoblasts); Albumin, AAT (Mature Hepatocytes) [69] | Glycogen storage, Albumin/urea production, CYP450 activity [69] |
| Cardiomyocytes | Information not explicitly available in search results | Information not explicitly available in search results | Information not explicitly available in search results |
| Motor Neurons | Information not explicitly available in search results | Information not explicitly available in search results | Information not explicitly available in search results |
Note: Data for cardiomyocyte and motor neuron differentiation efficiency and markers were not explicitly detailed in the provided search results. The table reflects the most relevant available information.
Hepatocyte Differentiation
Experimental Protocol Overview: The differentiation of iPSCs into functional hepatocytes is a multi-stage process that mimics in vivo liver development [69].
- Definitive Endoderm (DE) Induction: iPSCs are treated with activin A (a TGF-β family member) and Wnt activators (like CHIR99021) for approximately 5 days to drive differentiation into the definitive endoderm [69]. Some protocols use small molecule cocktails (e.g., CHIR99021, IDE1, sodium butyrate) as a cost-effective and stable alternative to growth factors [69].
- Hepatic Specification: The DE is then patterned into hepatic endoderm/hepatoblasts using signaling molecules such as BMP4, FGF2, and FGF4 over about 7 days [69]. Key transcription factors like HNF4α and GATA4 are critical at this stage.
- Hepatic Maturation: Immature hepatocytes are matured over several weeks using a combination of Hepatocyte Growth Factor (HGF), Oncostatin M (OSM), dexamethasone, and other factors. This step induces the expression of mature markers and functions [69].
Key Signaling Pathways in Hepatocyte Differentiation: The following diagram illustrates the core signaling pathways involved in directing iPSCs through the stages of hepatocyte differentiation.
Cardiomyocyte and Motor Neuron Differentiation
While detailed protocols for motor neurons were not available in the search results, the general principle for directed differentiation of pluripotent stem cells involves sequential treatment with specific signaling molecules based on developmental biology [70]. A powerful alternative strategy is transcription factor-directed differentiation, where forced expression of key lineage-specific transcription factors can directly reprogram PSCs into desired cell types like neurons, muscle, liver, and pancreatic endocrine cells, potentially offering higher efficiency and purity [70].
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful differentiation and validation require a suite of reliable reagents and tools. The following table details essential components for iPSC differentiation workflows.
Table 2: Key Research Reagent Solutions for iPSC Differentiation and Validation
| Reagent Category | Specific Examples | Function in Differentiation/Validation |
|---|---|---|
| Reprogramming Factors | Yamanaka Factors (OSKM) [1] [11], Thomson Factors (OSNL) [1] [2] | Foundation for creating integration-free, clinically relevant iPSC lines. |
| Key Growth Factors & Cytokines | Activin A [69], BMP4 [69], FGF2 [69], HGF [69], Oncostatin M [69] | Mimic developmental signaling to direct cell fate through specific stages. |
| Small Molecule Inducers | CHIR99021 (Wnt activator) [69], A83-01 (TGF-β inhibitor), Sodium Butyrate [69] | Cost-effective, stable alternatives to growth factors for modulating key signaling pathways. |
| Cell Culture Supplements | B-27 Supplement, N-2 Supplement | Provide optimized mixtures of hormones, proteins, and lipids to support survival and maturation of specialized cells like neurons. |
| Characterization & Validation Antibodies | Anti-SOX17 (Endoderm) [69], Anti-HNF4α (Hepatoblasts) [69], Anti-Albumin (Mature Hepatocytes) [69] | Critical for immunocytochemistry and flow cytometry to assess differentiation efficiency and purity at each stage. |
| Functional Assay Kits | CYP450 Activity Assays, Albumin ELISA Kits [69] | Quantify tissue-specific functional capacity of differentiated cells (e.g., hepatocytes). |
The functional validation of iPSC-derived cells is a cornerstone of their utility in biomedical research. Data shows that differentiation efficiency varies significantly by cell type, with hepatocyte protocols achieving notable milestones through well-defined, multi-stage processes [69]. The choice of initial reprogramming factors, such as the use of OSKM versus alternative cocktails, can set the foundation for this differentiation potential [1] [22].
The ongoing development of more complex models, like 3D organoids, and the refinement of direct differentiation protocols using transcription factors [70] promise even more physiologically relevant results. For researchers, selecting a differentiation strategy involves balancing efficiency, purity, functional maturity, and scalability. The experimental protocols and reagent solutions detailed herein provide a framework for the rigorous validation required to generate high-quality, reproducible data for drug discovery and therapeutic development.
Long-Term Stability: Karyotype and Epigenetic Profile Analysis Across Different Combinations
The discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) using transcription factors has revolutionized regenerative medicine. The original Yamanaka factors—OCT4, SOX2, KLF4, and c-MYC (OSKM)—established the foundation for resetting cellular age and developmental status [66]. However, the transition from research discovery to clinical therapy depends on thoroughly evaluating the long-term genomic and epigenetic stability of reprogrammed cells, as these characteristics directly impact safety and therapeutic potential [71].
This guide objectively compares the stability outcomes across different reprogramming factor combinations by synthesizing current experimental data. A critical analysis reveals that while the full OSKM combination achieves robust reprogramming, the omission of the oncogene c-MYC and the development of non-integrating delivery methods significantly enhance karyotypic stability and reduce tumorigenic risk, without fully compromising rejuvenative potential [14] [71].
Core Concepts and Assessment Methodologies
Defining Stability in Reprogrammed Cells
For iPSCs and their derivatives, long-term stability refers to the maintenance of a normal diploid karyotype and a predictable, functional epigenetic profile over extended periods of cell culture and proliferation. Karyotype stability ensures the absence of large-scale chromosomal abnormalities, such as aneuploidy or translocations, which can lead to tumorigenesis [71]. Epigenetic stability involves the faithful preservation of DNA methylation patterns, histone modifications, and gene expression profiles that are appropriate for the target cell type, preventing aberrant differentiation or malignant transformation [72].
Essential Analytical Techniques
Researchers employ a standardized toolkit to assess the stability of reprogrammed cells, with key methodologies detailed in the table below.
Table 1: Key Methodologies for Assessing Karyotypic and Epigenetic Stability
| Method | Target Aspect | Key Outcome Measures | Protocol Highlights |
|---|---|---|---|
| Karyotype Analysis (G-banding) [73] | Chromosomal number and structure | Diploid chromosome count; absence of rearrangements | Cells treated with colcemid → fixed and spread on slides → stained with Giemsa → microscopic analysis of ≥20 metaphase spreads. |
| In Vitro Differentiation & Teratoma Assay [74] [25] | Functional pluripotency and safety | Differentiation into three germ layers; formation of structured, non-malignant tissues in vivo | Cells aggregated into embryoid bodies or injected into immunodeficient mice → tissues analyzed histologically for ectoderm, mesoderm, and endoderm. |
| DNA Methylation Analysis [12] [72] | Epigenetic age and integrity | "Epigenetic clock" reversal; restoration of youthful transcriptome | Bisulfite sequencing of age-associated CpG sites; genome-wide transcriptome profiling via RNA-seq. |
| Immunofluorescence & RT-PCR [74] | Pluripotency marker expression | Presence of OCT4, SOX2, NANOG, SSEA1 | Fixed cells stained with fluorophore-conjugated antibodies; RNA extracted and analyzed via PCR for pluripotency genes. |
Comparative Analysis of Factor Combinations
The choice of reprogramming factors is a critical determinant of the resulting cells' stability. Comparisons of common combinations reveal distinct safety and efficacy profiles.
Table 2: Stability and Efficiency Profile of Different Reprogramming Factor Combinations
| Factor Combination | Reported Reprogramming Efficiency | Key Findings on Karyotype Stability | Key Findings on Epigenetic Rejuvenation | Associated Tumorigenic Risk |
|---|---|---|---|---|
| OSKM (Original Yamanaka) [66] [71] | High | Higher incidence of abnormalities; c-MYC is a known oncogene. | Effective but accompanied by full dedifferentiation. | High (teratoma formation in mice) [66]. |
| OSK (Excluding c-MYC) [14] [12] | Moderate | Significantly improved; exclusion of c-MYC reduces oncogenic pressure. | Effective age reversal in vivo; restores youthful DNA methylation and transcriptome. | Substantially reduced. |
| Chemical Cocktails (Non-Genetic) [14] [12] | Low to Moderate | Avoids risks of genetic integration; no evidence of genomic instability from method. | Reverses transcriptomic age and restores nuclear compartmentalization in human cells. | Low (no genetic manipulation). |
| OSK with AAV9 Delivery (In Vivo) [14] | N/A (In vivo application) | No teratomas reported in mouse studies. | Reversed epigenetic age, improved healthspan, and extended lifespan in aged mice. | Low with cyclic, controlled expression. |
The Gold Standard and Its Drawbacks
The original OSKM combination remains a benchmark for high-efficiency reprogramming. However, the inclusion of c-MYC, a potent oncogene, directly contributes to a high risk of tumorigenesis and karyotypic instability [66] [71]. Studies have shown that mice transplanted with iPSCs generated using c-MYC frequently developed teratomas [66]. This significant safety concern has driven the search for safer alternatives.
Safer Alternatives: OSK and Chemical Reprogramming
Omitting c-MYC from the cocktail (using only OSK) substantially reduces tumorigenic risk. Research demonstrates that OSK-mediated partial reprogramming can reverse the epigenetic clock and restore youthful gene expression patterns in vivo without causing teratomas [14] [12]. For instance, in vivo delivery of OSK via an AAV9 vector in aged mice extended their remaining lifespan by 109% and reduced frailty, without evidence of cancer formation [14].
Notably, a study on Werner syndrome (a premature aging disorder) cells demonstrated that reprogramming with OSKM not only suppressed premature senescence but also maintained a stable karyotype over a two-year long-term culture [73]. This suggests that the reprogramming process itself, when followed by stable pluripotency, can confer genomic stability, possibly through the reactivation of telomerase [73].
Fully chemical reprogramming represents the safest approach from a genetic integrity standpoint, as it avoids exogenous genetic material entirely. Specific chemical cocktails, such as the "7c" cocktail, have been shown to rejuvenate human cells, reversing transcriptomic age and restoring youthful nucleocytoplasmic compartmentalization [14] [12].
The Scientist's Toolkit: Essential Research Reagents
Successful reprogramming and stability analysis require a suite of specialized reagents and tools.
Table 3: Essential Reagents and Tools for Reprogramming and Stability Research
| Category / Reagent | Specific Examples | Function in Reprogramming & Analysis |
|---|---|---|
| Core Transcription Factors | OCT4, SOX2, KLF4, c-MYC | Master regulators that initiate epigenetic remodeling and dedifferentiation. |
| Delivery Vectors | Episomal vectors, Sendai virus, mRNA [71] | Methods to introduce reprogramming factors into somatic cells; non-integrating vectors are preferred for safety. |
| Cell Culture Supplements | Basic FGF, Knockout Serum Replacement [74] | Supports the survival and proliferation of pluripotent stem cells. |
| Senescence & Aging Reporters | NCC Assay (NLS-mCherry, NES-eGFP) [12] | Quantifies age-related breakdown in nuclear integrity; used for high-throughput screening of rejuvenating compounds. |
| Pluripotency Reporters | Oct4-EGFP transgenic system [74] | Allows real-time, non-invasive monitoring of pluripotency status in live cells. |
Visualizing Workflows and Pathways
The following diagrams summarize the core experimental workflows and biological pathways discussed in this guide.
In Vitro Reprogramming and Stability Assessment Workflow
Diagram Title: In Vitro Reprogramming and Stability Assessment Workflow
Partial vs. Full Reprogramming Pathways
Diagram Title: Partial vs. Full Reprogramming Pathways
The pursuit of clinically viable cell reprogramming therapies necessitates a careful balance between efficiency and safety. The experimental data clearly indicate that moving away from the original OSKM combination toward c-MYC-free protocols like OSK and fully non-integrating chemical methods provides a more favorable path for achieving long-term karyotypic and epigenetic stability [14] [12] [71]. The emergence of partial reprogramming strategies, which reverse hallmarks of aging without erasing cellular identity, is particularly promising for treating age-related diseases [14] [72].
Future work must focus on standardizing reprogramming protocols and stability assessment criteria to enable direct comparisons between studies. Furthermore, the development of more precise inducible systems and the discovery of novel, safer reprogramming factors—such as the single-gene target SB000 reported by Shift Bioscience—will be crucial for translating the revolutionary potential of cellular reprogramming into safe and effective human therapies [75].
The strategic formulation of optimized biological cocktails represents a paradigm shift in treating complex genetic and neurodegenerative diseases. This guide compares the application of these sophisticated combinations in two distinct areas: amyotrophic lateral sclerosis (ALS) modeling using stem cell-based cocktails and thalassemia treatment employing antioxidant and gene-editing formulations. The core thesis demonstrates that moving beyond single-target therapies to multi-component, rationally designed cocktails significantly enhances efficacy, addressing the multifaceted pathology of these diseases. The approaches in both fields share a common principle derived from a broader context of efficiency comparisons in biological factor combinations, such as Yamanaka factor research, highlighting how synergistic component interactions can yield outcomes superior to the sum of individual parts.
Comparative Efficacy of Cocktail Formulations
Table 1: Performance Comparison of Cocktail Therapies in Thalassemia and ALS
| Disease Area | Cocktail Type | Key Components | Primary Efficacy Outcomes | Study Phase |
|---|---|---|---|---|
| Transfusion-Dependent Beta Thalassemia (TDT) | CRISPR/Cas9 Gene-Editing ( [76] [77]) | CASGEVY (exagamglogene autotemcel) - autologous hematopoietic stem cells edited at BCL11A gene | 98.2% (54/55) achieved transfusion independence for ≥12 months; Mean duration: 40.5 months; Improved iron overload parameters [77] | Approved Therapy (2025 Long-Term Data) |
| β-Thalassemia/Hemoglobin E | Antioxidant + Iron Chelator ( [78] [79]) | Cocktail A: Curcuminoids (500 mg/day) + N-acetylcysteine (200 mg/day) + Deferiprone (50 mg/kg/day) [78] | Responders (Cocktail A): 11% max. Hb increase at 4 months; Responders (Cocktail B): 10% max. Hb increase at 10 months; Reduced oxidative stress & improved hypercoagulable state [78] | Clinical Trial (60 patients) |
| β-Thalassemia/Hemoglobin E | Antioxidant + Iron Chelator ( [78]) | Cocktail B: Vitamin E (400 IU/day) + N-acetylcysteine (200 mg/day) + Deferiprone (50 mg/kg/day) [78] | Same as above - differentiation by responder status rather than cocktail type [78] | Clinical Trial (60 patients) |
| ALS (Stem Cell Therapy) | Mesenchymal Stem Cells (MSCs) ( [80]) | Autologous bone marrow-derived MSCs delivered via intrathecal injection | Improved neuroprotection and regenerative qualities; Limited by small sample sizes and non-randomized designs in trials [80] | First Licensed Therapy (2015, South Korea) |
| ALS (Stem Cell Therapy) | Induced Pluripotent Stem Cells (iPSCs) ( [80]) | Patient-specific somatic cells reprogrammed using defined factors | Potential for neuronal replacement/support; Addresses ethical concerns of embryonic stem cells; Challenges: genotoxicity, epigenetic irregularities [80] | Preclinical/Research |
Experimental Protocols & Methodologies
Protocol for Antioxidant Cocktail Treatment in β-Thalassemia/HbE
Objective: Evaluate the effects of two antioxidant cocktails on iron load, oxidative stress, antioxidant status, blood coagulation potential, and anemia in patients with β-thalassemia/HbE [78].
Patient Selection:
- Genotype βâ°/βᴇ, age 18-50 years, Hb concentration 50–90 g/L.
- Excluded transfusion-dependent patients to eliminate impact of donor blood on parameters.
- 60 patients divided into two groups (n=30 each) using allocation concealment [78].
Intervention:
- CUR Group: Received 500 mg/day curcuminoids + 200 mg/day N-acetylcysteine + 50 mg/kg/day deferiprone.
- Vit-E Group: Received 400 IU/day vitamin E + 200 mg/day N-acetylcysteine + 50 mg/kg/day deferiprone.
- Treatment duration: 12 months [78].
Outcome Measurements:
- Hematological Parameters: Complete blood count (automated analyzer).
- Iron Load: Serum ferritin level, non-transferrin-bound iron (NTBI).
- Oxidative Stress: Blood ROS levels (flow cytometry), RBC malondialdehyde (MDA) concentration.
- Antioxidant Parameters: Superoxide dismutase (SOD) activity, glutathione peroxidase (GPx) activity, glutathione (GSH) levels.
- Coagulation Parameters: Platelet factor 3-like activity, phosphatidylserine-exposed RBC and platelets (flow cytometry) [78].
Responder Classification: Patients classified as "responders" if showing >20% decrease in both serum ferritin and RBC MDA levels after 4 months of treatment [78].
Protocol for CRISPR/Cas9 Gene Therapy in Thalassemia
Objective: Assess safety and efficacy of CASGEVY single-dose infusion in patients with transfusion-dependent beta thalassemia (TDT) [77].
Study Design:
- Ongoing Phase 1/2/3 open-label trials (CLIMB-111, CLIMB-121, CLIMB-131).
- Patients aged 12-35 years with TDT or SCD.
- Long-term follow-up trial (CLIMB-131) designed to follow patients for up to 15 years post-infusion [77].
Intervention:
- CASGEVY Manufacturing: Patient's own hematopoietic stem and progenitor cells edited ex vivo at erythroid-specific enhancer region of BCL11A gene using CRISPR/Cas9.
- Conditioning: Myeloablative conditioning with busulfan.
- Infusion: Single dose of autologous CRISPR-edited cells [77].
Primary Efficacy Endpoint:
- Transfusion independence for at least 12 consecutive months (TI12) with weighted average hemoglobin ≥9 g/dL [77].
Safety Assessment:
- Generally consistent with myeloablative conditioning with busulfan and autologous hematopoietic stem cell transplant [77].
Protocol for Stem Cell Therapy in ALS Modeling
Objective: Compare efficacy of three stem cell types for ALS treatment: mesenchymal stem cells (MSCs), neural stem cells (NSCs), and induced pluripotent stem cells (iPSCs) [80].
Cell Source and Preparation:
- MSCs: Derived from various tissues, most frequently from bone marrow stem cells (BMSCs).
- NSCs: Isolated from fetal spinal cord or brain.
- iPSCs: Created by reprogramming patient-specific somatic cells using defined factors, potentially including Yamanaka factors (Oct4, Sox2, Klf4, c-Myc) [80].
Delivery Method:
- Intrathecal injection for MSCs (as in first licensed therapy by CORESTEM).
- Direct transplantation to target areas for NSCs.
- Differentiation into motor neurons or supportive glial cells for iPSCs [80].
Outcome Assessment:
- Functional Integration: Markers like microtubule-associated protein 2 (MAP2) to assess neuronal maturation and dendritic growth.
- Safety Monitoring: Biomarkers including p53 and c-Myc to evaluate genetic stability and tumor formation risks.
- Disease Biomarkers: Neurofilament light (NfL) and glial fibrillary acidic protein (GFAP) to track neurodegeneration and glial activation [80].
Signaling Pathways and Molecular Mechanisms
CASGEVY (CRISPR/Cas9) Mechanism of Action in Thalassemia
This CRISPR-based approach targets the fundamental genetic pathology of thalassemia by disrupting the BCL11A gene, a key repressor of fetal hemoglobin (HbF) [77]. The edit results in sustained production of high levels of HbF in red blood cells, which compensates for the deficient adult hemoglobin, ultimately reducing or eliminating transfusion requirements [76] [77].
Antioxidant Cocktail Mechanism in Thalassemia Pathophysiology
The antioxidant cocktails target multiple pathways in the thalassemia pathophysiology cascade. The combination of hydrophobic antioxidants (curcuminoids/vitamin E), hydrophilic antioxidant (N-acetylcysteine), and iron chelator (deferiprone) simultaneously addresses ROS production, iron-mediated oxidative stress, and cellular damage [78]. This multi-target approach results in decreased oxidative stress, increased hemoglobin concentration, and improvement of the hypercoagulable state [78] [79].
Stem Cell Cocktail Mechanisms in ALS Modeling
The stem cell approaches in ALS employ different but complementary mechanisms. MSCs primarily provide neuroprotective and immunomodulatory support through paracrine signaling. iPSCs offer the potential for personalized neuronal replacement and disease modeling. NSCs aim to directly integrate into existing neural circuits to replace damaged motor neurons [80]. The efficiency and safety of these approaches, particularly iPSCs, can be influenced by the specific combination of reprogramming factors used, drawing from research on Yamanaka factor optimization [18] [80].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for Cocktail Optimization Studies
| Reagent Category | Specific Examples | Research Function | Application Context |
|---|---|---|---|
| Reprogramming Factors | Oct4, Sox2, Klf4, c-Myc (OSKM) [66] [18] | Somatic cell reprogramming to iPSCs | ALS modeling, disease mechanism studies [80] |
| Gene Editing Tools | CRISPR/Cas9 systems [76] [77] | Precise genetic modification | BCL11A editing for thalassemia (CASGEVY) [77] |
| Antioxidants (Hydrophobic) | Curcuminoids, Vitamin E (α-tocopherol) [78] | Lipid-soluble ROS scavengers, membrane protection | Thalassemia antioxidant cocktails [78] |
| Antioxidants (Hydrophilic) | N-acetylcysteine (NAC) [78] | Precursor for glutathione synthesis, direct ROS neutralization | Thalassemia antioxidant cocktails [78] |
| Iron Chelators | Deferiprone [78] | Reduction of iron overload, decreased NTBI | Thalassemia combination therapy [78] |
| Stem Cell Culture Supplements | Growth factors, small molecules [80] | Maintenance of pluripotency, directed differentiation | ALS stem cell therapy production [80] |
| Vector Systems | Lentiviral, episomal, adenoviral vectors [66] [80] | Delivery of genetic material | Factor expression in reprogramming & gene therapy [80] |
The comparative analysis of optimized cocktail applications in thalassemia and ALS research reveals several universal principles for complex disease targeting:
Synergistic Component Selection: Successful cocktails combine agents with complementary mechanisms—such as hydrophilic and hydrophobic antioxidants in thalassemia or neuroprotective and immunomodulatory approaches in ALS—that target multiple disease pathways simultaneously [78] [80].
Precision Molecular Targeting: The most transformative outcomes emerge from cocktails designed with precise molecular knowledge, exemplified by CRISPR targeting of BCL11A in thalassemia and factor-based reprogramming for ALS modeling [76] [77].
Patient Stratification Strategies: Efficacy optimization often requires identifying "responder" populations based on biomarkers, as demonstrated in the thalassemia antioxidant trials where patients with improved oxidative stress markers showed the most significant hemoglobin increases [78].
The evolution of cocktail optimization continues to advance with newer technologies, including third-generation sequencing for precise mutation detection [76] and refined factor combinations that improve the developmental potential of generated cells [18]. These approaches, built on the foundational research of factor combinations like the Yamanaka factors, provide a roadmap for developing increasingly effective multi-target therapies for complex genetic and neurodegenerative disorders.
Conclusion
The comparative analysis of Yamanaka factor combinations reveals a clear trade-off between reprogramming efficiency and clinical safety. While the original OSKM cocktail remains highly efficient, the omission of the oncogene c-MYC and the use of alternative factors like L-MYC or NANOG significantly enhance the safety profile for therapeutic applications. The choice of delivery system, particularly non-integrating methods like Sendai virus, and the supplementation with small molecules are critical for optimizing outcomes. Future directions should focus on standardizing reprogramming protocols, developing more precise temporal control over factor expression for partial reprogramming, and advancing non-integrating, factor-free chemical reprogramming methods. The continued refinement of these combinations is paramount for unlocking the full potential of iPSCs in regenerative medicine, personalized drug screening, and the treatment of complex diseases.
References
- 1. A comprehensive analysis of induced pluripotent stem cell ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1593207/full]
- 2. Induced pluripotent stem cells (iPSCs) [https://www.nature.com/articles/s41392-024-01809-0]
- 3. Cell reprogramming: methods, mechanisms and applications [https://cellregeneration.springeropen.com/articles/10.1186/s13619-025-00229-x]
- 4. Comparison of reprogramming factor targets reveals both ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6303873/]
- 5. Accelerating life sciences research [https://openai.com/index/accelerating-life-sciences-research-with-retro-biosciences/]
- 6. Advances in Induced Pluripotent Stem Cell Reprogramming ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12577567/]
- 7. Reprogramming to restore youthful epigenetics of ... [https://www.nature.com/articles/s41413-025-00416-1]
- 8. Sequential factor delivery enables efficient workflow for ... [https://www.nature.com/articles/s41598-025-17876-4]
- 9. Ethical and Safety Issues of Stem Cell-Based Therapy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5765738/]
- 10. Comparative study of human-induced pluripotent stem ... [https://pubmed.ncbi.nlm.nih.gov/22798560/]
- 11. A new kind of stem cell is revolutionizing regenerative ... [https://www.asbmb.org/asbmb-today/science/040125/stem-cells-revolutionize-regenerative-medicine]
- 12. Chemically induced reprogramming to reverse cellular aging [https://pmc.ncbi.nlm.nih.gov/articles/PMC10373966/]
- 13. Exploring 7c versus 2c Small Molecule Reprogramming ... [https://www.fightaging.org/archives/2025/07/exploring-7c-versus-2c-small-molecule-reprogramming-combinations-for-rejuvenation/]
- 14. The long and winding road of reprogramming-induced ... [https://www.nature.com/articles/s41467-024-46020-5]
- 15. SB000: a safer path to anti-aging therapies [https://www.drugtargetreview.com/news/165239/sb000-a-safer-path-to-anti-aging-therapies/]
- 16. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in ... [https://genomebiology.biomedcentral.com/articles/10.1186/gb-2012-13-10-251]
- 17. Cell-type-specific functionality encoded within the ... [https://www.nature.com/articles/s41467-025-63806-3]
- 18. Excluding Oct4 from Yamanaka Cocktail Unleashes the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6900749/]
- 19. The reprogramming factor KLF4 in normal and malignant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12206627/]
- 20. Insights into cancer, stem cell biology, and PPI networks [https://www.sciencedirect.com/science/article/abs/pii/S0378111924003287]
- 21. A comprehensive analysis of induced pluripotent stem cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12095295/]
- 22. Optimized Yamanaka factors combined with TERT gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12617746/]
- 23. Using human pluripotent stem cells to untangle ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11115022/]
- 24. A Review of the Present State of Epigenetic ... [https://www.fightaging.org/archives/2025/10/a-review-of-the-present-state-of-epigenetic-reprogramming-to-treat-aging/]
- 25. Current advances and future prospects of cell ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1546423/full]
- 26. Optimizing viral transduction in immune cell therapy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12046913/]
- 27. Human iPSC Reprogramming Success: The Impact of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11756937/]
- 28. Epigenetic-scale comparison of human iPSCs generated ... [https://www.sciencedirect.com/science/article/pii/S2352320418300324]
- 29. A comparison of non-integrating reprogramming methods [https://pmc.ncbi.nlm.nih.gov/articles/PMC4329913/]
- 30. Human iPSC Reprogramming Success: The Impact ... - PubMed [https://pubmed.ncbi.nlm.nih.gov/39850337/]
- 31. Comparison of the Efficiency of Viral Transduction and ... [https://link.springer.com/article/10.1007/s10517-015-3112-5]
- 32. Reprogramming Fibroblasts with the CytoTune®-iPS ... [https://www.thermofisher.com/us/en/home/references/protocols/cell-culture/stem-cell-protocols/ipsc-protocols/reprogramming-fibroblasts-with-cytotune-ips-reprogramming-it.html]
- 33. A Review of the Methods for Human iPSC Derivation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4176696/]
- 34. Development of a High-Efficacy Reprogramming Method ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7555779/]
- 35. Sendai virus persistence questions the transient naive ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10979911/]
- 36. Chemical reprogramming of human blood cells to ... [https://www.sciencedirect.com/science/article/abs/pii/S1934590925002607]
- 37. NEUROD1 efficiently converts peripheral blood cells into ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12067290/]
- 38. Generation of hypoimmunogenic universal iPS cells ... [https://www.nature.com/articles/s12276-025-01422-3]
- 39. Progress in Separating Rejuvenation from Pluripotency in ... [https://www.fightaging.org/archives/2025/06/progress-in-separating-rejuvenation-from-pluripotency-in-cell-reprogramming/]
- 40. Small molecules for cell reprogramming: a systems biology ... [https://www.aging-us.com/article/203791/text]
- 41. Chemical reprogramming ameliorates cellular hallmarks of ... [https://pubmed.ncbi.nlm.nih.gov/40588563/]
- 42. Potential Breakthrough in Cell Reprogramming as OpenAI ... [https://www.biopharmatrend.com/news/potential-breakthrough-in-cell-reprogramming-as-openai-and-retro-biosciences-report-50x-pluripotency-marker-expression-gains-1357/]
- 43. Induced Pluripotent Stem Cell (iPSC) Industry Trends for ... [https://bioinformant.com/ipsc-industry-trends/]
- 44. The Twin Potentials of iPS Research: Rejuvenation ... [https://www.nippon.com/en/japan-topics/c15102/the-twin-potentials-of-ips-research-rejuvenation-promise-and-cancer-risk.html]
- 45. Human Stem Cells in Disease Modelling and Treatment [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467989/]
- 46. Rejuvenation Roundup September 2025 [https://www.lifespan.io/news/rejuvenation-roundup-september-2025/]
- 47. Possible Strategies to Reduce the Tumorigenic Risk of ... [https://www.mdpi.com/1422-0067/25/10/5177]
- 48. MYC as a Target for Cancer Treatment: from Undruggable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12454499/]
- 49. Fueling the fire–a pan-cancer analysis of MYC-regulated ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1669544/full]
- 50. A cyclic AMP analog, 8-Br-cAMP, enhances the induction ... [https://pubmed.ncbi.nlm.nih.gov/21120637/]
- 51. Role of small molecules as drug candidates for ... [https://www.sciencedirect.com/science/article/abs/pii/S0010482524007467]
- 52. Roles of small molecules in somatic cell reprogramming [https://www.nature.com/articles/aps201373]
- 53. 8-Bromo-cAMP [https://www.stemcell.com/products/8-bromo-camp.html]
- 54. Chemicals as the Sole Transformers of Cell Fate - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4961099/]
- 55. MicroRNA Roles in Cell Reprogramming Mechanisms [https://www.mdpi.com/2073-4409/11/6/940]
- 56. Elevated p53 Activities Restrict Differentiation Potential ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5688240/]
- 57. Mechanisms, pathways and strategies for rejuvenation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11058000/]
- 58. Partial cellular reprogramming: A deep dive into an emerging ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10861195/]
- 59. Sequential addition of reprogramming factors improves ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5842428/]
- 60. Progress in understanding reprogramming to the induced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3273493/]
- 61. Facilitators and Impediments of the Pluripotency ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3508134/]
- 62. Oct4, Sox2, Klf4, c-My (OSKM) gene therapy in ... [https://www.aging-us.com/article/206191/text]
- 63. Oct4, Sox2, Klf4, c-My (OSKM) gene therapy in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11810065/]
- 64. The occurrence and development of induced pluripotent ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2024.1389558/full]
- 65. Genomic integrity of human induced pluripotent stem cells [https://pmc.ncbi.nlm.nih.gov/articles/PMC6828592/]
- 66. Application of the Yamanaka Transcription Factors Oct4 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10531188/]
- 67. Differentiation Potential of Induced Pluripotent Stem Cells [https://www.rndsystems.com/resources/articles/differentiation-potential-induced-pluripotent-stem-cells]
- 68. Pluripotency factors are repurposed to shape the epigenomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9743141/]
- 69. Human-Induced Pluripotent Stem Cells (iPSCs) for Disease ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12345660/]
- 70. Directed Differentiation of Pluripotent Stem Cells by ... [https://www.sciencedirect.com/science/article/pii/S1016847823003898]
- 71. The Challenges to Advancing Induced Pluripotent Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10768945/]
- 72. Cellular reprogramming and epigenetic rejuvenation [https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-021-01158-7]
- 73. Reprogramming Suppresses Premature Senescence ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0112900]
- 74. A real-time pluripotency reporter for the long-term and ... [https://www.aging-us.com/article/204083/text]
- 75. Novel single-gene target reverses cellular ageing [https://www.ddw-online.com/novel-single-gene-target-reverses-cellular-ageing-35271-202506/]
- 76. Advanced molecular approaches to thalassemia disorder and the selection of molecular-level diagnostic testing in resource-limited settings - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12206028/]
- 77. Vertex Presents Longer-Term Data at the 2025 European Hematology Association (EHA) Congress Demonstrating Durability of CASGEVY® and Provides Update on Expanding Global Access to CASGEVY | Vertex Pharmaceuticals [https://investors.vrtx.com/news-releases/news-release-details/vertex-presents-longer-term-data-2025-european-hematology]
- 78. Treatment of β-Thalassemia/Hemoglobin E with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4452506/]
- 79. Treatment of β-Thalassemia/Hemoglobin E with ... [https://pubmed.ncbi.nlm.nih.gov/26078808/]
- 80. Stem Cell Therapy for the Treatment of Amyotrophic Lateral ... [https://www.mdpi.com/2227-9059/13/1/35]