Preventing Exosome Aggregation: A Comprehensive Guide to Stable Storage and Freeze-Thaw Protocols for Translational Research
This article provides researchers, scientists, and drug development professionals with a systematic framework for preserving exosome integrity during storage and processing.
Preventing Exosome Aggregation: A Comprehensive Guide to Stable Storage and Freeze-Thaw Protocols for Translational Research
Abstract
This article provides researchers, scientists, and drug development professionals with a systematic framework for preserving exosome integrity during storage and processing. It synthesizes the latest evidence on the fundamental causes of exosome aggregation and instability, presents optimized methodological protocols for both short- and long-term preservation, offers troubleshooting strategies for common challenges, and establishes validation criteria for assessing exosome quality post-storage. The guidance is designed to enhance reproducibility and accelerate the clinical translation of exosome-based diagnostics and therapeutics.
Understanding Exosome Aggregation: The Science of Instability During Storage
Frequently Asked Questions (FAQs)
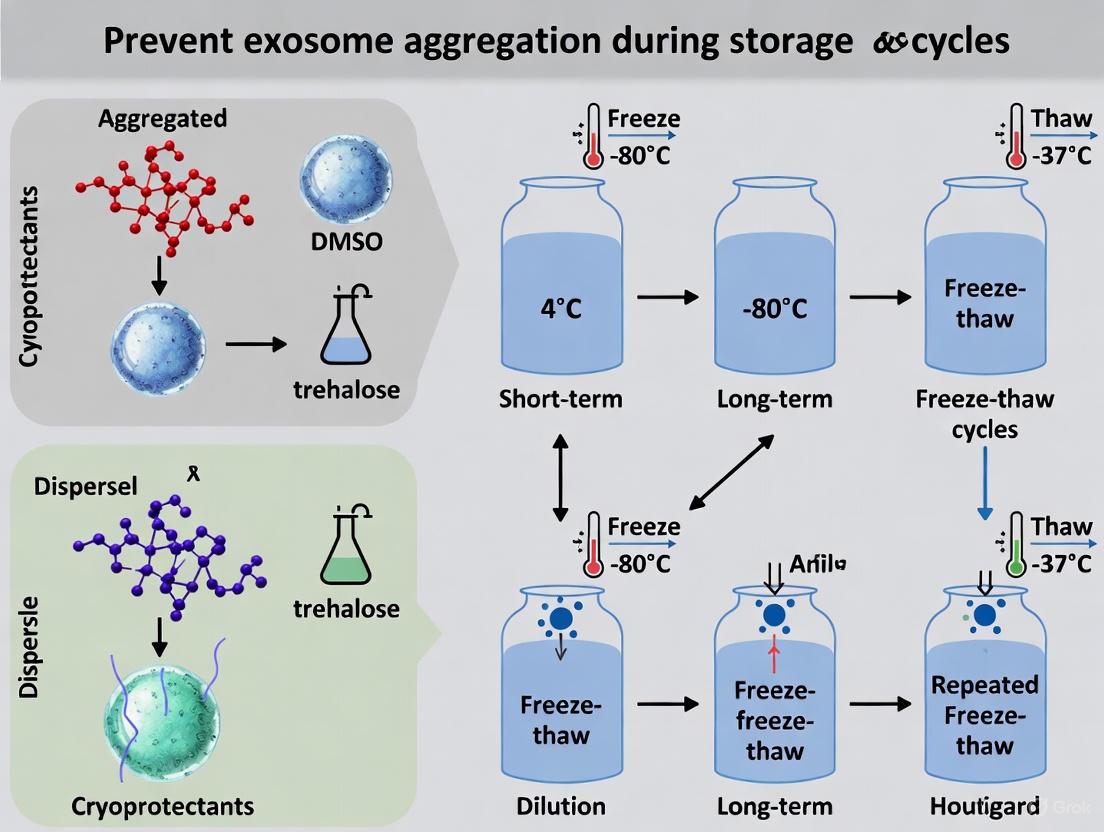
What are the primary causes of exosome aggregation during storage? Exosome aggregation occurs primarily due to insufficient electrostatic repulsion between particles and physical stresses induced by freezing. When the absolute value of the surface charge (zeta potential) approaches zero, electrostatic repulsion fails to counteract natural attractive forces, leading to clumping [1]. Furthermore, during freezing, the formation of ice crystals can disrupt vesicle membranes and force particles into close proximity, promoting fusion and aggregation [2] [3] [4].
How does aggregation affect the biological function of my exosome samples? Aggregation directly compromises exosome function by reducing their bioavailability and altering their interaction with recipient cells. Studies show that aggregated exosomes exhibit inconsistent biological activity. For instance, one study demonstrated that aggregated beta-cell extracellular vesicles showed lower TNF-alpha cytokine secretion stimulation indexes in macrophage immune assays compared to well-dispersed samples, indicating impaired bioactivity [1]. Furthermore, aggregation can change cellular uptake patterns and biodistribution, fundamentally altering experimental outcomes [5].
Can I simply vortex or pipette my exosome samples to reverse aggregation? Forceful mechanical disruption like vortexing is not recommended as it can damage exosome integrity through shear forces, potentially causing membrane rupture and cargo leakage. While gentle pipetting might disperse loose aggregates, it is ineffective for fusion-induced aggregates. The preferred approach is preventive—using appropriate buffers and storage conditions to avoid aggregation from the outset [1] [2].
How do freeze-thaw cycles contribute to exosome aggregation? Each freeze-thaw cycle subjects exosomes to substantial physical stress. Research demonstrates that multiple freeze-thaw cycles lead to a significant decrease in particle concentration, an increase in average particle size, and a wider size distribution—all indicative of aggregation and fusion events [5] [4]. One study employing fluorescently tagged exosomes provided direct evidence of fusion phenomena after freeze-thaw cycles, observing a new population of double-positive particles that did not exist in fresh samples [4].
Is storing exosomes in pure PBS sufficient to prevent aggregation? No, storage in phosphate-buffered saline (PBS) alone is suboptimal for preventing aggregation. PBS lacks protective agents to shield exosomes from freezing-induced damage and aggregation. Multiple studies have shown that exosomes stored in PBS exhibit increased particle size and aggregation after freezing compared to those stored in specialized buffers containing cryoprotectants like trehalose [1] [4].
Troubleshooting Guides
Problem: Increased Particle Size After Storage
Symptoms:
- Nanoparticle Tracking Analysis (NTA) shows an increase in mean and mode particle size.
- Broader size distribution (increased polydispersity index).
- Visible clumping or precipitation in the sample vial.
Solutions:
- Revise Storage Buffer: Add 25 mM trehalose to your PBS storage buffer. Research shows this natural disaccharide narrows particle size distribution and maintains individual particle integrity by preventing fusion and aggregation [1].
- Optimize Freezing Protocol: Implement rapid freezing rates (snap-freezing in liquid nitrogen or dry ice-ethanol baths) to minimize ice crystal formation that promotes aggregation [2] [3].
- Avoid Freeze-Thaw Cycles: Aliquot exosomes into single-use volumes to avoid repeated freezing and thawing, which significantly increases aggregation [5] [4].
Problem: Loss of Biological Activity After Freezing
Symptoms:
- Reduced efficacy in functional assays (e.g., cell uptake, immune response activation).
- Inconsistent results between freshly isolated and frozen batches.
- Decreased therapeutic potency in in vivo models.
Solutions:
- Use Cryoprotectants: Incorporate 25-50 mM trehalose into isolation and storage buffers. Studies confirm that trehalose preserves exosome functionality, as evidenced by consistently higher TNF-alpha stimulation indexes in immune assays [1].
- Control Storage Temperature: For long-term preservation, store exosomes at -80°C. Evidence indicates that -80°C storage better preserves biological functionality compared to -20°C [5] [3].
- Consider Lyophilization: For certain applications, lyophilization (freeze-drying) in the presence of trehalose can provide excellent stability while maintaining function, though this requires optimization for different exosome sources [2].
Problem: Low Particle Recovery After Thawing
Symptoms:
- Significant decrease in particle concentration measured by NTA after thawing.
- High loss of sample during post-thaw handling.
- Increased protein-to-particle ratio, indicating preferential particle loss.
Solutions:
- Minimize Freeze-Thaw Cycles: Subjecting exosomes to multiple freeze-thaw cycles dramatically reduces particle concentration. One study showed significant losses after just the first cycle [4].
- Use Appropriate Containers: Store exosomes in siliconized low-protein-binding vials to minimize adhesion to container walls [4].
- Add Stabilizing Agents: Beyond trehalose, consider other stabilizers like human serum albumin (HSA) or polyethylene glycol (PEG) to protect against surface adsorption and aggregation [2].
Data Presentation Tables
Table 1: Impact of Storage Temperature on Exosome Integrity Over Time
Data compiled from multiple studies [5] [3]
| Storage Temperature | Storage Duration | Particle Concentration Recovery | Mean Size Change | RNA Content Preservation | Recommended Use Case |
|---|---|---|---|---|---|
| 4°C | 7 days | ~60-70% | +15-25% | ~50-60% | Short-term experiments (<1 week) |
| -20°C | 28 days | ~50-60% | +30-50% | ~40-50% | Temporary storage (2-4 weeks) |
| -80°C | 28 days | ~85-90% | +10-15% | ~80-85% | Long-term preservation (>1 month) |
| -80°C with Trehalose | 28 days | ~90-95% | +5-10% | ~85-90% | Critical long-term applications |
Table 2: Effects of Multiple Freeze-Thaw Cycles on Exosome Quality
Quantitative data showing degradation trends [5] [4]
| Number of Freeze-Thaw Cycles | Particle Recovery (%) | Mean Size Increase | RNA Integrity | Functional Activity Retention |
|---|---|---|---|---|
| 0 (Fresh) | 100% | Baseline | 100% | 100% |
| 1 | 70-80% | 15-20% | 80-85% | 75-80% |
| 3 | 40-50% | 35-50% | 50-60% | 40-50% |
| 5 | 20-30% | 60-80% | 20-30% | 15-25% |
Experimental Protocols
Protocol: Evaluating Exosome Stability with Trehalose
Objective: To assess the protective effect of trehalose on exosome integrity during storage and freeze-thaw cycles.
Materials:
- Purified exosome sample
- Phosphate-buffered saline (PBS)
- D-(+)-Trehalose dihydrate
- Ultracentrifuge and tubes
- Nanoparticle Tracking Analysis instrument
- Materials for functional assay (e.g., macrophage TNF-α secretion assay)
Methodology:
- Sample Preparation: Isolate exosomes using your standard method (e.g., differential ultracentrifugation). Split the purified exosome sample into two equal aliquots.
- Buffer Exchange: Resuspend one aliquot in PBS (control) and the other in PBS containing 25 mM trehalose (experimental) [1].
- Baseline Characterization: Analyze both samples using NTA to determine initial particle concentration, size distribution, and zeta potential. Assess biological activity using your relevant functional assay.
- Storage Intervention: Divide each sample further into multiple aliquots. Subject aliquots to:
- Short-term storage: 4°C for 1 week
- Long-term storage: -80°C for 4 weeks
- Freeze-thaw stress: 1, 3, and 5 cycles between -80°C and room temperature
- Post-Storage Analysis: After each storage condition, repeat the characterization in step 3. Compare the results to baseline measurements and between PBS and trehalose groups.
Expected Outcomes: Exosomes stored in trehalose should demonstrate higher particle recovery, minimal size increase, narrower size distribution, and better preservation of biological function compared to PBS-stored controls, particularly after freeze-thaw cycles [1].
Protocol: Testing Exosome Fusion During Storage
Objective: To detect fusion events between distinct exosome populations during storage using fluorescent tagging.
Materials:
- Two cell lines expressing different fluorescent membrane proteins (e.g., GFP and mCherry)
- Standard exosome isolation equipment
- Flow cytometer equipped for vesicle analysis
- Cryogenic vials for storage
Methodology:
- Fluorescent Exosome Production: Culture two separate populations of donor cells—one expressing GFP-tagged membrane protein and another expressing mCherry-tagged membrane protein.
- Isolation and Mixing: Isolve exosomes from each cell line separately using standard methods. Mix the two exosome populations in equal particle concentrations to create a fresh control sample [4].
- Storage and Analysis: Split the mixed sample into aliquots and subject them to different storage conditions (e.g., -80°C in PBS, -80°C with cryoprotectant). Analyze by flow cytometry immediately after mixing (fresh control) and after each storage condition.
- Fusion Detection: In fresh samples, expect two distinct populations: GFP-positive and mCherry-positive. The appearance of a double-positive (GFP+mCherry+) population after storage indicates fusion events between vesicles from the two different sources [4].
Interpretation: The percentage of double-positive events quantifies the extent of fusion occurring during storage. Effective cryoprotectants should minimize the emergence of this population.
Signaling Pathways, Workflows, and Relationships
Diagram: Experimental Workflow for Testing Storage Conditions
Diagram: Mechanisms of Exosome Aggregation and Protection
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for Preventing Exosome Aggregation
| Reagent | Function/Mechanism | Application Protocol | Key References |
|---|---|---|---|
| Trehalose | Natural cryoprotectant that stabilizes membranes through water replacement and vitrification mechanisms; prevents fusion and aggregation | Add to PBS at 25-50 mM final concentration for exosome resuspension and storage | [1] [2] |
| Dimethyl Sulfoxide (DMSO) | Penetrating cryoprotectant that reduces ice crystal formation; use at low concentrations to minimize toxicity | 6-10% in PBS; requires removal before functional assays | [4] |
| Protease Inhibitor Cocktails | Prevent protein degradation that can expose hydrophobic regions and promote aggregation | Add according to manufacturer instructions during isolation and storage | [4] |
| Siliconized Low-Binding Tubes | Minimize surface adhesion and particle loss during storage and handling | Use for all exosome storage and processing steps | [4] |
| Glycerol | Non-penetrating cryoprotectant that provides extracellular protection; less effective than trehalose for exosomes | 30% in PBS; may interfere with some downstream applications | [4] |
Frequently Asked Questions: Managing Exosome Aggregation
What are the primary triggers for exosome aggregation during storage? The main triggers are multiple freeze-thaw cycles and storage in simple buffers like PBS without protective additives. Freeze-thaw cycles cause ice crystal formation, which can mechanically damage exosome membranes and promote fusion. Storage at standard freezer temperatures (e.g., -20°C or -80°C) can still lead to slow aggregation and a reduction in sample purity over time [4].
How do freeze-thaw cycles damage my exosome samples? Each freeze-thaw cycle inflicts cumulative damage. The freezing process leads to ice crystal formation, which can pierce and disrupt the exosome lipid bilayer. Upon thawing, this damage manifests as vesicle fusion, increased particle size, and a loss of individual particles. Multiple cycles significantly decrease particle concentration and increase sample heterogeneity [6] [4].
What is the best temperature for short-term storage of exosomes? For short-term storage (e.g., 24 hours), 4°C has been shown to maintain exosome concentration and marker proteins better than -80°C, -20°C, or higher temperatures [6]. For any storage beyond a few days, freezing at -80°C with a cryoprotectant is recommended [2].
Can I simply avoid aggregation by storing my samples at -80°C? While -80°C is better than -20°C, it is not a perfect solution. Storage at -80°C still leads to a time-dependent reduction in particle concentration and sample purity, and can still increase particle size and variability [4]. The use of a cryoprotectant like trehalose is critical to mitigate these effects.
Are there any additives that can prevent aggregation? Yes, the non-reducing disaccharide trehalose has been extensively documented as an effective stabilizer. When added to storage buffers at concentrations such as 25 mM, it narrows the exosome size distribution, increases particle yield, and helps maintain biological activity by preventing aggregation and fusion during freeze-thaw cycles [1] [2] [4].
Table 1: Impact of Freeze-Thaw Cycles on Exosome Integrity
| Freeze-Thaw Cycles | Particle Concentration | Mean Particle Size | Exosomal Markers (ALIX, TSG101, HSP70) |
|---|---|---|---|
| 1 Cycle | Decrease [4] | Increase [4] | Decrease [6] |
| Multiple Cycles (1-5) | Cycle-dependent decrease [6] [4] | Cycle-dependent increase [6] [4] | Cycle-dependent decrease [6] |
Table 2: Effect of Short-Term (24-hour) Storage Temperature
| Storage Temperature | Particle Concentration | Cellular Uptake | Notes |
|---|---|---|---|
| 4°C | Highest concentration maintained [6] | Lower compared to frozen samples [6] | Best for very short-term preservation of concentration [6] |
| -80°C | Decrease [6] | Increase [6] | |
| -20°C | Decrease [6] | Increase [6] | |
| 37°C | Decrease [6] | Increase [6] | Significant degradation [6] |
Table 3: Effect of Cryoprotectants on Exosome Stability
| Storage Condition | Particle Aggregation | Particle Count per μg Protein | Preservation of Function |
|---|---|---|---|
| PBS (control) | High [1] | Low [1] | Reduced biological activity [1] |
| Trehalose 25 mM | Reduced [1] | 3x higher than PBS control [1] | Improved preservation of TNF-α stimulation [1] |
Experimental Protocol: Evaluating Aggregation Using Nanoparticle Tracking Analysis (NTA)
This protocol allows you to quantitatively assess the impact of different storage conditions on your exosome preparations.
- Exosome Isolation: Isolate exosomes from your cell culture medium or biofluid using your standard method (e.g., differential ultracentrifugation, size-exclusion chromatography, or precipitation). Note the isolation method, as it can affect initial exosome stability [1] [6] [4].
- Sample Aliquot and Treatment:
- Divide the freshly isolated exosome sample into multiple aliquots.
- Control: Resuspend one aliquot in PBS.
- Stabilized: Resuspend another aliquot in PBS containing 25 mM Trehalose [1].
- Subject aliquots to your chosen stress conditions (e.g., 1-5 freeze-thaw cycles, storage at different temperatures for set durations).
- NTA Measurement:
- Dilute each exosome sample in sterile, particle-free PBS or water to achieve a concentration within the ideal detection range of your NTA instrument (typically 10^8-10^9 particles/mL).
- Load the sample into the instrument chamber using a sterile syringe.
- Perform measurements with consistent settings (camera level, detection threshold) across all samples. Capture three 60-second videos for each sample.
- Ensure the environment is vibration-free and at a stable temperature.
- Data Analysis:
- Use the built-in NTA software to analyze the videos and generate data for particle concentration (particles/mL) and mode/mean particle size (nm).
- Compare the particle size distribution and concentration between the fresh sample and treated aliquots. An increase in mean/mode size and a widening of the size distribution (increased standard deviation) indicates aggregation and fusion [1] [4].
- Statistical analysis (e.g., student's t-test) should be performed to confirm the significance of observed differences.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Reagents for Exosome Storage and Stability Studies
| Reagent / Material | Function / Application | Key Details |
|---|---|---|
| Trehalose | Cryoprotectant & Aggregation Inhibitor | Stabilizes lipid membranes and proteins via water replacement and vitrification mechanisms; use at 25 mM in PBS [1]. |
| Phosphate-Buffered Saline (PBS) | Standard Storage Buffer | Serves as a control and base buffer; alone, it offers poor protection against aggregation and freeze-thaw damage [1] [4]. |
| Dimethyl Sulfoxide (DMSO) | Cryoprotectant | Can be used at 6-10% concentration; however, potential cytotoxicity and interference with downstream applications should be considered [2] [4]. |
| Protease Inhibitor Cocktail | Prevent Protein Degradation | Added to storage buffers to prevent proteolytic degradation of exosomal surface and cargo proteins during storage [4]. |
| Size-Exclusion Chromatography (SEC) Columns | Exosome Isolation | Provides a gentle method for isolating exosomes with high purity, minimizing co-isolation of contaminants that could affect stability [4]. |
Diagram: Mechanisms of Exosome Aggregation and Stabilization
Troubleshooting Guide: Freeze-Thaw Damage in EV Experiments
FAQ: What are the specific detrimental effects of freeze-thaw cycles on my EV samples?
Repeated freeze-thaw cycles cause measurable damage to extracellular vesicles across multiple parameters. The primary mechanisms include particle aggregation, membrane deformation, cargo loss, and reduced bioactivity [3] [2].
Aggregation and Size Changes: Multiple freeze-thaw cycles significantly decrease particle concentrations while increasing average EV size due to aggregation. Studies consistently show the proportion of particles larger than 400 nm (indicative of aggregates) increases substantially after freezing at -70°C [7] [2].
Membrane Integrity Compromise: Electron microscopy reveals vesicle enlargement, fusion, and membrane deformation following suboptimal freezing protocols. Membrane disruption is particularly evident in EVs frozen in liquid nitrogen followed by storage at -80°C [3].
Cargo and Functional Loss: Freeze-thaw cycles decrease RNA content and impair biological functionality. The structural damage directly correlates with reduced cellular uptake and therapeutic efficacy [3] [7] [2].
Table 1: Quantitative Impacts of Freeze-Thaw Cycles on EV Parameters
| Parameter Measured | Impact of Freeze-Thaw Cycles | Experimental Evidence |
|---|---|---|
| Particle Concentration | Significant decrease | NTA shows lower total particle counts after freezing [7] |
| Size Distribution | Increase in particles >400 nm | Aggregation measured by NTA and FE-SEM [7] |
| RNA Content | Decreased content | Reduced RNA yield and integrity after multiple cycles [3] [2] |
| Membrane Morphology | Deformation, enlargement, fusion | Electron microscopy observations [3] [2] |
| Biological Function | Impaired bioactivity | Reduced cellular uptake and functional efficacy [3] [7] |
FAQ: What methods can effectively reverse EV aggregation after freeze-thaw cycles?
Water-Bath Sonication effectively disperses aggregated EVs. Experimental data demonstrates that sonication at power level 3 (40 kHz, 100 W) for 15 minutes significantly increases detectable EV concentration and reduces aggregation [7]. This treatment restores cellular uptake efficiency comparable to fresh EVs in vivo.
Critical Note on Pipetting: Regular tip-based pipetting does not effectively disperse aggregated EVs and may promote re-aggregation in previously sonicated samples [7].
Molecular Dynamics Evidence: Simulations confirm that sonication provides sufficient energy to overcome adhesion forces between EV membranes, quantified at approximately 0.167 J/m² for phospholipid bilayers [7].
FAQ: What are the optimal storage conditions to prevent freeze-thaw damage?
Temperature Optimization: Constant storage at -80°C provides the best preservation of EV quantity, cargo integrity, and bioactivity across most EV types and sources [3]. Storage at -20°C shows significant particle aggregation and size increase compared to -80°C [3] [2].
Stabilization Strategies:
- Cryoprotectants: Addition of stabilizers like trehalose helps maintain EV integrity during freezing [3] [2].
- Native Environment: EVs stored in native biofluids show improved stability over purified EVs in buffers [3] [2].
- Formulation Advances: Encapsulation in hyaluronic acid-based microneedles or supplementation with trehalose and cellulose enables EV preservation for up to 12 months at room temperature [2].
Table 2: EV Preservation Under Different Storage Conditions
| Storage Condition | Preservation Performance | Recommended Application |
|---|---|---|
| -80°C (long-term) | Optimal for particle concentration, RNA content, morphology, and bioactivity | Standard storage for most EV types; suitable for >1 week to long-term [3] |
| -20°C | Significant particle aggregation and size increase; suboptimal | Not recommended for critical applications [3] [2] |
| Liquid Nitrogen (-196°C) | Less commonly used; may cause membrane disruption | Not generally recommended; limited comparative data [3] |
| 4°C (short-term) | Moderate stability for limited durations | Suitable for very short-term storage only [7] |
| With Stabilizers (trehalose) | Improved integrity maintenance | Recommended for sensitive applications [3] [2] |
Experimental Protocols for Freeze-Thaw Studies
Protocol: Evaluating Freeze-Thaw Impact on EV Integrity
Sample Preparation:
- Isolate EVs using preferred method (ultracentrifugation, SEC, or other)
- Divide into equal aliquots for experimental groups
- Use fresh EVs as control group (no freezing)
Freeze-Thaw Cycling:
- Freezing condition: -80°C for minimum 3 hours
- Thawing: Room temperature water bath (5-10 minutes)
- Repeat cycles as needed (1, 3, 5, 10 cycles)
- Include stabilizer-treated groups (e.g., 5-10% trehalose)
Assessment Methods:
- Nanoparticle Tracking Analysis (NTA): Measure concentration and size distribution pre- and post-freezing [7]
- Electron Microscopy: Evaluate morphology and aggregation state via FE-SEM [7]
- RNA/Protein Analysis: Quantify cargo preservation
- Functional Assays: Test cellular uptake and bioactivity [7]
Protocol: Sonication-Mediated Dispersion of Aggregated EVs
Equipment Setup:
- Water-bath sonicator (40 kHz frequency, 100 W power)
- Temperature control maintained at 20-25°C
- Timer
Procedure:
- Thaw frozen EV samples completely at room temperature
- Mix gently by hand swirling (avoid pipetting)
- Place sample tube in sonication water bath
- Sonicate at power level 3 for 15 minutes
- Remove sample and proceed immediately to experiments
Validation Steps:
- Confirm dispersion efficiency by NTA (reduction in >400nm particles)
- Test cellular uptake in relevant models
- Compare to fresh EV controls for functionality [7]
Research Reagent Solutions for EV Freeze-Thaw Studies
Table 3: Essential Reagents and Materials for EV Freeze-Thaw Research
| Reagent/Material | Function/Application | Usage Notes |
|---|---|---|
| Trehalose | Cryoprotectant that stabilizes EV membranes during freezing | Use at 5-10% concentration; improves integrity preservation [3] [2] |
| Dimethyl Sulfoxide (DMSO) | Traditional cryoprotectant | Use with caution due to potential cytotoxicity [3] |
| Phosphate Buffered Saline (PBS) | Common EV suspension buffer | Suboptimal for freezing; native biofluids provide better stability [3] [2] |
| Hyaluronic Acid (HA) | Matrix for EV encapsulation in stabilization systems | Enables room temperature storage in microneedle formats [2] |
| Cellulose-based Stabilizers | Structural support for EV preservation | Used in combination with trehalose for long-term stability [2] |
| Water-bath Sonicator | Dispersion of aggregated EVs post-thaw | Optimal at 40 kHz, 100 W power for 15 minutes [7] |
| POPC Lipids | Model membranes for molecular dynamics studies | Representative phospholipid for adhesion energy calculations [7] |
Key Technical Considerations for EV Storage
Critical Parameter: Freezing Rate and Temperature Consistency
Rapid freezing procedures and maintaining constant subzero temperatures are critical for optimal EV preservation. Temperature fluctuations during storage or processing can accelerate degradation processes [3] [2].
Solution: Minimize Freeze-Thaw Cycles Through Aliquot Strategy
The most effective approach is to avoid repeated freezing and thawing through proper experimental planning:
- Divide EV samples into small single-use aliquots
- Use cryoprotectants in storage buffers
- Characterize fresh samples whenever possible
- Implement proper inventory tracking systems
The evidence consistently demonstrates that careful attention to freezing protocols, stabilization strategies, and post-thaw processing methods can significantly mitigate the damaging effects of freeze-thaw cycles on EV samples, enabling more reproducible research outcomes and maintaining therapeutic efficacy [3] [7] [2].
How do different storage temperatures affect the stability and functionality of extracellular vesicles (EVs)?
The stability of extracellular vesicles (EVs) is highly dependent on storage temperature. Based on a systematic review of current evidence, the following table summarizes the effects of different storage conditions on EV integrity:
| Storage Condition | Impact on EV Concentration | Impact on EV Size & Morphology | Impact on Molecular Cargo & Bioactivity |
|---|---|---|---|
| -80 °C (Constant) | Appropriate preservation of particle quantity [2]. | Maintains size distribution; appropriate preservation of morphology [2]. | Appropriate preservation of RNA and protein content [2]. |
| Repeated Freeze-Thaw Cycles | Decreased particle concentration [2] [1]. | Increased particle size and aggregation; vesicle enlargement, fusion, and membrane deformation observed [2] [1]. | Decreased RNA content; impaired bioactivity [2]. |
| Room Temperature (with lyophilization & trehalose) | Maintained count for up to 12 months in microneedles [2]. Lyophilization with trehalose preserves particle concentration [8]. | Maintained size for up to 12 months in microneedles; prevents aggregation during lyophilization [2] [8]. | Protein and RNA content, as well as cargo function, preserved after lyophilization with trehalose [8]. |
| 4 °C (with stabilizers) | Significant decrease in EVs stored in PBS over time; negligible decrease when encapsulated in hyaluronic acid microneedles for up to 6 months [2]. | Not specified in results. | Protein activity lost in PBS within 2 weeks; preserved over 99% in hyaluronic acid microneedles at 4°C for 6 months [2]. |
Key Experimental Protocol: To assess stability, researchers often isolate EVs via methods like size exclusion chromatography or tangential flow filtration. The EVs are then aliquoted and stored under different conditions (e.g., -80°C, with/without multiple freeze-thaw cycles, or lyophilized). Key parameters measured post-storage include:
- Concentration & Size: Using Nanoparticle Tracking Analysis (NTA) or Tunable Resistive Pulse Sensing [2] [1].
- Morphology: Using transmission electron microscopy (TEM) or cryo-electron tomography to visualize membrane integrity and aggregation [2] [1] [8].
- Cargo Integrity: Using techniques like qRT-PCR for RNA content, Western blot for protein markers, and functional immune assays (e.g., TNF-α secretion) for bioactivity [2] [1] [8].
What is the role of trehalose in preventing exosome aggregation during freeze-thaw cycles?
Trehalose is a natural, non-toxic disaccharide that acts as a highly effective cryoprotectant and stabilizer for exosomes. Its role is crucial in mitigating the damage caused by freezing and thawing.
Mechanism of Action: Trehalose protects exosomes through multiple proposed mechanisms. It can physically shield fragile vesicle membranes by replacing water molecules around the lipid bilayer, a process known as the "water replacement" theory. It can also form a stable, glassy matrix (vitrification) that immobilizes the exosomes and prevents ice crystal formation that could pierce and fuse vesicles [1] [8].
Experimental Evidence: A key study demonstrated that adding 25 mM trehalose to phosphate-buffered saline (PBS) used for exosome isolation and storage had several positive effects compared to PBS alone [1]:
- Reduced Aggregation: Trehalose narrowed the particle size distribution and reduced the mean particle size, indicating less aggregation.
- Increased Yield: A three-fold increase in the number of individual particles per microgram of protein was observed.
- Preserved Integrity: During repeated freeze-thaw cycles, exosomes in PBS showed an increase in particle concentration and wider size distribution (indicating aggregation and fragmentation), while exosomes in trehalose showed no significant changes.
- Maintained Bioactivity: Macrophage immune assays showed that exosomes stored in trehalose consistently stimulated higher TNF-α secretion, indicating better preservation of biological activity.
Detailed Protocol for Using Trehalose:
- Solution Preparation: Prepare your storage buffer, such as PBS, and supplement it with 25 mM trehalose [1].
- Isolation/Resuspension: Isolate exosomes via your standard method (e.g., differential centrifugation) and resuspend the final pellet in the trehalose-supplemented buffer [1].
- Freezing: Aliquot the exosome suspension to avoid repeated freeze-thaw cycles and freeze at -80°C [2].
- Thawing: When needed, thaw aliquots rapidly in a 37°C water bath and mix gently by pipetting before use.
My exosome samples are aggregating after lyophilization. How can I prevent this?
Aggregation after lyophilization is a common issue caused by stresses during the freezing and drying process. The most effective preventive strategy is the use of cryoprotectants.
Primary Solution: Use of Trehalose in Lyophilization Research has shown that lyophilizing exosomes in the presence of trehalose successfully prevents aggregation. One study found that while exosomes lyophilized without a cryoprotectant formed large aggregates, those lyophilized with trehalose maintained a dispersed state and showed no signs of aggregation under transmission electron microscopy [8].
Lyophilization Workflow with Trehalose:
Detailed Lyophilization Protocol:
- Prepare Exosome Sample: Isolate exosomes using your preferred method (e.g., ultracentrifugation, size-exclusion chromatography).
- Add Cryoprotectant: Mix the exosome suspension with a solution of trehalose. The specific concentration can be optimized, but studies have used it successfully as a cryoprotectant [8].
- Freeze: Place the mixture in a freezer (-80°C) or on a shelf of a freeze-dryer for freezing.
- Lyophilize: Transfer the frozen sample to a lyophilizer. The process involves a primary drying phase to sublimate ice under vacuum, followed by a secondary drying phase to remove residual water.
- Store: The resulting lyophilized powder can be stored sealed at room temperature or 4°C. When needed, rehydrate with sterile water or an appropriate buffer by gentle pipetting or vortexing.
How does the composition of a buffer solution impact cellular and molecular outcomes in experiments like dielectrophoresis (DEP)?
Buffer composition is a critical, yet often overlooked, factor that can significantly influence cell viability, morphology, and gene expression, thereby affecting the outcome and interpretation of sensitive experiments.
Key Findings from DEP Buffer Studies: A study investigating the impact of four different buffers on two cancer cell lines (Caco-2 and K562) revealed that even buffers that support good cell viability can induce significant molecular changes [9].
Summary of Buffer Composition Impact:
| Parameter Assessed | Influence of Buffer Composition |
|---|---|
| Cell Viability & Growth Recovery | Buffer composition differently influenced the viability of Caco-2 and K562 cells. Some buffers maintained viability and allowed growth recovery after 24 hours, while others were cytotoxic [9]. |
| Cell Morphology & Size | NaCl concentration in the buffer was found to influence both flow cytometry outcomes and cell size. Morphology was stable for up to 1 hour in certain buffers but not others [9]. |
| Gene Expression | Buffer composition significantly modulated the expression of inflammation (IL-6), oxidative stress (iNOS), and metabolism (GAPDH) markers in both cell lines, even under apparently non-cytotoxic conditions [9]. |
Experimental Protocol for Buffer Evaluation: To assess the impact of a buffer for your application, the following methodology can be employed:
- Buffer Preparation: Prepare the buffers of interest, measuring and adjusting their conductivity to the required level (e.g., 33 mS/m for DEP) using solutions like KCl [9].
- Cell Treatment: Incubate the target cells in the different buffers for a set duration (e.g., 1 hour).
- Viability & Morphology Assay: Use the MTT assay to evaluate cell viability and flow cytometry to analyze cell size and granulocyte stress formation [9].
- Molecular Analysis: Perform quantitative real-time PCR (qRT-PCR) to evaluate the gene expression levels of key markers relevant to your study, such as IL-6, iNOS, and GAPDH [9].
Interpretation: The key takeaway is that a buffer should be selected not only based on its electrokinetic performance but also on its ability to preserve the native biochemical and molecular state of the cells. A buffer that induces stress responses can confound experimental results.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Experimentation |
|---|---|
| Trehalose | A non-reducing disaccharide used as a cryoprotectant and stabilizer to prevent exosome/EV aggregation during freezing, thawing, and lyophilization [1] [8]. |
| Phosphate Buffered Saline (PBS) | A common isotonic buffer for washing cells and diluting biological samples. It often serves as a base solution, but may require additives like trehalose for sensitive applications [1]. |
| Fetal Bovine Serum (FBS) | Provides a rich mixture of growth factors, hormones, and lipids to support the growth of cells in culture. It is often used as a supplement in cell culture media [9]. |
| Bovine Serum Albumin (BSA) | Used as a protein stabilizer and to block non-specific binding in assays like Western blotting and immunoassays. It is also added to buffer solutions to reduce surface adhesion [9]. |
| Sucrose | Used to create isotonic or hypertonic conditions in buffers, helping to maintain osmotic balance and protect cells and vesicles from osmotic shock [9]. |
| Radioimmunoprecipitation Assay (RIPA) Buffer | A harsh lysis buffer containing detergents and salts, used to lyse cells and vesicles for protein extraction, particularly for Western blotting [10]. |
| β-mercaptoethanol | A reducing agent that breaks disulfide bonds between cysteine residues in proteins, aiding in protein denaturation for Western blot analysis [10]. |
What are the best practices for preparing biofluid samples to ensure analytical reproducibility?
Proper sample preparation is fundamental for obtaining accurate and reproducible data, especially when working with complex matrices like biofluids.
Core Principles of Biofluid Sample Prep: The goal is to extract and concentrate the analyte while removing sample constituents that might interfere with the analysis. Key considerations are outlined in the following workflow:
Detailed Methodologies:
- General Handling for Serum/Plasma:
- Thawing: Thaw frozen samples completely, then vortex mix thoroughly.
- Clarification: Centrifuge samples at a minimum of 10,000 x g for 5-10 minutes to remove particulates, lipids, and cells. For viscous samples, centrifugation may need to be repeated [11].
- Protein Precipitation (PPT): Primarily used for blood-based samples. Adding excess organic solvent (e.g., acetonitrile) precipitates proteins, which are then removed by centrifugation or filtration. This is quick and simple but only removes proteins, not other interferents like phospholipids [12].
- Phospholipid Depletion (PLD): A crucial step for LC-MS/MS analysis of blood samples. Phospholipids can cause significant ion suppression. They are removed using a specialized scavenging adsorbent, often following PPT [12].
- Supported Liquid Extraction (SLE): A targeted extraction technique based on the same principle as liquid-liquid extraction (LLE) but is easier to automate and avoids emulsion formation. An aqueous sample is loaded onto an inert solid support, and an immiscible organic solvent is passed through to elute the analytes [12].
FAQs on Sample Preparation:
- Q: Can I use a "dilute and shoot" approach for urine samples?
- A: Yes, for urine, a simple 1:10 dilution with water or buffer can sometimes be sufficient. However, this approach negatively impacts the limit of detection and does not remove matrix components that can foul instrumentation or suppress ionization [12].
- Q: Why is it important to remove phospholipids specifically?
- A: Phospholipids elute at various points in a chromatographic run and cause significant ion suppression in the mass spectrometer, leading to inaccurate quantification and loss of sensitivity [12].
Optimized Storage Protocols: Practical Strategies for Preserving Exosome Integrity
Technical Support Center: Preventing Exosome Aggregation
Troubleshooting Guides
Problem: High Particle Count and Polydispersity Post-Thaw
- Question: My exosome samples show a significant increase in particle size and polydispersity index (PDI) after a single freeze-thaw cycle from -80°C. What is the cause and how can I prevent it?
- Answer: This is a classic sign of freeze-thaw-induced aggregation. The formation of ice crystals during freezing can disrupt exosome membranes, leading to fusion and aggregation upon thawing.
- Solution 1: Implement a Controlled, Slow Freezing Rate. Use an isopropanol-filled "Mr. Frosty" or a programmable freezer to cool samples at approximately -1°C per minute before transferring to -80°C. This reduces ice crystal formation.
- Solution 2: Introduce a Cryoprotectant. Add a non-penetrating cryoprotectant like 5-10% (w/v) trehalose to your exosome suspension. It forms a stable glassy matrix that separates exosomes and protects membranes.
- Solution 3: Aliquot Samples. Avoid repeated freeze-thaw cycles by aliquoting exosomes into single-use volumes.
Problem: Poor Recovery After Lyophilization
- Question: I am experiencing low particle recovery and loss of functional biomarkers after lyophilizing my exosome sample. What steps am I likely missing?
- Answer: Lyophilization without a proper lyoprotectant causes massive aggregation and membrane damage due to mechanical and osmotic stress.
- Solution 1: Optimize the Lyoprotectant Formulation. Use a combination of cryo- and lyo-protectants. A standard formulation is 5% trehalose + 1% sucrose in your resuspension buffer prior to freezing and lyophilization.
- Solution 2: Control the Lyophilization Cycle. Ensure a primary drying phase that is long enough to remove all ice without collapsing the cake structure. Use a pilot study to optimize time and temperature.
- Solution 3: Validate Reconstitution. Rehydrate the lyophilized cake gently with the original volume of a compatible buffer (e.g., PBS or 0.9% saline). Avoid vortexing; use gentle pipetting or slow rotation.
Problem: Rapid Degradation at 4°C
- Question: My exosomes are stable for less than a week at 4°C, showing a drop in CD63 expression. Is this expected?
- Answer: Yes, this is expected. 4°C is not suitable for long-term storage. Hydrolytic and enzymatic degradation processes remain active, and exosomes can sink and aggregate in the tube over time.
- Solution: For any storage beyond 48-72 hours, move to -80°C or lyophilization. If you must use 4°C for short-term experiments, ensure your buffer contains protease inhibitors and use low-protein-binding tubes to minimize surface adsorption.
Frequently Asked Questions (FAQs)
Q: What is the single most important factor for preserving exosome function during freeze-thaw?
- A: The use of a cryoprotectant, specifically trehalose, is the most critical factor. It stabilizes the lipid bilayer without being internalized, significantly reducing aggregation and preserving surface protein integrity.
Q: Can I store my exosomes at -20°C for a few months?
- A: It is not recommended. The -20°C environment is within the eutectic point range where recrystallization can occur, causing more damage than -80°C storage. For any storage beyond a few weeks, -80°C is the minimum standard.
Q: How does lyophilization prevent aggregation when it removes water?
- A: Lyophilization prevents aggregation by immobilizing the exosomes in a rigid, amorphous glassy matrix formed by the lyoprotectants (e.g., sugars). This matrix physically separates the exosomes, preventing their membranes from contacting and fusing, both during the dried state and upon rehydration.
Q: My downstream application is RNA sequencing. Which storage method is best?
- A: For RNA integrity, rapid freezing and storage at -80°C with a cryoprotectant like trehalose is superior. Lyophilization can also be effective but requires rigorous validation, as the process can sometimes induce minor RNA degradation if not perfectly optimized.
Table 1: Impact of Storage Conditions on Exosome Integrity
| Parameter | 4°C (7 days) | -20°C (30 days) | -80°C (6 months) | Lyophilization (12 months) |
|---|---|---|---|---|
| Particle Concentration Recovery | 60-75% | 50-70% | 80-95%* | 70-90% |
| Mean Particle Size Increase | 15-30% | 20-50% | 5-15%* | 10-20% |
| Polydispersity Index (PDI) Change | +0.08 to +0.15 | +0.10 to +0.25 | +0.02 to +0.08* | +0.05 to +0.12 |
| Surface Marker Preservation (e.g., CD81) | Low | Moderate | High* | High |
| Functional Cargo Retention (e.g., miRNA) | Low | Moderate | High* | Moderate-High |
*With use of 5-10% trehalose as a cryoprotectant.
Table 2: Recommended Applications and Limitations
| Storage Method | Recommended Storage Duration | Best For | Key Limitations |
|---|---|---|---|
| 4°C | < 72 hours | Immediate use, in-process handling | Rapid degradation, aggregation, bacterial growth. |
| -20°C | < 2 weeks | Temporary holding | High risk of ice crystal damage; not for long-term. |
| -80°C with Cryoprotectant | 6 months - 2 years | Long-term biobanking, functional assays | Requires reliable freezer; dependent on freeze-thaw protocol. |
| Lyophilization | > 2 years | Shipping, room-temperature storage | Complex process; potential for oxidative damage; requires reconstitution. |
Experimental Protocols
Protocol 1: Assessing Cryoprotectant Efficacy for -80°C Storage
- Isolate Exosomes via ultracentrifugation or SEC and resuspend in PBS.
- Aliquot into 100 µL portions.
- Add Cryoprotectants: To separate aliquots, add trehalose (5%, 10%), sucrose (5%), or DMSO (5%).
- Control: Leave one aliquot in PBS only.
- Freeze: Place all aliquots in a Mr. Frosty freezing container at -80°C for 24 hours.
- Thaw: Rapidly thaw in a 37°C water bath with gentle agitation.
- Analyze: Use Nanoparticle Tracking Analysis (NTA) to measure particle concentration, size, and PDI. Validate with Western Blot for marker proteins (CD9, CD63, CD81).
Protocol 2: Lyophilization of Exosomes for Long-Term Stability
- Prepare Exosome Formulation: Dialyze the exosome pellet against a solution of 5% trehalose and 1% sucrose in ultrapure water to remove salts.
- Aliquot: Dispense 200 µL volumes into sterile lyophilization vials.
- Snap Freeze: Place vials in a bath of liquid nitrogen or a -80°C freezer for 2 hours.
- Lyophilize: Transfer vials to a pre-cooled (-40°C) lyophilizer. Run a primary drying cycle at -40°C for 24 hours under vacuum (< 100 mTorr). Follow with a secondary drying cycle, ramping to 25°C over 8 hours.
- Store: Seal vials under inert gas (e.g., Argon) if possible and store at 4°C or room temperature, protected from light.
- Reconstitute: Add 200 µL of nuclease-free water or PBS and allow to rehydrate for 30 minutes with gentle inversion.
Visualizations
Title: Exosome Storage Decision Guide
Title: Freeze-Thaw Aggregation Pathway
The Scientist's Toolkit
Table 3: Essential Reagents for Exosome Storage
| Reagent/Material | Function | Key Consideration |
|---|---|---|
| Trehalose | Non-penetrating cryo-/lyo-protectant. Forms a glassy state to separate and stabilize exosomes. | Preferred over sucrose due to higher glass transition temperature (Tg) and stability. |
| Sucrose | Lyoprotectant used in combination with trehalose to enhance matrix formation during lyophilization. | Can be more susceptible to hydrolysis than trehalose. |
| Mr. Frosty / Nalgene Freezing Container | Provides a consistent -1°C/minute cooling rate for controlled freezing to -80°C. | Essential for standardizing the freezing process across samples. |
| Low-Protein-Bind Microtubes | Minimizes exosome adhesion to tube walls, maximizing recovery. | Critical for all steps, especially with low-concentration samples. |
| Protease Inhibitor Cocktails | Prevents proteolytic degradation of exosome surface markers during short-term storage at 4°C. | Must be added fresh to buffers. |
Extracellular vesicles (EVs), including exosomes, are promising tools in regenerative medicine and drug delivery. However, their clinical translation faces significant challenges, particularly in maintaining stability during storage. The nanoscale properties of EVs make them sensitive to environmental conditions, leading to aggregation, cargo loss, and functional impairment during freeze-thaw cycles. Optimal cryoprotectants are therefore crucial for preserving EV structural, molecular, and functional integrity. This guide evaluates the efficacy of trehalose, sucrose, Human Serum Albumin (HSA), and Dimethyl Sulfoxide (DMSO) to help you select the right protocol for your research.
Troubleshooting Guide: Cryoprotectant Performance and Common Issues
Problem 1: EV Aggregation After Thawing
- Potential Cause: Inadequate cryoprotection leading to ice crystal formation and membrane fusion.
- Solution: Incorporate non-reducing disaccharides like trehalose or sucrose at 25-50 mM into your storage buffer. These sugars act as water substitutes and form stable glassy matrices that prevent vesicle-vesicle contact [1] [13].
Problem 2: Loss of Biological Activity in Functional Assays
- Potential Cause: Cryodamage to membrane proteins or leakage of bioactive cargo.
- Solution: Use trehalose (25 mM) as a cryoprotectant. Studies demonstrate that EVs cryopreserved with trehalose maintain their ability to stimulate immune responses and support the expansion of hematopoietic stem cells, outperforming PBS-only controls [1] [14].
Problem 3: Decreased Particle Concentration and Increased Size After Freeze-Thaw Cycles
- Potential Cause: Vesicle rupture and the formation of aggregates from multiple fusion events.
- Solution: Avoid multiple freeze-thaw cycles. Aliquot EVs in a 5% sucrose solution. Research shows sucrose provides superior preservation of EV size distribution and concentration compared to PBS after storage at -80°C [15] [13].
Problem 4: Concerns About Cytotoxicity or Introduction of Exogenous Contaminants
- Potential Cause: Use of cryoprotectants like DMSO, which can inhibit specific downstream processes, or HSA, which may introduce unknown variables.
- Solution: Opt for trehalose or sucrose. These are natural, non-toxic sugars widely used in food and drug industries. They do not require a washing step post-thaw and avoid potential cytotoxicity associated with DMSO [1] [14].
Quantitative Comparison of Cryoprotectants
The following table summarizes key experimental data on the performance of different cryoprotectants for EV storage.
Table 1: Efficacy of Cryoprotectants in EV Preservation
| Cryoprotectant | Typical Concentration | Key Findings on EV Integrity | Impact on Biological Function |
|---|---|---|---|
| Trehalose | 25 mM | • Narrowed particle size distribution & reduced aggregation [1]• Higher particle concentration per μg of protein [1]• Maintained integrity over 12 freeze-thaw cycles in microneedles [2] | • Consistently higher TNF-α stimulation in macrophages [1]• Maintained HSC-supportive potential [14] |
| Sucrose | 5% (w/v) | • Better preservation of size/concentration vs. PBS at -80°C [15] [13]• More prevalent molecular surface protrusions & transmembrane proteins [13] | Data specific to functional assays not provided in available sources. |
| HSA (Human Serum Albumin) | Not Specified | • Improved EV quality after short & long-term storage vs. PBS alone [16] | Data specific to functional assays not provided in available sources. |
| DMSO | 6% (v/v) | • Increased EV yield & procoagulant activity from platelets [17] | • Potential cytotoxicity & inhibition of downstream processes [2] |
| Control (PBS) | N/A | • Particle aggregation & increased size after freeze-thaw [2] [1]• Decreased particle concentration & RNA content [2] | • Rapid loss of protein activity & impaired bioactivity [2] |
Experimental Protocols for Cryoprotectant Evaluation
Protocol 1: Assessing the Anti-Aggregation Efficacy of Trehalose
This protocol is adapted from a study demonstrating that trehalose prevents the aggregation of pancreatic beta-cell exosome-like vesicles (beta-ELVs) [1].
- EV Isolation and Preparation: Isolate EVs from your cell culture supernatant (e.g., MIN6 beta-cells) using differential centrifugation and ultrafiltration.
- Buffer Exchange: Resuspend the final EV pellet in two different buffers:
- Experimental Buffer: Phosphate-Buffered Saline (PBS) supplemented with 25 mM Trehalose (TRE).
- Control Buffer: PBS alone.
- Freeze-Thaw Cycling: Subject the EV suspensions to repeated freeze-thaw cycles (e.g., 3-5 cycles). Freeze at -80°C and thaw rapidly at 37°C.
- Characterization and Analysis:
- Nanoparticle Tracking Analysis (NTA): Measure the particle concentration and size distribution (mode, mean, and standard deviation) after freeze-thaw cycles. A lower standard deviation and span in the TRE group indicates a narrower size distribution and less aggregation [1].
- Cryo-Electron Tomography: Confirm the presence of individual, circular nanovesicles and the absence of large aggregates in the TRE group.
Protocol 2: Testing Functional Preservation in Hematopoietic Stem Cell (HSC) Expansion
This protocol is based on research showing that MSC-derived EVs cryopreserved with trehalose retain their ability to expand HSCs in vitro [14].
- EV Cryopreservation: Isolate Microvesicles (MVs) and exosomes from Mesenchymal Stromal Cell (MSC) conditioned medium. Aliquot the EVs and cryopreserve them at -80°C in:
- Experimental Buffer: PBS supplemented with 25 mM Trehalose.
- Control Buffer: PBS alone.
- HSC Co-culture: After thawing, co-culture freshly isolated mouse bone marrow HSCs with the cryopreserved EVs (MVs or exosomes) in a suitable expansion medium for 5-7 days.
- Functional Assessment:
- Stemness Maintenance: Analyze the co-cultured cells for the preservation of stem cell markers (e.g., Sca-1, c-Kit) using flow cytometry. EVs stored with trehalose should better maintain the stem cell population [14].
- Clonogenic Assay: Plate the cells in methylcellulose-based media to assess colony-forming unit (CFU) potential. A higher number of colonies indicates retained EV functionality [14].
- Migration Assay: Perform a transwell migration assay towards a SDF-1α gradient to evaluate the homing capacity of the expanded HSCs, a key functional outcome [14].
Experimental Workflow for EV Cryoprotectant Testing
Cryoprotectant Mechanism of Action
Cryoprotectant Mechanisms and Outcomes
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for EV Cryopreservation Studies
| Reagent / Material | Function in Protocol | Example Usage & Rationale |
|---|---|---|
| Trehalose | Non-reducing disaccharide cryoprotectant | Used at 25 mM in PBS to prevent aggregation and preserve biological activity during freeze-thaw cycles and long-term storage [1] [14]. |
| Sucrose | Cryoprotectant and buffer component | Used as a 5% (w/v) solution for -80°C storage to better maintain EV size distribution, concentration, and membrane integrity compared to PBS [15] [13]. |
| Human Serum Albumin (HSA) | Protein-based stabilizer | Added to storage buffers to improve EV quality and stability over time by reducing vesicle adhesion and surface-induced stress [16]. |
| Dimethyl Sulfoxide (DMSO) | Penetrating cryoprotectant | Used at ~6% for freezing platelet-derived EVs to increase yield and procoagulant activity, but carries risk of cytotoxicity for sensitive applications [17]. |
| Phosphate-Buffered Saline (PBS) | Standard ionic storage buffer | Serves as a common control and base buffer for cryoprotectant studies, though alone it often leads to aggregation and damage [2] [1]. |
| Nanoparticle Tracking Analysis (NTA) | Instrument for particle characterization | Measures hydrodynamic diameter and concentration of EVs to quantify aggregation (increased size) and particle loss after thawing [1]. |
| Cryo-Electron Microscope | High-resolution imaging instrument | Visualizes EV morphology and membrane integrity directly, confirming the presence of intact, non-aggregated vesicles post-thaw [1] [13]. |
Frequently Asked Questions (FAQs)
Q1: Can I use sucrose instead of trehalose for lyophilizing exosomes? Yes, both sucrose and trehalose are effective lyoprotectants. They work by forming a stable, amorphous glassy matrix that immobilizes the EVs and protects their membrane during the freeze-drying process and subsequent storage at room temperature. The choice may depend on optimization for your specific EV type, but both have been shown to outperform buffers like PBS [16] [18].
Q2: How many freeze-thaw cycles can exosomes tolerate when stored with trehalose? The number of safe freeze-thaw cycles is limited even with cryoprotectants. One study incorporating EVs into a hyaluronic acid-based microneedle formulation with trehalose showed negligible degradation after 10 freeze-thaw cycles [2]. However, for EVs in liquid suspension, it is strongly recommended to avoid multiple freeze-thaw cycles. Best practice is to aliquot your EV samples into single-use volumes to minimize repetitive freezing and thawing.
Q3: Why is DMSO less preferred than trehalose for EV cryopreservation? While DMSO is a highly effective cryoprotectant for whole cells, its use for EVs is limited due to potential cytotoxicity and its tendency to inhibit specific downstream biological processes or assays [2]. In contrast, trehalose is a natural, non-toxic sugar that does not require a washing step after thawing and does not interfere with cellular functions, making it a safer and more practical choice for most EV-based applications [1] [14].
Q4: Does storing exosomes in their native biofluid offer any advantage? Yes. Evidence suggests that storing EVs in their native biofluid (e.g., plasma, serum) offers improved stability compared to storing purified EVs in buffers like PBS. The native environment likely contains natural stabilizing factors that help protect the vesicles from degradation and aggregation [2].
Troubleshooting Guides
FAQ 1: How can I prevent exosome aggregation and function loss during long-term storage?
Problem: Exosomes rapidly lose structural integrity and biological function when stored in standard buffers like PBS, even at recommended freezing temperatures [2] [19].
Solutions:
- Incorporate trehalose: Add 25 mM trehalose to your storage buffer. This natural disaccharide acts as a cryoprotectant by stabilizing lipid bilayers and preventing fusion [1] [8].
- Utilize hyaluronic acid microneedles (EV@MN): Encapsulate exosomes within dissolvable hyaluronic acid-based microneedles. This matrix maintains exosome bioactivity for over six months at 4°C [19].
- Avoid repeated freeze-thaw cycles: Aliquot exosomes to minimize freeze-thaw cycles, which cause irreversible damage including particle aggregation, cargo loss, and membrane deformation [2] [4].
Table 1: Comparison of Exosome Storage Strategies
| Storage Method | Storage Duration | Key Findings | Reference |
|---|---|---|---|
| PBS at -80°C | 6 months | Significant decrease in EVs, protein activity lost within 2 weeks | [2] [19] |
| PBS with 25 mM Trehalose at -80°C | - | Prevents aggregation, maintains particle concentration and biological activity | [1] [8] |
| Hyaluronic Acid Microneedles (EV@MN) at 4°C | 6 months | >85% particles remained, >99% protein activity preserved | [19] |
| Lyophilization with Trehalose at RT | 1 week | Preserved physical properties, protein/RNA content, and functionality | [8] |
FAQ 2: What is the optimal strategy for transdermal delivery of exosomes while maintaining stability?
Problem: The skin's stratum corneum barrier limits exosome penetration for topical applications, and conventional storage methods degrade exosome functionality before application [19] [20].
Solutions:
- Fabricate hyaluronic acid dissolving microneedles: Use low molecular weight HA (30-50 kDa) to form dissolvable microneedle tips that encapsulate exosomes [19] [20].
- Utilize micromolding technique: Centrifuge HA-exosome mixture into PDMS molds to form needle structures, then air-dry at 25°C with desiccant [19].
- Characterize microneedle performance: Verify skin penetration capability, exosome release profile, and biological activity post-fabrication [19].
Experimental Protocol: Fabrication of HA Microneedles Loaded with Exosomes
- Isolate exosomes from your cell source of interest using standard methods (ultracentrifugation, size exclusion chromatography)
- Prepare 30% (w/v) hyaluronic acid (MW 30-50 kDa) aqueous solution
- Mix exosomes with HA solution at appropriate ratio for your application
- Cast mixture onto polydimethylsiloxane (PDMS) micromolds
- Centrifuge at 2380× g for 15 minutes to remove air bubbles and fill mold cavities
- Dry in oven at 25°C with dry silica gel for complete solidification
- Demold carefully and store in sealed container with desiccant [19]
FAQ 3: How does trehalose prevent exosome damage during freezing and lyophilization?
Problem: Conventional freezing without cryoprotectants causes exosome damage through ice crystal formation, osmotic stress, and membrane phase transitions [2] [1].
Solutions:
- Leverage multiple protective mechanisms: Trehalose protects through water replacement, vitrification, and preferential exclusion [1] [21].
- Optimize concentration: Use 25 mM trehalose for liquid storage or 100-250 mM for lyophilization protocols [1] [8] [22].
- Control freezing rate: Implement rapid freezing for trehalose-containing formulations to enhance glass formation [2].
Experimental Protocol: Lyophilization of Exosomes with Trehalose
- Isolate and concentrate exosomes using standard methods
- Add trehalose to exosome suspension to achieve final concentration of 100-250 mM
- Aliquot into lyophilization vials and freeze at -80°C for 12 hours
- Transfer to freeze-dryer with condenser temperature maintained at -54°C
- Perform primary drying at 54×10⁻³ bar for 36 hours without heating
- Complete secondary drying to reduce residual moisture
- Store lyophilized exosomes at room temperature with desiccant [8] [22]
Table 2: Troubleshooting Common Exosome Stabilization Issues
| Problem | Possible Cause | Solution | Preventive Measures |
|---|---|---|---|
| Exosome aggregation in storage | Insufficient electrostatic repulsion, buffer incompatibility | Add 25 mM trehalose to storage buffer | Avoid phosphate buffers; use HEPES or trehalose solutions |
| Loss of biological activity after freezing | Ice crystal damage, membrane phase separation | Use rapid freezing rates, incorporate cryoprotectants | Aliquot to avoid repeated freeze-thaw cycles |
| Poor skin penetration | Stratum corneum barrier function | Incorporate into dissolving microneedles | Optimize microneedle length (600μm) and shape |
| Low microneedle mechanical strength | Suboptimal polymer concentration or molecular weight | Increase HA concentration to 30%, use appropriate MW HA | Ensure complete drying with desiccant |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Exosome Stabilization Research
| Reagent/Material | Function/Application | Key Considerations |
|---|---|---|
| Hyaluronic Acid (30-50 kDa) | Base polymer for dissolving microneedles | Provides mechanical strength while maintaining biocompatibility and dissolution rate |
| Trehalose | Cryoprotectant and lyoprotectant | 25 mM for liquid storage, 100-250 mM for lyophilization; non-reducing properties prevent browning reactions |
| Polydimethylsiloxane (PDMS) Molds | Microneedle fabrication | Reusable molds with specific needle dimensions (e.g., 600μm height, 300μm base diameter) |
| Size Exclusion Chromatography Columns | Exosome isolation | Maintains exosome integrity compared to precipitation methods; reduces contaminant proteins |
| Protease Inhibitor Cocktails | Prevention of proteolytic degradation | Particularly important for long-term storage of complex biofluids |
| Dimethyl Sulfoxide (DMSO) | Alternative cryoprotectant | Use at 6-10% concentrations; may interfere with downstream biological assays |
| Hydroxyethyl Starch | Macromolecular cryoprotectant | Increases glass transition temperature when combined with trehalose |
Frequently Asked Questions (FAQs)
Q1: Why is buffer selection critical for exosome storage? The buffer composition is a primary factor in maintaining exosome integrity. Suboptimal buffers can lead to particle aggregation, a drastic loss in concentration, and damage to the lipid bilayer, ultimately compromising the exosomes' biological activity and function [16] [23]. The ionic strength, pH, and presence of cryoprotectants in the buffer directly influence exosome stability during both freezing and refrigeration.
Q2: Is PBS always the best choice for storing exosomes? While Phosphate-Buffered Saline (PBS) is the most commonly used buffer, research shows its performance is highly variable and often suboptimal. Studies indicate that PBS can cause significant damage to EVs during storage, leading to a drastic loss in particle recovery [16] [23]. Its suitability depends on the storage duration and method. One study found PBS superior for maintaining exosome concentration in short-term storage (≤2 weeks), but it is often outperformed by specialized buffers for long-term preservation [16] [24].
Q3: What are the observed drawbacks of using Normal Saline (NS) or 5% Glucose Solution (5%GS)? Comparative studies have shown that exosomes stored in Normal Saline (NS) and 5% Glucose Solution (5%GS) can exhibit specific physical drawbacks:
- Normal Saline (NS): Exosomes in NS can display shriveled morphology and are prone to progressive aggregation, especially under lyophilization [24].
- 5% Glucose Solution (5%GS): Similar to NS, exosomes in 5%GS show aggregation and less of the characteristic biconcave shape compared to those in PBS [24]. These observations suggest that while isotonic, these solutions may lack components that prevent membrane stress and particle-particle interactions.
Q4: How do freeze-thaw cycles affect exosomes, and how can this be mitigated? Multiple freeze-thaw cycles are particularly detrimental to exosomes. They can lead to decreased particle concentrations, loss of RNA content, impaired bioactivity, and an increase in vesicle size due to aggregation and fusion [2] [3]. To mitigate this damage, you should:
- Aliquot exosome samples into single-use volumes to avoid repeated freezing and thawing.
- Use cryoprotectants like trehalose, which has been shown to prevent aggregation and preserve biological activity across freeze-thaw cycles [1].
- Employ rapid freezing procedures to minimize the formation of damaging ice crystals.
Q5: What are the advantages of lyophilization (freeze-drying) for long-term exosome storage? Lyophilization offers significant logistical advantages by enabling the direct preservation of exosomes at room temperature, thereby reducing costs associated with ultra-low temperature freezers and simplifying transportation [16]. However, the process itself poses risks, including ice crystal formation and osmotic stress, which can compromise exosomal structure. The success of lyophilization is highly dependent on using appropriate lyoprotectants (e.g., trehalose, sucrose) in the buffer to maintain size distribution, morphological integrity, and cargo content [16].
Troubleshooting Guides
Problem: Low Exosome Recovery After Thawing
Possible Causes and Solutions:
- Cause 1: Storage in plain PBS. PBS alone is known to cause a significant drop in particle recovery after freezing.
- Cause 2: Multiple freeze-thaw cycles.
- Cause 3: Slow or inconsistent freezing.
- Solution: Use a controlled-rate freezer or snap-freeze aliquots in a slurry of dry ice and ethanol before transferring to -80°C for consistent and rapid freezing [3].
Problem: Exosome Aggregation and Size Increase
Possible Causes and Solutions:
- Cause 1: Unsuitable buffer ionic strength or composition.
- Solution: If aggregation is observed in PBS, consider testing buffers with additives. Trehalose has been demonstrated to narrow exosome size distribution and prevent aggregation by providing a physical shield to the vesicle membrane [1].
- Cause 2: Storage temperature is too high.
- Solution: For long-term storage, ensure a constant temperature of -80°C. Storage at -20°C has been shown to induce significant aggregation and size increase compared to -80°C [3].
- Cause 3: Lyophilization without protectants.
- Solution: When using lyophilization, incorporate lyoprotectants like trehalose or sucrose into the buffer formulation to protect against dehydration and membrane damage that leads to aggregation [16].
The following table summarizes key quantitative findings from a comparative study on the stability of MSCs-derived exosomes in different buffers under cryopreservation and lyophilization [16] [24].
Table 1: Comparative Stability of Exosomes in Different Buffers Over a 4-Week Period
| Storage Buffer | Storage Method | Short-Term Concentration (≤2 wk) | Long-Term Concentration (4 wk) | Size Homogeneity | Key Morphological Observations |
|---|---|---|---|---|---|
| PBS | Cryopreservation (-80°C) | Superior maintenance | Progressive loss | Moderate | More uniform distribution, biconcave-disk shape |
| Normal Saline (NS) | Cryopreservation (-80°C) | Significant loss | Significant loss | Low | Shriveled and aggregated vesicles |
| 5% Glucose (5%GS) | Cryopreservation (-80°C) | Significant loss | Significant loss | Low | Shriveled and aggregated vesicles |
| PBS | Lyophilization | Induced concentration loss | Induced concentration loss | High | Maintained size integrity despite concentration loss |
| Normal Saline (NS) | Lyophilization | N/A | N/A | Low | Progressive aggregation |
| 5% Glucose (5%GS) | Lyophilization | N/A | N/A | Low | Progressive aggregation |
Experimental Protocols
Protocol 1: Assessing Exosome Stability in Different Buffers
This protocol is adapted from studies comparing PBS, NS, and 5% GS for exosome storage [16] [24].
Key Research Reagent Solutions:
- Purified Exosomes: Isolated from cell culture media (e.g., MSCs) via ultracentrifugation or tangential flow filtration.
- Storage Buffers: Sterile PBS (pH 7.4), 0.9% Normal Saline, 5% Glucose Solution.
- Cryoprotectant Solution: 25mM - 100mM Trehalose in PBS.
- Characterization Instruments: Nanoparticle Tracking Analyzer (NTA), Transmission Electron Microscope (TEM), Western Blot apparatus.
Methodology:
- Isolate and Purify Exosomes from your chosen cell source using your standard method (e.g., differential ultracentrifugation).
- Resuspend and Divide the final exosome pellet equally into three aliquots.
- Centrifuge again if needed to replace the original medium with the test buffers.
- Resuspend one aliquot in PBS, one in Normal Saline, and one in 5% Glucose Solution.
- Optional: Add a cryoprotectant like trehalose to a subset of each buffer to test its effect.
- Store Aliquots at your desired temperatures (e.g., -80°C, 4°C) and timepoints (e.g., Fresh, 1-week, 2-week, 4-week).
- Characterize the exosomes at each timepoint.
- Concentration & Size: Use NTA to measure particle concentration and size distribution.
- Morphology: Use TEM to visually assess structural integrity and aggregation.
- Marker Integrity: Use Western Blot to confirm the presence of exosomal markers (e.g., CD63, TSG101).
Protocol 2: Testing the Efficacy of Cryoprotectants
This protocol is based on research demonstrating the protective effect of trehalose [1] [23].
Methodology:
- Prepare your purified exosome sample as in Protocol 1.
- Divide the sample into two equal aliquots.
- Resuspend one aliquot in standard PBS. Resuspend the other aliquot in PBS supplemented with 25mM trehalose.
- Subject both samples to multiple (e.g., 3-5) freeze-thaw cycles. For each cycle, freeze at -80°C for at least 1 hour and thaw at room temperature.
- Analyze the samples after the final thaw.
- Use NTA to compare particle concentration and size distribution between the PBS and PBS-Trehalose groups.
- For functional assessment, treat recipient cells with the exosomes and measure a relevant bioactivity (e.g., macrophage immune response, cytokine secretion).
Visualization of Workflows and Relationships
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Exosome Storage Buffer Optimization
| Reagent / Solution | Function / Rationale | Key Findings / Considerations |
|---|---|---|
| Phosphate-Buffered Saline (PBS) | Isotonic, pH-stabilizing buffer; the current most common choice. | Superior for short-term (≤2 wk) concentration stability, but can lead to major particle loss and function impairment over time [16] [23]. |
| Normal Saline (0.9% NaCl) | Isotonic solution providing basic osmotic balance. | Observed to cause exosome shrinkage and aggregation; not recommended for purified exosomes [24]. |
| 5% Glucose Solution (5%GS) | Isotonic sugar solution providing osmotic balance. | Similar to NS, leads to aggregation and is less effective than PBS for maintaining stability [24]. |
| Trehalose | Non-reducing disaccharide cryo- & lyo-protectant. | Prevents aggregation, narrows size distribution, preserves biological activity across freeze-thaw cycles, and serves as a lyoprotectant [1] [23]. |
| Human Serum Albumin (HSA) / Bovine Serum Albumin (BSA) | Protein additive that stabilizes vesicles. | Prevents adsorption to tube walls and improves recovery. PBS-HAT buffer (with HSA & Trehalose) shows drastically improved preservation [23] [25]. |
| Specialized EV Storage Buffer | Optimized, pre-formulated buffer (e.g., with Trehalose & BSA). | Shown to better protect EV cargo (e.g., DNA), maintain particle numbers, and preserve targeting functionality compared to PBS [25]. |
Technical Support Center
Frequently Asked Questions (FAQs)
Q1: Why is it critical to minimize freeze-thaw cycles for exosome samples? A: Each freeze-thaw cycle induces mechanical and osmotic stress, leading to exosome membrane rupture, loss of cargo (e.g., RNA, proteins), and increased aggregation. This compromises downstream experimental results, including functional assays and biomarker profiling.
Q2: What is the optimal aliquot volume to prepare? A: The optimal volume is the exact amount required for a single experiment. Common practice is 10-100 µL, depending on the assay. This minimizes the volume of sample subjected to repeated thawing and avoids the need for re-freezing any leftover material.
Q3: What is the recommended cooling rate for "rapid" freezing? A: A cooling rate of -1°C to -3°C per minute is often recommended until the sample passes the freezing point, after which it can be transferred to long-term storage. This controlled rate minimizes ice crystal formation that can damage exosomes.
Q4: Can I use liquid nitrogen for flash-freezing my exosome aliquots? A: Direct immersion in liquid nitrogen is not recommended for small aqueous volumes in standard tubes due to the risk of tube rupture and potential sample contamination. Using a pre-cooled rack in a -80°C freezer or a specialized controlled-rate freezer is safer and more effective.
Troubleshooting Guide
Problem: Low exosome recovery post-thaw.
- Potential Cause: Exosomes are adhering to the tube walls.
- Solution: Use low protein-binding tubes (e.g., siliconized tubes). Briefly centrifuge the tube before opening to collect the entire sample.
Problem: Increased particle size and polydispersity after thawing.
- Potential Cause: Exosome aggregation due to slow freezing or the absence of a cryoprotectant.
- Solution: Ensure rapid freezing protocols are followed. Incorporate a cryoprotectant like trehalose (e.g., 5-10% w/v) or HSA (0.5-1%) into the resuspension buffer.
Problem: Loss of biological activity in functional assays.
- Potential Cause: Damage to surface proteins from ice crystal formation during freezing.
- Solution: Implement single-use aliquoting strictly. Verify the use of a cryoprotectant and avoid any refreezing of thawed samples.
Experimental Protocols
Protocol 1: Single-Use Aliquoting and Rapid Freezing
- Preparation: Pre-chill a box of 1.5 mL or 2.0 mL low-protein-binding microcentrifuge tubes on ice.
- Mixing: Gently vortex the purified exosome suspension to ensure a homogeneous solution. Avoid foaming.
- Aliquoting: Using a calibrated pipette, dispense the desired single-experiment volume into each pre-chilled tube. Work quickly to minimize time at room temperature.
- Cryoprotection (Optional but Recommended): If using, add trehalose from a sterile stock solution to a final concentration of 5-10% and mix gently by pipetting.
- Freezing:
- Place the aliquots in a pre-chilled (4°C) isopropanol freezing jar or a passive cooling device.
- Immediately transfer the jar to a -80°C freezer for a minimum of 2 hours. This ensures a controlled cooling rate of approximately -1°C/min.
- Long-Term Storage: After rapid freezing, transfer the aliquots to a designated rack in the -80°C freezer for long-term storage. Maintain a detailed inventory to track aliquot usage.
Protocol 2: Assessing Exosome Integrity Post-Thaw (NTA and Protein Assay)
- Thawing: Remove one single-use aliquot from -80°C and thaw it rapidly in a 37°C water bath for 60-90 seconds. Gently mix the tube by inversion.
- Nanoparticle Tracking Analysis (NTA):
- Dilute the thawed aliquot in sterile, particle-free PBS to a concentration within the ideal detection range of the NTA instrument (e.g., 10^8-10^9 particles/mL).
- Load the sample and perform particle sizing and concentration analysis according to the manufacturer's instructions. Compare the mean/median particle size and mode size to a freshly prepared sample.
- Protein Assay (e.g., BCA):
- Lyse a separate portion of the thawed aliquot with RIPA buffer.
- Perform a BCA protein assay according to the kit protocol.
- Compare the total protein yield to a freshly prepared sample to assess cargo retention.
Data Presentation
Table 1: Impact of Freeze-Thaw Cycles on Exosome Integrity
| Freeze-Thaw Cycles | Mean Particle Size (nm) | Particle Concentration (particles/mL) | Total Protein Recovery (%) |
|---|---|---|---|
| 0 (Fresh) | 125 ± 5 | 3.5 x 10^10 ± 0.2 x 10^10 | 100 ± 3 |
| 1 | 130 ± 7 | 3.2 x 10^10 ± 0.3 x 10^10 | 95 ± 4 |
| 3 | 155 ± 12 | 2.1 x 10^10 ± 0.4 x 10^10 | 78 ± 6 |
| 5 | 210 ± 25 | 1.3 x 10^10 ± 0.3 x 10^10 | 60 ± 8 |
Table 2: Efficacy of Cryoprotectants in Preventing Aggregation
| Cryoprotectant Condition | Mean Particle Size Post-Thaw (nm) | % Aggregates (>300nm) |
|---|---|---|
| None (PBS only) | 155 ± 12 | 25% |
| 5% Trehalose | 128 ± 6 | 5% |
| 1% HSA | 132 ± 8 | 7% |
| 5% DMSO | 140 ± 10 | 15% |
Diagrams
Title: Exosome Aliquoting and Freezing Workflow
Title: Pathways of Freeze-Thaw Induced Damage
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions
| Item | Function & Rationale |
|---|---|
| Low-Protein-Bind Microcentrifuge Tubes | Minimizes adhesion of exosomes to tube walls, maximizing recovery post-thaw. |
| D-(+)-Trehalose Dihydrate | A non-reducing disaccharide cryoprotectant that stabilizes membranes and proteins by forming a glassy state and replacing water molecules. |
| Human Serum Albumin (HSA) | Acts as a bulking agent and cryoprotectant, reducing surface-induced stress and aggregation. |
| Particle-Free PBS | Used for dilution and buffer exchange; the absence of contaminating particles is critical for accurate NTA. |
| Controlled-Rate Freezing Jar | Provides a consistent cooling rate of ~-1°C/min, preventing the damaging effects of slow or flash freezing. |
Troubleshooting Common Pitfalls and Advanced Optimization Strategies
What are the key indicators that my exosome sample has degraded during storage?
The primary indicators of exosome storage failure are measurable changes in physical characteristics, which can be diagnosed using specific instrumentation. The table below summarizes the key signs of degradation and the appropriate methods for detecting them.
| Sign of Failure | Diagnostic Method | Observation in Degraded Samples |
|---|---|---|
| Particle Aggregation | Nanoparticle Tracking Analysis (NTA), Tunable Resistive Pulse Sensing (TRPS) | Increased standard deviation and span of size distribution; visible aggregates [1]. |
| Transmission Electron Microscopy (TEM) | Visual observation of fused vesicles or clusters of particles [2]. | |
| Increase in Particle Size | Dynamic Light Scattering (DLS), NTA | Increase in mean particle size, hydrodynamic diameter, and modal size [16] [4]. |
| Loss of Particle Concentration | NTA, TRPS | Reduction in the number of particles per milliliter or per microgram of protein [16] [4]. |
| Change in Surface Charge | Zeta Potential Measurement | Shift in zeta potential towards zero, reducing electrostatic repulsion and increasing aggregation propensity [4] [1]. |
What is the experimental protocol for systematically assessing exosome stability?
A robust protocol for diagnosing storage failure involves characterizing samples before and after storage using a multi-modal approach.
Methodology for Stability Assessment
Baseline Characterization: Isolate exosomes (e.g., via size-exclusion chromatography or ultracentrifugation) and perform a full characterization of the fresh sample. This includes NTA for concentration and size, TEM for morphology, and Western Blot for marker expression (e.g., CD63, TSG101) [16] [4].
Storage Intervention: Aliquot the exosomes into the desired storage buffer (e.g., PBS, PBS with trehalose). Subject the aliquots to the storage condition being tested (e.g., -80°C, freeze-thaw cycles, lyophilization) [16] [1].
Post-Storage Analysis: After the storage period, thaw and reconstitute the samples appropriately.
- Concentration & Size Distribution: Analyze using NTA. A significant increase in mean/median size and a broadening of the size distribution indicate aggregation. A drop in particle concentration suggests fragmentation or adsorption to tubes [16] [4] [1].
- Morphology: Use TEM to visually confirm the integrity of the lipid bilayer and look for fused, ruptured, or irregularly shaped vesicles that are not present in fresh samples [2].
- Surface Charge: Measure zeta potential. A shift towards neutral values (e.g., from -20 mV to -10 mV) is a quantitative indicator of decreased colloidal stability [4] [1].
- Cargo Integrity (Optional): For a comprehensive analysis, extract and quantify RNA or protein content to confirm cargo retention [2].
Diagram 1: Experimental workflow for diagnosing exosome storage failure, showing the key comparison between pre- and post-storage analytical data.
What underlying mechanisms cause these signs of storage failure?
The observable signs of degradation are the result of physical and biochemical processes.
Aggregation and Size Increase: During freezing, ice crystal formation creates osmotic and mechanical stress that can compromise the exosome membrane. Upon thawing, this can lead to vesicle fusion, creating larger, artefactual particles. Evidence from flow cytometry experiments with differently tagged exosomes shows a population of double-positive particles after freeze-thaw, strongly suggesting membrane fusion [4]. When electrostatic repulsion (measured by zeta potential) is insufficient, exosomes can also aggregate due to attractive van der Waals forces [1].
Concentration Loss: Freeze-thaw cycles can cause irreversible lysis or rupture of exosomes, releasing their cargo and causing the particle count to drop [2] [4]. Furthermore, fragmented or damaged vesicles may fall below the detection limit of characterization instruments. Multiple freeze-thaw cycles are particularly detrimental, with the most significant particle loss occurring after the first cycle [4].
Diagram 2: Causal pathways linking storage stress to observable signs of exosome failure.
How can I prevent aggregation and concentration loss during storage?
Prevention strategies focus on mitigating the damaging effects of freezing and storage.
Use Cryoprotectants: Add trehalose (e.g., 25 mM) to the storage buffer. Trehalose functions through the "water replacement" theory, stabilizing the phospholipid bilayer and preventing aggregation during freezing and freeze-drying. Studies show it narrows particle size distribution and maintains particle concentration after freeze-thaw cycles [1]. Other agents like human serum albumin (HSA) have also shown protective effects [16].
Avoid Repeated Freeze-Thaw Cycles: Always aliquot exosomes into single-use volumes before freezing. Each freeze-thaw cycle induces particle loss and size increase [4] [26].
Optimize Storage Temperature and Buffer:
- For long-term storage (months to years), -80°C is the preferred temperature [26] [17].
- For short-term storage (≤72 hours), refrigeration at 4°C may be superior to freezing, as it avoids damage from the freezing process itself [16].
- Consider lyophilization (freeze-drying) with trehalose as a lyoprotectant for room-temperature storage, which can prolong shelf-life and reduce costs [16].
Research Reagent Solutions
| Reagent | Function in Exosome Storage | Key Findings |
|---|---|---|
| Trehalose | Cryoprotectant & Lyoprotectant | Prevents aggregation, maintains size distribution, and preserves biological activity during freezing and lyophilization [1]. |
| Phosphate-Buffered Saline (PBS) | Common Storage Buffer | Standard buffer, but may cause vesicle damage and aggregation during storage without additives [16]. |
| Human Serum Albumin (HSA) | Stabilizing Additive | Improves exosome quality following short-term and long-term storage when added to PBS [16]. |
| Dimethyl Sulfoxide (DMSO) | Cryoprotectant | Used in some protocols, but potential cytotoxicity and interference with downstream processes should be considered [2] [4]. |
Troubleshooting Guides
Guide 1: Addressing Exosome Aggregation and Loss of Function After Freezing and Thawing
Problem: After thawing frozen exosome samples, you observe an increase in particle size, a decrease in particle concentration, and a loss of biological activity in functional assays.
Explanation: Multiple freeze-thaw cycles are highly detrimental to exosome integrity. The formation of ice crystals during slow freezing can physically damage the lipid bilayer, leading to membrane deformation, vesicle fusion, and rupture. This damage results in the leakage of cargo (like RNA and proteins) and the loss of surface markers crucial for function [2] [17]. Each cycle increases the degree of aggregation and functional impairment [2].
Solution:
- Implement Single-Use Aliquots: Divide the exosome preparation into small, single-use volumes before the initial freeze to completely avoid repeated thawing [27] [28].
- Optimize Freezing Rate: Use a controlled-rate freezer or an isopropanol freezing chamber (e.g., "Mr. Frosty") placed at -80°C overnight. This ensures a slow, controlled cooling rate of approximately -1°C per minute, which reduces ice crystal formation [29] [28].
- Incorporate Cryoprotectants: Add stabilizers like trehalose (e.g., 25 mM) to the storage buffer. Trehalose acts as a water replacement and stabilizes the lipid membrane, preventing aggregation and preserving biological activity during freezing and thawing [2] [1].
Guide 2: Inconsistent Experimental Results from Exosomes Stored in PBS
Problem: Cell uptake assays or functional studies using exosomes stored in phosphate-buffered saline (PBS) show high variability and inconsistent results between batches.
Explanation: While PBS is a commonly used storage buffer, it can be suboptimal for exosome stability. Storage in PBS alone, especially at -80°C, can lead to vesicle aggregation, fragmentation, and damage to the exosome membrane over time, compromising their function [16]. The lack of protective agents makes exosomes suspended in PBS particularly susceptible to damage from freeze-thaw cycles.
Solution:
- Modify Storage Buffer: Supplement PBS with protective additives.
- Consider Alternative Buffers: For certain applications, storing exosomes in a sucrose solution or even in their native biofluid (e.g., plasma) has been shown to offer superior preservation of ultrastructure, size distribution, and surface protein expression compared to PBS [2] [17] [16].
- Avoid Freezing Altogether for Short-Term Use: For experiments conducted within 72 hours, storage at 4°C may be a viable option to avoid freeze-thaw damage entirely [16].
Guide 3: Selecting a Storage Strategy for Clinical Applications
Problem: Developing a robust, scalable, and compliant storage protocol for exosome-based therapeutics or diagnostics.
Explanation: Clinical translation requires protocols that ensure exosome stability over long periods, minimize batch-to-batch variation, and are compatible with Good Manufacturing Practices (GMP). Reliance on -80°C freezers alone presents risks from equipment failure and requires a continuous cold chain, which complicates logistics and distribution [16].
Solution:
- Adopt Lyophilization: Freeze-drying (lyophilization) enables long-term storage of exosomes at room temperature, dramatically simplifying logistics and reducing costs [16].
- Use Lyoprotectants: Lyophilization without protectants can damage exosomes. The use of trehalose or sucrose as lyoprotectants is critical to maintain exosome size, morphology, and cargo content during the freeze-drying process [16].
- Implement Automated Cryogenic Systems: For liquid nitrogen storage, automated systems (e.g., at -135°C to -190°C) enhance sample integrity by minimizing transient warming events during retrieval, providing rigorous inventory management, and ensuring full traceability—key elements for regulatory compliance [30] [31].
Frequently Asked Questions (FAQs)
FAQ 1: What is the single most effective step I can take to protect my exosome samples from freeze-thaw damage?
The most critical step is to aliquot your exosome preparation into single-use volumes before the initial freeze [27] [28]. This simple practice eliminates the need for repeated thawing of a master stock and is the most effective way to mitigate the damaging effects of multiple freeze-thaw cycles on particle concentration, size, and bioactivity [2].
FAQ 2: Besides avoiding multiple freeze-thaw cycles, what are the optimal short-term and long-term storage temperatures for exosomes?
For short-term storage (≤72 hours), 4°C is a viable option and avoids freezing-related damage altogether [16]. For long-term storage, -80°C is the most commonly recommended and studied temperature [2] [17]. For maximum long-term stability (years), storage in the vapor phase of liquid nitrogen (below -135°C) is considered the gold standard, as it effectively suspends all biochemical activity [30] [28].
FAQ 3: How do cryoprotectants like trehalose work to protect exosomes?
Trehalose, a non-reducing sugar, protects exosomes through multiple mechanisms. It acts as a cryoprotectant by forming a glassy matrix during freezing, which reduces mechanical stress from ice crystals. It also functions as a lyoprotectant during freeze-drying by stabilizing the exosome's lipid bilayer through the "water replacement" theory, preventing membrane fusion and aggregation during both freezing and drying processes [1].
FAQ 4: We are considering freeze-drying our exosome product. What are the key challenges?
The primary challenge is preventing damage during the ice crystal formation and dehydration phases, which can compromise structural integrity and cargo [16]. Success hinges on using an optimized formulation with lyoprotectants like trehalose or sucrose. These agents help maintain the exosome's spherical morphology, particle concentration, and protein/RNA content after reconstitution [16].
Experimental Data & Protocols
Table 1: Impact of Storage Conditions on Exosome Integrity
Summary of quantitative data on key exosome parameters under different storage conditions.
| Storage Condition | Particle Concentration | Size Distribution / Aggregation | Biological Activity | Key Findings |
|---|---|---|---|---|
| Multiple Freeze-Thaw Cycles | Decreased [2] | Increased size & aggregation [2] | Impaired [2] | Rapid loss of RNA content and biofunction; membrane deformation observed [2]. |
| -80°C with Trehalose | Maintained [1] | Narrower distribution; less aggregation [1] | Better preserved [1] | Higher particle count per μg protein; consistent stimulation of immune responses [1]. |
| Lyophilization with Trehalose | Some loss possible [16] | Size integrity maintained [16] | Varies | Enables room-temperature storage; buffer-dependent effects on concentration [16]. |
| 4°C (Short-Term) | Varies | Increased over time [32] | Gradual decline [32] | Viable alternative to avoid freeze-thaw damage for very short-term use [16]. |
| Storage in Native Biofluid | N/A | Improved stability [2] | Better preserved [2] | Superior stability vs. purified EVs in PBS buffers [2]. |
Protocol 1: Isolating and Freezing Exosomes with Trehalose for Optimal Stability
This protocol outlines a method for isolating exosomes and resuspending them in a trehalose-containing buffer to maximize stability during freezing [2] [1].
Materials:
- Cryoprotectant: D-(+)-Trehalose dihydrate
- Sterile Phosphate-Buffered Saline (PBS)
- Cryogenic vials
- Controlled-rate freezing container (e.g., "Mr. Frosty") or -80°C freezer
Procedure:
- Isolate exosomes from your cell culture supernatant or biofluid using your preferred method (e.g., differential ultracentrifugation, size-exclusion chromatography).
- Prepare a 25 mM trehalose solution in sterile PBS. Filter-sterilize through a 0.22 μm membrane.
- After the final isolation wash, thoroughly resuspend the exosome pellet in the trehalose/PBS solution.
- Immediately aliquot the exosome suspension into single-use cryogenic vials.
- Place the vials in a controlled-rate freezing container and transfer them to a -80°C freezer for overnight freezing.
- For long-term storage, transfer the frozen vials to a liquid nitrogen tank for storage in the vapor phase (-135°C to -196°C).
Protocol 2: Evaluating the Stability of Stored Exosomes via Nanoparticle Tracking Analysis (NTA)
This protocol describes how to assess the impact of storage on exosome concentration and size.
Materials:
- Nanoparticle Tracking Analysis (NTA) instrument (e.g., Malvern Nanosight)
- Sterile PBS or water for dilution
Procedure:
- Rapidly thaw the frozen exosome sample in a 37°C water bath and keep it on ice.
- Dilute the sample appropriately in sterile PBS or filtered water to achieve an ideal concentration for NTA measurement (e.g., 20-100 particles per frame).
- Inject the sample into the NTA instrument and perform the analysis according to the manufacturer's instructions. Ensure consistent measurement settings (e.g., camera level, detection threshold) across all samples for valid comparison.
- Record the particle concentration (particles/mL) and mode/mean particle size.
- Compare the results from stored samples (or those subjected to freeze-thaw cycles) with data from freshly isolated exosomes to determine the extent of aggregation and particle loss.
Visualizations
Exosome Storage Stability Workflow
Damage from Improper Storage
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Exosome Storage workflows
| Item | Function & Rationale | Example |
|---|---|---|
| Cryoprotectant | Stabilizes the exosome lipid bilayer during freezing by preventing ice crystal formation and reducing membrane fusion. Trehalose is preferred for its proven efficacy and low toxicity [1]. | D-(+)-Trehalose dihydrate |
| Lyoprotectant | Protects exosomes during the freeze-drying (lyophilization) process by forming a stable, glassy matrix that prevents dehydration-induced damage, enabling room-temperature storage [16]. | Sucrose, Trehalose |
| Controlled-Rate Freezer | Ensures a consistent, slow freezing rate (approx. -1°C/min), which is critical to minimize cellular and vesicular damage from rapid ice crystal formation [29] [28]. | Mr. Frosty (Nalgene), Corning CoolCell |
| Cryogenic Vials | Specially designed tubes that withstand extreme temperatures of liquid nitrogen storage without cracking, ensuring sample integrity and safety [29] [28]. | Internal-threaded cryovials |
| Serum Albumin | Acts as a bulking agent and stabilizer in storage buffers, helping to preserve exosome quality and function during storage [16]. | Human Serum Albumin (HSA), Bovine Serum Albumin (BSA) |
| Automated Storage System | Provides secure, traceable, and consistent cryogenic storage while minimizing transient warming events caused by manual handling, crucial for clinical-grade samples [30] [31]. | Azenta CryoArc |
FAQs on Preventing Exosome Aggregation
FAQ 1: What are the most critical factors to prevent aggregation during exosome storage? The most critical factors are consistent sub-zero storage temperature, the use of appropriate cryoprotectant buffers, and minimizing freeze-thaw cycles. Storage at -80°C is widely recommended for long-term stability [2] [4]. The choice of storage buffer significantly impacts stability; phosphate-buffered saline (PBS) is commonly used, but adding cryoprotectants like trehalose or human serum albumin (HSA) can better preserve exosome integrity by preventing ice crystal formation and membrane fusion [2] [4] [16]. Avoiding repeated freeze-thaw cycles is crucial, as each cycle can lead to particle loss, increased average size, and aggregation [4].
FAQ 2: How should storage protocols be adapted for exosomes from different sources? Optimal storage conditions can vary depending on the exosome source:
- MSC-EVs: Isolated MSC-EVs in PBS show better concentration retention over short-term storage (≤2 weeks) compared to other buffers like normal saline (NS) or 5% glucose solution (GS) [16].
- Biofluid-Derived Exosomes: For exosomes isolated from plasma or serum, storage in the native biofluid itself offers improved stability over purified exosomes resuspended in buffers like PBS. This is likely due to the natural protein and lipid environment which acts as a stabilizer [2] [4].
- Engineered Exosomes: For sensitive engineered exosomes, more advanced strategies like embedding them in a stabilizing matrix (e.g., hyaluronic acid-based microneedles) can maintain their count, size, and bioactivity for up to 12 months at room temperature [2].
FAQ 3: Does lyophilization (freeze-drying) help prevent aggregation in long-term storage? Lyophilization is a promising strategy for enabling room-temperature storage, but it requires careful optimization to prevent aggregation. While lyophilization can help maintain exosome size distribution, it may lead to a significant loss in particle concentration upon reconstitution unless protective agents are used [16]. The addition of lyoprotectants like trehalose or sucrose before lyophilization is essential to protect exosomal membranes from ice crystal damage and maintain the integrity of their cargo [2] [16].
FAQ 4: What quality control checks should be performed after storage and thawing? After thawing, you should characterize your exosomes to check for aggregation and integrity loss. Key analyses include:
- Concentration and Size Distribution: Use Nanoparticle Tracking Analysis (NTA) or Tunable Resistive Pulse Sensing (TRPS) to detect changes in particle concentration, average size increases, or a wider size distribution, which are indicators of aggregation [33] [4].
- Morphology: Use Transmission Electron Microscopy (TEM) to visually inspect for membrane damage, vesicle fusion, or clumping [2] [4].
- Cargo Integrity: Perform Western Blot to check for the presence and integrity of protein markers (e.g., CD63, TSG101) and use techniques like RNA electrophoresis to assess RNA content [2] [10].
- Functionality: When possible, perform a functional assay (e.g., cell uptake or a bioactivity assay) to confirm that storage has not impaired biological function [2].
Table 1: Impact of Storage Conditions on Exosome Integrity
| Storage Factor | Key Findings | Recommendation |
|---|---|---|
| Temperature | -80°C is standard for long-term storage; 4°C may be superior for short-term (≤72h) to avoid freeze-thaw damage [2] [16]. | Use -80°C for archives; 4°C for frequently used samples. |
| Freeze-Thaw Cycles | Each cycle decreases particle concentration, increases size, and impairs bioactivity. The first cycle causes the most significant damage [2] [4]. | Aliquot exosomes to avoid multiple freeze-thaw cycles. |
| Storage Buffer | PBS is standard but suboptimal. Additives like trehalose (25-50mM) or HSA improve stability. Native biofluids offer natural protection [2] [4] [16]. | Use cryoprotectant-enriched buffers or store in native biofluid if possible. |
| Lyophilization | Enables room-temperature storage but can cause concentration loss. Lyoprotectants are critical for success [16]. | Implement with trehalose for specific applications needing ambient storage. |
Table 2: Protocol Adaptation for Different Exosome Sources
| Exosome Source | Isolation Considerations | Recommended Storage Strategy | Key Stability Metrics |
|---|---|---|---|
| MSC-EVs | Often isolated via Ultracentrifugation or TFF [34] [35]. | PBS with trehalose at -80°C; avoid lyophilization without optimization [16]. | Concentration, CD63/CD81 expression, pro-angiogenic function. |
| Biofluid-Derived (e.g., Plasma) | SEC is effective for removing contaminants like lipoproteins [4]. | Small aliquots in native plasma or PBS with HSA at -80°C [2] [4]. | Particle size distribution, absence of apolipoproteins. |
| Engineered Exosomes | May require specialized purification (e.g., SEC, immunoaffinity) [33]. | Matrix-embedded (e.g., microneedles) for RT storage; trehalose in PBS for -80°C [2]. | Cargo integrity (e.g., loaded RNA), targeting ligand functionality. |
Experimental Protocols for Stability Assessment
Protocol 1: Evaluating Buffer Additives for Cryopreservation
This protocol assesses different buffers and additives for their ability to preserve exosome integrity during frozen storage.
1. Sample Preparation:
- Isolate exosomes from your source (e.g., MSC culture supernatant) using a consistent method like Size-Exclusion Chromatography (SEC) or Tangential Flow Filtration (TFF) [33] [35].
- Divide the purified exosome preparation into equal volumes and resuspend them in the following buffers:
- PBS (Control)
- PBS + 25mM Trehalose
- PBS + 0.1% Human Serum Albumin (HSA)
- Normal Saline (NS)
- 5% Glucose Solution (GS)
- Create small, single-use aliquots (e.g., 20-50 µL) for each condition to avoid freeze-thaw cycles.
2. Storage and Analysis:
- Store the aliquots at -80°C for a defined period (e.g., 2 weeks, 1 month, 6 months).
- After storage, thaw an aliquot of each condition on ice and characterize using:
Protocol 2: Testing the Impact of Freeze-Thaw Cycles
This protocol systematically quantifies the damage caused by repeated freezing and thawing.
1. Baseline Characterization:
- Take a fresh, purified exosome sample and perform a baseline analysis of particle concentration (by NTA), size, and marker expression (by Western Blot) [4].
2. Cycling and Measurement:
- Subject the remaining sample to sequential freeze-thaw cycles. For each cycle:
- Freeze completely at -80°C for at least 4 hours.
- Thaw completely in a room-temperature water bath or on ice.
- After 1, 3, and 5 cycles, remove an aliquot and analyze particle concentration and size using NTA or TRPS.
- After the final cycle, perform Western Blot analysis to compare marker presence and integrity with the baseline sample.
Protocol 3: Functional Assay After Storage
This protocol verifies that storage has not compromised exosome bioactivity.
1. Cell-Based Assay Setup:
- Choose a relevant functional assay. For example, if studying MSC-EVs for wound healing, use a fibroblast proliferation/migration assay [34] [36].
- Culture fibroblasts in a standard medium until they reach 70-80% confluence.
2. Treatment and Analysis:
- Create a scratch (gap) in the cell monolayer.
- Treat the cells with:
- Freshly isolated exosomes (positive control).
- Exosomes that have been stored under test conditions.
- PBS only (negative control).
- Monitor and quantify the closure of the scratch over 24-48 hours using live-cell imaging or endpoint staining. Comparable scratch closure between fresh and stored exosomes indicates preserved bioactivity [2].
The Scientist's Toolkit
Table 3: Essential Reagents and Materials for Exosome Storage Research
| Item | Function/Application |
|---|---|
| Trehalose | A non-reducing disaccharide that stabilizes lipid bilayers during freezing and desiccation by forming a glassy state, preventing ice crystal formation [2] [16]. |
| Dimethyl Sulfoxide (DMSO) | A common cryoprotectant that penetrates cells and vesicles, but its use for EVs is debated due to potential cytotoxicity and effects on downstream applications [2] [4]. |
| Human Serum Albumin (HSA) | Acts as a bulking agent and stabilizer in solution, reducing aggregation and surface adsorption of exosomes during storage [16]. |
| Protease Inhibitor Cocktail | Prevents proteolytic degradation of exosomal surface and internal proteins during storage, especially important for biofluid-derived samples [4]. |
| Size-Exclusion Chromatography (SEC) Columns (e.g., qEV) | For high-purity isolation of exosomes from biofluids and cell culture media with minimal damage, which is a critical first step before storage studies [33] [4]. |
| Nanoparticle Tracking Analyzer (NTA) | Instrument for measuring particle concentration and size distribution before and after storage to quantify aggregation and loss [33] [4]. |
Workflow Diagrams for Experimental Planning
Diagram 1: Overall workflow for optimizing exosome storage protocols.
Diagram 2: A decision tree for selecting a storage strategy based on research goals.
Lyophilization, or freeze-drying, is a sophisticated dehydration process that removes water from a frozen material via sublimation under reduced pressure. For researchers working with extracellular vesicles (EVs) like exosomes, this technique offers a promising solution for achieving room-temperature storage and enhancing shelf-life stability. However, the process carries inherent risks of structural compromise that must be carefully managed. This technical support center provides comprehensive guidance for scientists navigating the complexities of exosome lyophilization, with specific protocols to prevent aggregation and maintain functional integrity.
Frequently Asked Questions (FAQs)
Q1: What are the primary advantages of lyophilizing exosomes compared to standard frozen storage? Lyophilization offers three key advantages: (1) Significant shelf-life extension, potentially for years, by enabling room-temperature storage and eliminating cold chain requirements [37] [38]; (2) Enhanced transport logistics due to reduced weight and stability at ambient temperatures [37] [39]; and (3) Microbial growth inhibition by removing water necessary for metabolic activity [37] [40].
Q2: What are the critical risks of structural compromise during exosome lyophilization? The main risks are (1) Membrane deformation and vesicle fusion leading to increased particle size and aggregation [2] [3]; (2) Cargo loss or degradation of functional RNA, proteins, and lipids due to process stresses [2] [41]; and (3) Irreversible aggregation during reconstitution, particularly after multiple freeze-thaw cycles or suboptimal freezing [3] [1].
Q3: Which formulation additives help stabilize exosomes during lyophilization? Trehalose (25-100 mM) is the most documented cryoprotectant, acting through water replacement and vitrification mechanisms to shield exosome membranes and prevent aggregation [1]. Sucrose and mannitol are also used as bulking agents and cryoprotectants in pharmaceutical lyophilization [42], though specific exosome data is less extensive than for trehalose.
Q4: What is the optimal storage temperature after lyophilization? While lyophilization enables room-temperature storage, for maximum long-term stability (years), storing desiccated products at 2-8°C protects against thermal degradation. Critical storage parameters include protection from oxygen, moisture, and light through hermetic sealing with proper primary packaging [38].
Q5: How do multiple freeze-thaw cycles affect lyophilized exosomes? Multiple freeze-thaw cycles progressively degrade exosome quality, causing decreased particle concentrations, reduced RNA content, impaired bioactivity, and increased aggregation [2] [3] [41]. Aliquotting into single-use portions is strongly recommended to avoid repeated cycling.
Troubleshooting Common Lyophilization Issues
Problem 1: Exosome Aggregation After Reconstitution
Potential Causes:
- Improper freezing rate leading to large ice crystal formation that damages exosome membranes [37]
- Inadequate cryoprotectant concentration or type [1]
- Residual moisture too high or too low after secondary drying [43]
- Osmotic shock during reconstitution [41]
Solutions:
- Implement controlled nucleation during freezing to ensure uniform ice crystal structure [42]
- Optimize trehalose concentration between 25-100 mM based on your exosome source and concentration [1]
- Validate residual moisture with Karl Fischer titration, targeting <1% for most formulations [43]
- Use phosphate-buffered saline (PBS) for gradual rehydration instead of pure water [41]
Problem 2: Loss of Biological Activity Post-Lyophilization
Potential Causes:
- Temperature excursion during primary drying exceeding the product's collapse temperature [43]
- Oxidative damage during storage [38]
- Protein denaturation or RNA degradation due to insufficient stabilization [2] [3]
Solutions:
- Determine critical formulation parameters including eutectic point (Teu) and glass transition temperature (Tg') before cycle development [39]
- Use antioxidant additives or inert gas (nitrogen) headspace flushing during vial stoppering [39]
- Implement functional potency assays (e.g., cytokine response in recipient cells) rather than just physical characterization [1]
Problem 3: Inconsistent Results Across Batches
Potential Causes:
- Variable freezing rates between vial positions in the lyophilizer [43]
- Inadequate process control and monitoring [42]
- Formulation heterogeneity between preparations [39]
Solutions:
- Employ thermal mapping to identify shelf temperature variations and implement a standardized loading pattern [43]
- Install Process Analytical Technology (PAT) tools for real-time monitoring of critical process parameters [42]
- Standardize pre-lyophilization steps including exosome isolation, concentration, and buffer exchange [3]
Lyophilization Process Optimization Guide
Comparative Analysis: Lyophilization Advantages vs. Risks
Table 1: Comprehensive benefits and challenges of exosome lyophilization
| Advantages | Associated Risks & Mitigation Strategies |
|---|---|
| Room Temperature Storage: Enables shipping and storage without cold chain, potentially for 10-25 years [38] | Structural Damage Risk: Implement controlled nucleation and optimize cryoprotectants [37] [42] |
| Microbial Stability: Low water activity prevents bacterial growth and degradation [37] [40] | Aggregation During Reconstitution: Use optimized rehydration protocols with PBS [41] [1] |
| Convenient Handling: Lightweight, portable samples with reduced storage space requirements [37] | Process Complexity: Requires specialized equipment and extensive process development [39] |
| Reduced Contaminants: Eliminates need for preservatives that might interfere with downstream applications [37] | High Capital Investment: Significant equipment costs and energy consumption [39] [40] |
Quantitative Impact of Storage Conditions on EV Integrity
Table 2: Effects of storage parameters on exosome quality metrics based on systematic review data [2] [3]
| Storage Condition | Particle Concentration | Size Distribution | RNA Content | Bioactivity |
|---|---|---|---|---|
| -80°C (Reference) | Minimal decrease | Stable | >90% preserved | Maintained |
| -20°C | Moderate decrease | Increased aggregation | ~70-80% preserved | Partial loss |
| 4°C (short-term) | Significant decrease over days | Progressive increase | Rapid degradation | Substantial loss |
| Multiple Freeze-Thaw Cycles | Decreases with each cycle | Marked increase & aggregation | Progressive loss | Significantly impaired |
| With Trehalose Stabilizer | Better preservation | Reduced aggregation | Improved protection | Enhanced maintenance |
Experimental Protocols for Exosome Lyophilization
Protocol 1: Trehalose-Stabilized Lyophilization
Background: Based on the systematic review by Saeed et al. (2024) demonstrating trehalose effectively prevents EV aggregation and cryodamage [2] [3], and the specific research of Krol et al. (2016) showing trehalose maintains exosome integrity through freeze-thaw cycles [1].
Materials:
- Purified exosome preparation
- Trehalose (pharmaceutical grade)
- Cryovials (2 mL)
- Lyophilizer with shelf temperature control capability
- Phosphate-buffered saline (PBS)
Methodology:
- Pre-formulation: Concentrate exosomes to target concentration (e.g., 1-5 × 10^10 particles/mL) via ultrafiltration.
- Buffer Exchange: Exchange storage buffer to PBS containing 25-100 mM trehalose using size exclusion chromatography or dialysis.
- Aliquoting: Dispense 1 mL volumes into sterile lyophilization vials under aseptic conditions.
- Freezing: Load vials onto pre-cooled lyophilizer shelves at -40°C and hold for 2 hours to ensure complete freezing.
- Primary Drying: Apply vacuum (≤100 mTorr) and maintain shelf temperature at -30°C for 24 hours to enable sublimation.
- Secondary Drying: Gradually raise shelf temperature to 25°C over 8 hours while maintaining vacuum to remove bound water.
- Sealing: Backfill with nitrogen gas and stopper vials under vacuum.
- Storage: Store at 2-8°C with desiccant until use.
Validation Assays:
- Nanoparticle Tracking Analysis (NTA) for concentration and size distribution
- Cryo-electron microscopy for morphological assessment
- Western blot for exosomal markers (CD63, CD81, TSG101)
- Functional assay (e.g., macrophage TNF-α secretion) for bioactivity [1]
Protocol 2: Controlled Nucleation for Improved Freezing Uniformity
Background: Temperature excursions during initial freezing can disrupt uniform ice crystal formation, leading to incomplete crystallization and heterogeneous moisture distribution [43]. Controlled nucleation standardizes the freezing step.
Materials:
- Formulated exosome sample (with appropriate cryoprotectants)
- Lyophilizer with controlled nucleation capability
- Temperature probes for product monitoring
Methodology:
- Sample Preparation: Load vials containing formulated exosomes onto lyophilizer shelves precooled to +4°C.
- Cooling Phase: Lower shelf temperature to -5°C at 1°C/min and hold for 30 minutes to equilibrate.
- Nucleation Induction: Apply a controlled nitrogen gas pulse or vacuum spike to induce simultaneous ice nucleation across all vials.
- Freezing Completion: After nucleation, immediately lower shelf temperature to -40°C at 0.5°C/min and hold for 2 hours.
- Proceed with Standard Lyophilization Cycle: Continue with primary and secondary drying phases as optimized for the formulation.
Quality Control Checkpoints:
- Visual inspection of cake structure (should be uniform with no melt-back)
- Residual moisture analysis by Karl Fischer (<1%)
- Reconstitution time (<3 minutes with gentle agitation)
Essential Research Reagent Solutions
Table 3: Key reagents and materials for exosome lyophilization research
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Trehalose | Cryoprotectant that shields membranes via water replacement | Use at 25-100 mM; superior to sucrose for preventing aggregation [1] |
| Dimethyl Sulfoxide (DMSO) | Cryoprotectant for pre-lyophilization freezing | Potential cytotoxicity; remove before lyophilization via buffer exchange [2] |
| Sucrose | Bulking agent and cryoprotectant | Common in pharmaceutical lyophilization; may be less effective than trehalose for exosomes [42] |
| Mannitol | Crystalline bulking agent | Provides elegant cake structure; can crystallize during freezing [42] |
| Poloxamer 188 | Surfactant for preventing surface-induced degradation | Minimizes interfacial stress during freezing and drying; use at 0.01-0.1% [39] |
Process Flow Visualization
Lyophilization Process Workflow: Critical unit operations for exosome preservation
Factors Affecting Lyophilized Exosome Stability: Key parameters influencing product quality
Successful lyophilization of exosomes requires meticulous attention to both process parameters and formulation components. While the benefits of room-temperature storage and extended shelf life are substantial, they must be balanced against the real risks of structural compromise and functional loss. The protocols and troubleshooting guides presented here provide a foundation for developing robust lyophilization processes that maintain exosome integrity. As research in this field evolves, continued refinement of stabilization strategies and process controls will further enhance the reliability of lyophilization as a preservation method for these critical therapeutic and diagnostic vesicles.
Frequently Asked Questions (FAQs)
Q1: How can I tell if my exosome sample has aggregated? You can identify exosome aggregation through several methods. Nanoparticle Tracking Analysis (NTA) will show a significant increase in mean particle size and a wider size distribution. Electron microscopy will reveal visible clumping, vesicle enlargement, and membrane deformation. Dynamic Light Scattering (DLS) may indicate decreased colloidal stability, and functional assays often demonstrate impaired bioactivity [2] [3] [1].
Q2: What are the main causes of exosome aggregation and precipitation? The primary causes include:
- Multiple freeze-thaw cycles: This is a major contributor to particle aggregation and size increase.
- Suboptimal storage conditions: Storage at -20°C instead of -80°C significantly promotes aggregation.
- Inappropriate storage buffers: PBS alone is prone to causing aggregation compared to buffers with cryoprotectants.
- High centrifugal forces: During isolation, ultracentrifugation can damage exosomes and promote clumping.
- Sample concentration: Highly concentrated exosome suspensions are more susceptible to aggregation [44] [2] [3].
Q3: Can aggregated exosomes be fully restored to their original state? While physical dispersion techniques can break up visible clumps, complete restoration of native morphology and, crucially, biological function is not always guaranteed. The primary goal of rescue protocols is to minimize further damage and recover as much functionality as possible for downstream applications. Prevention through optimized storage is always preferable to rescue [3] [1].
Q4: Does storage in native biofluid vs. purified buffer affect stability? Yes, evidence suggests that storing exosomes in their native biofluids (e.g., plasma, cell culture media) offers improved stability compared to purified and resuspended exosomes in simple buffers like PBS. The complex matrix of biofluids may have a natural stabilizing effect [3].
Troubleshooting Guide: Techniques for Sample Rescue
Problem: Aggregated Exosomes After Freeze-Thaw
Observation: Clumped particles, increased mean size on NTA, reduced biological activity.
Rescue Protocol:
- Gentle Vortexing: Briefly vortex the sample at a low to medium setting for 5-10 seconds.
- Pipette Mixing: Gently pipette the sample up and down with a standard or wide-bore pipette tip to physically disperse clumps.
- Sonication: Use a water bath sonicator (not a probe sonicator) for a short duration (30-60 seconds). Monitor closely to avoid overheating.
- Filtration: Pass the sample through a 0.22 µm or 0.45 µm syringe filter. This can remove large aggregates but may also result in the loss of some exosomes.
Prevention for Future:
- Aliquot: Divide the sample into single-use aliquots to avoid repeated freeze-thaw cycles [2] [3].
- Optimize Storage: Store at -80°C instead of -20°C [3] [26].
- Add Cryoprotectants: Use Trehalose (25 mM) in the storage buffer to prevent aggregation and cryodamage [1].
Problem: Precipitated Exosomes in Polymer-Based Kits
Observation: Visible pellet after low-speed centrifugation, low yield in supernatant.
Rescue Protocol:
- Thorough Resuspension: After the precipitation step, ensure the pellet is thoroughly resuspended in an appropriate buffer (e.g., PBS).
- Buffer Exchange: Perform a buffer exchange using size-exclusion chromatography (e.g., qEV columns) or ultrafiltration to remove the precipitating polymer (e.g., PEG), which can interfere with downstream applications [44] [45].
Prevention for Future:
- Consider alternative isolation methods like size-exclusion chromatography (SEC) or density gradient ultracentrifugation for higher purity and less aggregation [44] [45].
Quantitative Data on Storage Conditions and Aggregation
Table 1: Impact of Storage Conditions on Exosome Integrity
| Storage Condition | Impact on Particle Size | Impact on Concentration | Impact on Bioactivity | Key Evidence |
|---|---|---|---|---|
| Multiple Freeze-Thaw Cycles | Significant increase; aggregation | Decrease | Impaired | Systematic review of 50 studies [2] [3] |
| Storage at -20°C | Significant aggregation and size increase over time | Decrease over time | Impaired | EVs stored at -20°C showed aggregation vs. -80°C [3] |
| Storage at -80°C | Minimal change; best for integrity | Best preservation | Best preservation | Optimal for long-term preservation [3] [26] |
| Addition of 25mM Trehalose | Narrower size distribution; less aggregation | Higher particles/μg protein | Improved preservation of function (e.g., TNF-α stimulation) | Scientific study on beta-cell ELVs [1] |
Experimental Protocol: Evaluating Rescue Protocols with Trehalose
Objective: To assess the efficacy of trehalose in preventing and rescuing aggregated exosome samples.
Materials:
- Isolated exosomes (e.g., from cell culture medium via ultracentrifugation [46] [47] or SEC [44] [45])
- Trehalose solution (sterile)
- Phosphate-Buffered Saline (PBS)
- Nanoparticle Tracking Analysis (NTA) system
- Western blot reagents for markers (e.g., CD63, CD81) [10] [45]
- Functional assay reagents (e.g., macrophage TNF-α secretion assay [1])
Methodology:
- Exosome Isolation: Isolate exosomes from your chosen source using a standardized method [46] [44].
- Sample Preparation:
- Resuspend the exosome pellet in either PBS (control) or PBS containing 25mM Trehalose (experimental) [1].
- Induction of Stress:
- Subject aliquots from both groups to multiple freeze-thaw cycles (e.g., 3-5 cycles).
- Analysis:
- Physical Characterization: Use NTA to measure particle concentration and size distribution before and after stress [1].
- Molecular Characterization: Perform Western blotting for canonical exosome markers (CD9, CD63, CD81) to check for protein degradation or loss [10] [26].
- Functional Characterization: Apply the treated exosomes to a relevant bioassay (e.g., immune cell activation) to measure the preservation of biological activity [1].
Workflow Diagram for Rescue Protocol Evaluation
The Scientist's Toolkit: Essential Reagents for Recovery and Stabilization
Table 2: Key Research Reagent Solutions for Exosome Stabilization
| Reagent / Material | Function / Role | Example Usage in Protocol |
|---|---|---|
| Trehalose | Natural cryoprotectant; prevents aggregation and preserves functionality by stabilizing membranes. | Add to storage buffer at 25mM final concentration [1]. |
| Size-Exclusion Chromatography (SEC) Columns | For gentle buffer exchange to remove contaminants or precipitating polymers after rescue. | Use to replace aggregation-prone buffer with trehalose-containing buffer post-isolation [44] [45]. |
| Density Gradient Medium (e.g., Iodixanol) | Isolates high-purity exosomes based on buoyant density, reducing contaminants that can promote aggregation. | Use in density gradient ultracentrifugation as a high-purity isolation method [46] [44]. |
| Protease Inhibitors | Prevents degradation of exosomal proteins during processing and storage, which can exacerbate aggregation. | Add to lysis buffer or storage buffer when exosomes are resuspended [47] [10]. |
| Surface-Active Agents (e.g., BSA) | Can reduce nonspecific adhesion and aggregation of exosomes to tube walls. | Add at low concentrations (e.g., 0.1%) to storage buffers [48]. |
Validation and Quality Control: Ensuring Functional Integrity Post-Storage
Troubleshooting Guides
Nanoparticle Tracking Analysis (NTA) Troubleshooting
Problem: High background particle concentration in samples.
- Potential Cause: Contaminated buffers or improper system cleaning.
- Solution: Always filter buffers (e.g., PBS) before use. Perform thorough instrument cleaning: wash the syringe and glass chamber with 1 mL of distilled water 2-3 times until no particles are visible on the screen before sample injection [49].
- Prevention: Run a blank (diluent only) before sample measurements. A good blank should have a low particle concentration and display only 1-10 particles per screen in the live view [50].
Problem: Inconsistent particle concentration and size results between replicates.
- Potential Cause: Insufficient sample mixing or inhomogeneous sample.
- Solution: Use a magnetic stir bar inside the cuvette during analysis to maintain a homogeneous suspension [50]. For viscous samples, ensure adequate dilution in the appropriate buffer.
- Prevention: Prime the cuvette with your diluted sample before measurement. Load 400-500 μL of sample, discard it, and then load a fresh 400-500 μL for the actual measurement to wash out residues [50].
Dynamic Light Scattering (DLS) Troubleshooting
Problem: DLS results show high polydispersity or unexpected size distributions.
- Potential Cause: The presence of a small population of large aggregates can disproportionately skew the intensity-weighted size distribution due to the d^6 relationship between scattering intensity and particle size [51].
- Solution: Centrifuge the sample to remove large aggregates before analysis. Always perform measurements in triplicate on freshly prepared aliquots to distinguish true sample polydispersity from false positives caused by random sampling of large particles [51].
- Prevention: For samples with a polydispersity index (PDI) >0.7, the sample is likely unsuitable for DLS analysis as it will not give a stable distribution [51].
Problem: Periodic failures to meet size specifications despite a consistent process.
- Potential Cause: False positive results due to the intensity skew of DLS and normal sampling variation, especially with broader distributions [51].
- Solution: Follow ASTM E2490-09 guidelines by analyzing at least three separate aliquots for any material. Do not rely on a single measurement [51].
- Prevention: Understand that the intensity distribution is inherently skewed towards larger particles. Consider if a number-weighted distribution (from techniques like NTA) might provide a more relevant measurement for your application.
Transmission Electron Microscopy (TEM) Troubleshooting
Problem: TEM images show exosome aggregation or membrane damage.
- Potential Cause: Suboptimal storage conditions, such as multiple freeze-thaw cycles or storage in PBS without cryoprotectants, can cause fusion, enlargement, and membrane deformation [2] [4].
- Solution: Add stabilizers like trehalose to the storage buffer. Data indicate that rapid freezing and constant storage at -80 °C better preserve morphology [2] [1].
- Prevention: Avoid repeated freeze-thaw cycles. Process samples for TEM immediately after a single, careful thaw of a freshly isolated or cryopreserved sample.
Problem: Lack of clear, defined vesicular structures in TEM images.
- Potential Cause: Co-isolation of non-vesicular material, such as protein aggregates or lipoproteins, which is common with certain isolation methods like ultracentrifugation of complex biofluids [52].
- Solution: Optimize isolation protocols. Size-exclusion chromatography (SEC) can improve sample purity by removing contaminating proteins [52] [4].
- Prevention: Characterize your sample purity by measuring the ratio of particle concentration to protein concentration. A higher ratio indicates a purer EV preparation [1].
Frequently Asked Questions (FAQs)
Q1: What is the most stable way to store exosomes for long-term preservation? A1: The current evidence suggests that constant storage at -80 °C is a widely used and relatively stable method [2] [4]. However, for improved stability, consider adding cryoprotectants. Trehalose (25 mM) has been shown to effectively prevent aggregation and preserve biological activity during storage and freeze-thaw cycles [1] [8]. Another promising technique is lyophilization (freeze-drying) with trehalose, which allows for storage at room temperature while maintaining exosome integrity, protein content, and RNA content [8].
Q2: Why do my exosome size measurements differ between NTA and DLS? A2: This is a common occurrence due to the fundamental differences in how these techniques operate:
- DLS provides an intensity-weighted distribution. Because scattering intensity is proportional to the sixth power of the diameter (d^6), a few large particles or aggregates can dominate the signal, making the population appear larger and less polydisperse than it actually is [51].
- NTA directly tracks and sizes individual particles based on their Brownian motion. While it also has biases, it tends to provide a distribution that is often closer to a number-weighted distribution, offering a different perspective on the sample's heterogeneity [50].
The following table summarizes the core principles behind these differences:
Table: Core Differences Between DLS and NTA
| Feature | Dynamic Light Scattering (DLS) | Nanoparticle Tracking Analysis (NTA) |
|---|---|---|
| Weighting | Intensity-weighted | Number-weighted (closer to) |
| Size Principle | Fluctuations in scattered light intensity | Direct tracking of Brownian motion |
| Sensitivity to Aggregates | Very high (skews results significantly) | Moderate (aggregates are counted as single particles) |
| Ideal Polydispersity | Low (PDI < 0.2) for accurate results [51] | More suitable for polydisperse samples |
Q3: How do freeze-thaw cycles affect my exosome preparation? A3: Multiple freeze-thaw cycles are detrimental to exosome quality. The impacts are cumulative and include [2] [4]:
- Decreased particle concentration and RNA content.
- Increased average particle size and aggregation, observable under electron microscopy as vesicle enlargement and fusion.
- Impaired bioactivity in functional assays. It is strongly recommended to aliquot exosomes into single-use portions to avoid repeated freezing and thawing.
Q4: My exosomes have aggregated after storage. Can I use them for my experiment? A4: Using aggregated exosomes is not advisable for most quantitative or functional studies. Aggregation alters concentration measurements, size distribution, and may block injection needles in animal studies. More importantly, it can significantly impact the biological activity and uptake of exosomes by recipient cells [2] [1]. It is better to use a fresh aliquot or an aliquot stored with a cryoprotectant like trehalose to minimize aggregation.
Experimental Protocols
Detailed Protocol: Nanoparticle Tracking Analysis (NTA)
This protocol is adapted for the analysis of extracellular vesicles (EVs) [49] [50].
1. Sample Preparation:
- Dilute the isolated EV sample in filtered PBS to achieve a concentration within the instrument's optimal detection range (typically 10^7 - 10^9 particles/mL). The exact dilution factor must be determined empirically.
- Handle cuvettes with gloves to prevent fingerprints.
- Place a magnetic stir bar in the cuvette and use a hook tool to insert the cuvette insert.
- Slowly pipette 400-500 μL of the diluted sample into the cuvette, avoiding air bubbles. Gently pipette up and down to mix.
- Cap the cuvette and tap gently to remove any bubbles.
2. Instrument Operation & Measurement:
- Turn on the NTA instrument and computer. Launch the software.
- Follow the on-screen instructions to enter sample information.
- Start the camera and infuse the sample using the syringe pump.
- Adjust the camera focus and detection threshold until individual particles are clearly visible and tracked.
- Record multiple videos (e.g., 30-60 seconds each) from different segments of the sample.
- Process the recordings to generate data on particle concentration and size distribution.
- Export the data for further analysis.
3. System Cleaning:
- After measurements, wash the syringe and chamber with a total of 3 mL distilled water.
- Clean the glass chamber with ethanol to remove any residual contaminants [49].
Workflow Diagram: Impact of Storage on Exosome QC Metrics
The diagram below visualizes the relationship between storage conditions, their physical impact on exosomes, and the resulting changes in key QC metrics.
Impact of Trehalose on Exosome Stability
Trehalose, a non-reducing disaccharide, is a highly effective cryoprotectant for exosomes. The data below summarize its benefits as demonstrated in key studies.
Table: Efficacy of Trehalose in Preserving Exosome Quality
| Parameter | PBS (Control) | PBS with 25 mM Trehalose | Significance & Source |
|---|---|---|---|
| Particle Size Distribution | Wider spread (higher standard deviation and span) | Narrower size distribution; reduced mean particle size by ~13 nm [1] | p = 0.002 for standard deviation; p = 0.023 for span [1] |
| Freeze-Thaw Cycles | Particle concentration and size distribution width increase after cycles [1] | Particle concentration and size distribution remain stable after cycles [1] | Prevents degradation induced by freeze-thaw [2] [1] |
| Particle Yield | Lower particle concentration per μg of protein [1] | 3-fold higher particle concentration per μg of protein [1] | p = 0.001 [1] |
| Lyophilization | Causes severe aggregation [8] | Prevents aggregation; maintains structure and cargo [8] | Enables room temperature storage [8] |
Comparison of Storage and Cryoprotectant Strategies
The following table synthesizes findings from systematic reviews and studies comparing various storage strategies for isolated exosomes.
Table: Comparison of Exosome Storage Strategies
| Storage Condition | Impact on Particle Concentration | Impact on Size & Morphology | Impact on Cargo & Function |
|---|---|---|---|
| -80°C (no additive) | Time-dependent reduction [4] | Increased size and size variability; membrane deformation [2] [4] | Decreased RNA content and bioactivity after long-term storage [2] |
| -80°C with Trehalose | Better preservation of concentration [1] | Prevents aggregation; maintains size distribution [1] [8] | Preserves RNA and biological activity (e.g., immune stimulation) [1] |
| Lyophilization with Trehalose | Maintains concentration post-rehydration [8] | Prevents aggregation; maintains morphology [8] | Preserves protein and RNA content, and protein function [8] |
| Multiple Freeze-Thaw Cycles | Decreases after first cycle [2] [4] | Cycle-dependent increase in size and aggregation [2] [4] | Decreased RNA content and impaired bioactivity [2] |
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Materials for Exosome Storage and QC
| Reagent / Tool | Function / Application | Key Details |
|---|---|---|
| Trehalose | Cryoprotectant / Lyoprotectant | 25 mM in PBS is a common effective concentration. Prevents aggregation and cryodamage during freezing and lyophilization by stabilizing membranes [1] [8]. |
| Size Exclusion Chromatography (SEC) Columns | EV Isolation / Purification | Isolates EVs with high purity by separating them from contaminating proteins (e.g., albumin, IgGs). Crucial for clean downstream analysis [52] [4]. |
| Capturem EV Isolation Kit | EV Isolation / Purification | Membrane-based affinity isolation. Provides a quick (30 min) protocol for highly pure EVs from plasma, urine, and cell media, minimizing protein contamination [52]. |
| Phosphate-Buffered Saline (PBS) | Storage Buffer / Diluent | Standard buffer for suspending and diluting EVs. Always use filtered PBS to minimize background noise in NTA [49] [50]. |
| Protease Inhibitor Cocktail | Storage Additive | Added to storage buffers to prevent proteolytic degradation of exosomal proteins during storage [4]. |
Extracellular vesicles (EVs), including exosomes, have emerged as powerful tools in regenerative medicine, diagnostics, and drug delivery due to their ability to transfer functional biomolecules—including RNAs, proteins, and lipids—between cells. However, the nanoscale properties of EVs make them exceptionally sensitive to environmental conditions, and suboptimal storage can lead to rapid degradation of their valuable cargo. Maintaining the structural and functional integrity of RNA content, protein activity, and surface markers is essential for ensuring reproducible research outcomes and reliable clinical applications. The preservation of this cargo is particularly challenging during freeze-thaw cycles and long-term storage, where aggregation, degradation, and cryodamage frequently occur. This technical support center provides targeted troubleshooting guides and FAQs to help researchers overcome these challenges, with a specific focus on preventing exosome aggregation and preserving cargo integrity within the broader context of exosome research and therapeutic development.
Understanding Cargo Degradation Pathways and Risks
Major Threats to Cargo Integrity
RNA Degradation: RNA is particularly vulnerable to degradation due to its chemical structure. The presence of a 2'-hydroxyl group in the ribose sugar makes RNA susceptible to hydrolysis, especially at elevated temperatures or in the presence of divalent cations like Mg²⁺ that catalyze cleavage reactions [53]. Ribonucleases (RNases) represent another significant threat, as they are ubiquitous in the environment and require no cofactors to function.
Protein and Surface Marker Compromise: Protein activity can be diminished through denaturation, aggregation, or proteolytic cleavage. Surface markers on both exosomes and cells are especially vulnerable to enzymatic degradation during tissue dissociation procedures. Studies demonstrate that enzymatic digestion with collagenase or dispase can impair detection of essential markers such as CD4, CD8, CD62L, CD69, CD103, CD19, B220, CXCR5, and CD138 [54].
Exosome Membrane Damage and Aggregation: The lipid bilayer membrane of exosomes is sensitive to physical stress during freezing and thawing. This can lead to membrane deformation, vesicle fusion, and aggregation. When exosomes aggregate, it not only alters their size distribution but also obscures surface markers and can trap cargo within fused vesicles, making it inaccessible for downstream functions [2] [1].
Impact of Storage Conditions on Cargo Stability
The table below summarizes key findings from systematic reviews on how storage conditions affect various exosome parameters:
Table: Effects of Storage Conditions on Exosome Cargo Integrity
| Storage Condition | Impact on RNA Content | Impact on Protein/Surface Markers | Impact on Vesicle Integrity |
|---|---|---|---|
| Multiple freeze-thaw cycles | Decreased RNA content [2] | Altered protein detection [1] | Increased size, aggregation, membrane deformation [2] |
| Storage at -20°C | Significant degradation over weeks [3] | Protein activity loss [2] | Particle aggregation and size increase [3] |
| Storage at -80°C | Best preservation for long-term storage [3] | Improved preservation of protein activity [2] | Maintained size distribution and morphology [3] |
| Room temperature without stabilizers | Rapid degradation [53] | Not reported | Aggregation and cargo loss [1] |
| With trehalose stabilizer | Improved RNA protection [1] | Maintained biological activity [1] | Reduced aggregation, maintained individual particles [1] |
FAQs and Troubleshooting Guides
RNA Preservation FAQs
Q: Why does my RNA degrade even when I store exosomes at -80°C?
A: RNA degradation can occur due to several factors:
- Residual RNase activity: Even trace amounts of RNases can cause significant degradation. Ensure all buffers and equipment are certified RNase-free.
- Improper freezing/thawing: Repeated freeze-thaw cycles accelerate degradation. Always aliquot exosome preparations into single-use portions.
- Ineffective cryopreservation: Without appropriate stabilizers, ice crystal formation during freezing can damage exosome membranes, exposing RNA to degradative factors [53] [2].
Q: What RIN (RNA Integrity Number) value is acceptable for downstream RNA-seq applications?
A: While conventional wisdom suggested RIN >8 was necessary, recent evidence indicates that samples with RIN values >5.3 have minimal impact on quantitative RNA-seq results for blood samples. This broader threshold increases utility of samples that might otherwise be discarded [55].
Q: How can I preserve RNA in exosomes without constant cold chain storage?
A: Novel approaches include:
- Anhydrous storage: Drying RNA samples in the presence of stabilizers in air- and water-tight containers dramatically reduces degradation by eliminating hydrolytic reactions.
- Trehalose stabilization: This disaccharide protects RNA during freezing by forming a stable matrix that prevents molecular movement and degradation [56] [1].
Protein and Surface Marker Preservation FAQs
Q: Why do I detect lower levels of surface markers after freeze-thawing my exosomes?
A: Surface marker detection can decrease due to:
- Epitope masking: Aggregation during freezing can physically obscure binding sites for detection antibodies.
- Conformational changes: Ice formation can alter protein structure, affecting antibody binding.
- Actual degradation: Proteases released from damaged exosomes or present in contaminants can cleave surface markers [1] [54].
Q: How does tissue dissociation affect surface marker integrity in single-cell analyses?
A: Enzymatic digestion during tissue dissociation significantly compromises surface marker detection:
- Direct cleavage: Proteases like collagenase and dispase directly cleave extracellular epitopes, reducing antibody binding.
- Artifactual activation: Enzymatic treatment can induce stress responses that alter marker expression patterns, potentially misleading interpretations of cellular states [54].
Q: What methods best preserve surface epitopes for flow cytometry?
A: Non-enzymatic dissociation techniques such as acoustic dissociation better preserve surface marker integrity. Studies show significantly higher median fluorescence intensity for markers including CD19, CD138, and CD45 compared to enzymatic methods [54].
Exosome Integrity and Aggregation FAQs
Q: Why does exosome aggregation matter for cargo integrity assessment?
A: Aggregation creates multiple problems:
- Altered biodistribution: Aggregated exosomes behave differently in biological systems.
- Cargo trapping: Molecules trapped within aggregates may be inaccessible for detection or function.
- Size-based separation failures: Aggregates complicate purification and analysis techniques that rely on size, such as size exclusion chromatography or nanoparticle tracking analysis [1].
Q: How many freeze-thaw cycles can exosomes tolerate before significant cargo loss occurs?
A: Systematic evidence indicates that even a single freeze-thaw cycle can cause measurable damage, with significant deterioration in RNA content, bioactivity, and vesicle integrity after multiple cycles. It's strongly recommended to avoid freeze-thaw cycles altogether by using single-use aliquots [2] [3].
Q: What is the optimal storage temperature for long-term exosome preservation?
A: Most studies indicate -80°C provides superior preservation compared to -20°C for long-term storage. However, the addition of cryoprotectants like trehalose is equally important as temperature selection alone [3].
Experimental Protocols for Cargo Integrity Assessment
Protocol: Assessing RNA Integrity in Stored Exosomes
Principle: Evaluate RNA quality and quantity from exosomes subjected to different storage conditions to determine optimal preservation methods.
Materials:
- Qubit RNA HS Assay Kit (Thermo Fisher)
- Agilent 2100 Bioanalyzer with RNA Nano Kit
- RNase-free water and consumables
- TRIzol reagent for RNA extraction
Procedure:
- Isolate exosomes using your standard method (ultracentrifugation recommended)
- Divide exosome preparation into equal aliquots for different storage conditions
- Store aliquots under test conditions (e.g., -80°C without cryoprotectant, -80°C with trehalose, -20°C, room temperature with stabilizer)
- After predetermined timepoints (e.g., 1 week, 1 month, 3 months), extract RNA using TRIzol
- Quantify RNA using Qubit fluorometer
- Assess RNA integrity using Bioanalyzer to generate RNA Integrity Numbers (RIN)
- Compare RIN values and rRNA ratios between conditions
Expected Results: Properly preserved exosomes should maintain RIN values >7.0 and show intact ribosomal RNA profiles if derived from cells, or a characteristic small RNA profile for exosomal RNA [55].
Protocol: Evaluating Surface Marker Preservation
Principle: Compare detection levels of key surface markers after different storage or processing conditions.
Materials:
- Flow cytometer with appropriate lasers and filters
- Antibodies against target surface markers (e.g., CD63, CD81, CD9 for exosomes)
- Paraformaldehyde for fixation (if needed)
- Trehalose-containing preservation buffer
Procedure:
- Divide exosome preparation or cell suspension into equal aliquots
- Process aliquots with different preservation methods:
- Method A: Standard freezing at -80°C in PBS
- Method B: Freezing at -80°C in PBS with 25mM trehalose
- Method C: Acoustic dissociation (for cells) vs enzymatic dissociation
- For exosomes, bind to aldehyde/sulfate latex beads for flow cytometry analysis
- Stain samples with antibodies against target surface markers
- Analyze by flow cytometry, comparing median fluorescence intensity (MFI) between conditions
- Calculate percentage preservation compared to fresh controls
Expected Results: Samples preserved with trehalose should show significantly higher MFI values for surface markers, indicating better epitope preservation. Similarly, acoustically dissociated cells should show superior marker preservation compared to enzymatically dissociated counterparts [1] [54].
Protocol: Functional Assessment of Cargo Integrity Through Bioactivity Assays
Principle: Evaluate whether preserved exosomes maintain biological function, which depends on integrated integrity of all cargo components.
Materials:
- Reporter cell line appropriate for exosome function (e.g., macrophages for immune activation)
- Cell culture reagents and equipment
- ELISA kits for cytokine detection
- Trehalose-containing preservation buffer
Procedure:
- Iserve and divide exosomes as described in previous protocols
- Store aliquots under different test conditions
- Thaw stored exosomes and treat reporter cells with equal particle numbers
- Measure functional response:
- For immunomodulatory exosomes: measure cytokine secretion (e.g., TNF-α) by ELISA
- For regenerative exosomes: measure proliferation or migration assays
- For RNA transfer: measure reporter gene expression
- Compare bioactivity between storage conditions
Expected Results: Exosomes stored with cryoprotectants like trehalose should maintain significantly higher biological activity compared to those stored without protection, even when particle concentration appears similar [1].
Research Reagent Solutions for Cargo Integrity
Table: Essential Reagents for Preserving Exosome Cargo Integrity
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| Cryoprotectants | Trehalose, DMSO, glycerol | Stabilize membranes and proteins during freezing; reduce ice crystal formation [1] |
| RNase Inhibitors | RNaseOUT, SUPERase•In, DEPC | Prevent RNA degradation during processing and storage [53] |
| Protease Inhibitors | PMSF, Complete Mini tablets | Protect protein cargo and surface markers from proteolytic degradation [54] |
| Chelating Agents | EDTA, EGTA | Bind divalent cations that catalyze RNA hydrolysis [53] |
| Antioxidants | Ascorbic acid, glutathione | Protect against oxidative damage to lipids and proteins [53] |
| Non-Enzymatic Dissociation Reagents | Acoustic dissociation systems | Preserve surface epitopes during tissue processing for single-cell analyses [54] |
| Storage Buffers | RNAstable, RNAprotect | Chemically stabilize RNA for room temperature storage [53] [56] |
| Purification Kits | RNeasy Mini Kit, Exosome Isolation Kits | Provide optimized buffers for maintaining cargo integrity during processing [53] |
Visualization: Cargo Integrity Assessment Workflow
Diagram: Cargo integrity assessment workflow comparing preserved versus unpreserved samples across multiple analytical dimensions, with expected outcomes based on quantitative thresholds.
Advanced Techniques and Future Directions
Innovative Approaches for Room Temperature Storage
While ultra-low temperature storage remains the standard for exosome preservation, innovative technologies enable room temperature storage without compromising cargo integrity. One promising approach involves drying RNA samples in the presence of a stabilizer in stainless steel minicapsules that maintain an anhydrous and anoxic environment, effectively isolating RNA from atmospheric humidity [56]. Extrapolations from stability studies suggest that RNA stored under these conditions could remain intact for centuries, with an estimated degradation rate of 0.7-1.3 cuts per 1000 nucleotides per century [56]. For protein and surface marker preservation, advanced formulations combining trehalose with other stabilizers like hydroxyethyl cellulose in microneedle arrays have demonstrated excellent preservation of bioactivity for up to six months at 4°C or -20°C [2]. These emerging technologies offer promising alternatives to cold chain dependency, particularly for clinical applications and biobanking.
Integration of Multi-Parameter Assessment
Future directions in cargo integrity assessment emphasize integrated multi-parameter analysis rather than relying on single metrics. The most comprehensive approach simultaneously evaluates:
- Physical integrity through nanoparticle tracking analysis and electron microscopy
- RNA quality via RIN values and rRNA ratios
- Surface marker preservation through multi-parameter flow cytometry
- Functional bioactivity in biologically relevant assays
This integrated assessment recognizes that different cargo components may have varying stability profiles, and that physical integrity alone does not guarantee functional preservation. By implementing the troubleshooting guides, experimental protocols, and preservation strategies outlined in this technical support center, researchers can significantly improve the reliability and reproducibility of their exosome research, ultimately accelerating the translation of exosome-based discoveries into clinical applications.
Frequently Asked Questions (FAQs) on Exosome Handling and Assay Performance
Q1: What is the optimal storage condition for preserving exosome bioactivity? For long-term preservation, storing isolated exosomes at -80 °C is the most practical and commonly recommended option. This temperature best maintains particle concentration, RNA content, morphology, and biological functionality compared to higher temperatures like -20 °C [3]. For short-term storage (e.g., less than one week), 4 °C is a suitable alternative [57]. Note that storing exosomes in their native biofluid, rather than in purified buffers, can offer improved stability [3].
Q2: How does freezing cause exosome aggregation and how does it affect my assays? Freezing, particularly at -70 °C or -80 °C, can induce significant exosome aggregation [57]. This reduces the effective concentration of available vesicles and increases the measured particle size [57]. Functionally, this aggregation:
- Reduces cell uptake efficiency: Aggregated exosomes are less efficiently internalized by recipient cells (e.g., alveolar macrophages), which can lead to a significant underestimation of their bioactivity in functional assays [57].
- Impairs drug delivery efficiency: Aggregation directly reduces the effectiveness of exosomes used as drug delivery vehicles [57].
Q3: What is the best method to disperse aggregated exosomes after thawing? Water-bath sonication is a simple and effective method. One study demonstrated that sonication at 40 kHz, 100 W for 15 minutes significantly increased EV concentration and reduced aggregation [57]. In contrast, regular pipetting was not effective at dispersing aggregates and could even promote re-aggregation if performed after sonication [57].
Q4: I observe inconsistent results in my cell migration assays. What could be the cause? A common issue is the choice of fluorescent label for tracking cells. The frequently used calcein AM can exhibit time-dependent attenuation of fluorescence in certain cell types, such as macrophages, leading to unstable or decreasing signals and compromising quantification [58]. As an alternative, PKH26, which labels cell surfaces, provides a more stable fluorescent signal and more reliable results for migration assays [58].
Q5: Are there open-source software options for analyzing cell migration data? Yes, several open-source programs are available, providing free and accessible alternatives to costly proprietary software [59]. For example, MigraR (R-based) allows users to plot cell trajectories, quantify migration speed and straightness, and analyze directionality [59]. Another tool, CellTracksColab, is a user-friendly, web browser-based platform for cell migration analysis [59].
Troubleshooting Guides for Common Experimental Issues
Issue: Low or Inconsistent Signal in Cell Uptake Assays
| Possible Cause | Solution |
|---|---|
| Exosome Aggregation | Implement a water-bath sonication step (e.g., 40 kHz, 100 W for 15 min) post-thaw to disperse aggregates before adding them to cells [57]. |
| Improper Storage | Ensure exosomes are stored at -80 °C and avoid multiple freeze-thaw cycles. Aliquot exosomes to minimize repeated freezing and thawing [3]. |
| Incorrect Labeling | Verify the efficiency and stability of the fluorescent dye used to label exosomes. Ensure the dye is compatible with your detection system. |
Issue: High Background or No Signal in Cell Migration/Proliferation Assays
| Possible Cause | Solution |
|---|---|
| Unstable Fluorescent Label | If using calcein AM, switch to a more stable dye like PKH26 for consistent fluorescence during time-lapse experiments [58]. |
| Weak Chemoattractant Gradient | For migration assays, use a sustained-release system, such as a hydrogel, to maintain a stable gradient of the chemoattractant over time [58]. |
| Assay Reagent Issues | Equilibrate all reagents to the correct assay temperature. Check that reagents have been stored properly and have not expired [60]. Avoid bubbles in the wells, which can disrupt readings [60]. |
Summarized Quantitative Data from Key Studies
Table 1: Impact of Storage Conditions on Exosome Integrity
| Storage Condition | Impact on Concentration | Impact on Size/Aggregation | Functional Consequence |
|---|---|---|---|
| -80 °C (Long-term) | Preserved particle concentration [3] | Minimal size change; reduced aggregation [3] | Maintained bioactivity (e.g., chondrocyte proliferation, T-cell modulation) [61] |
| -20 °C | Significant decrease [3] | Significant aggregation & size increase [3] | Loss of pre-clinical efficacy in inflammatory disease models [61] |
| Multiple Freeze-Thaw Cycles | Decreased particle concentration [3] | Increased size and aggregation [3] | Reduced RNA content and impaired bioactivity [3] |
| Liquid Nitrogen (-196 °C) | Higher concentration loss vs. -80°C [3] | Can cause size reduction or membrane disruption [3] | Less commonly used; more data needed |
Table 2: Efficacy of Methods to Disperse Aggregated Exosomes
| Method | Conditions | Effect on Aggregation | Effect on Concentration |
|---|---|---|---|
| No Treatment (Control) | N/A | High level of aggregation [57] | Low [57] |
| Regular Pipetting | Repeated pipetting | Not effective [57] | No significant change [57] |
| Water-Bath Sonication | 40 kHz, 100 W, 15 min | Significantly reduced [57] | Significantly increased [57] |
| Sonication + Pipetting | Sonication followed by pipetting | High level of aggregation (re-aggregation) [57] | No significant benefit [57] |
Detailed Experimental Protocols
Protocol: Dispersing Exosome Aggregates via Water-Bath Sonication
This protocol is adapted from a 2025 study that effectively dispersed exosomes aggregated during storage at -70°C [57].
Principle: Low-power sonication generates vibrational energy that separates aggregated vesicles without causing significant damage to their integrity.
Materials:
- Water-bath sonicator (e.g., 40 kHz, 100 W)
- Thawed exosome sample
- Ice
Procedure:
- Thaw: Remove the exosome sample from -80 °C storage and thaw it on ice.
- Sonicate: Place the sample in a water-bath sonicator pre-set to power level 3 (approx. 100 W).
- Time: Sonicate the sample for 15 minutes.
- Use Immediately: Use the sonicated exosomes directly in downstream functional assays. Do not pipette vigorously after sonication, as this may cause re-aggregation [57].
Protocol: Validating Migratory Function with Stable Fluorescent Labeling
This protocol addresses the common problem of fluorescent signal decay in cell migration assays [58].
Principle: Using a stable lipophilic dye (PKH26) instead of calcein AM ensures consistent fluorescence for accurate quantification of migrated cells over time.
Materials:
- Motile cells (e.g., macrophages)
- PKH26 fluorescent cell linker kit
- Cell culture inserts with microporous membranes
- Fluorescence microscope
- Appropriate chemoattractant
Procedure:
- Harvest Cells: Collect and wash the cells in a serum-free medium.
- Label Cells: Resuspend the cell pellet in the PKH26 dye solution according to the manufacturer's instructions. Incubate for a specific duration (e.g., 2-5 minutes).
- Stop Staining: Add an equal volume of serum to bind the excess dye. Wash the labeled cells thoroughly to remove any unbound dye.
- Seed Cells: Resuspend the labeled cells in the appropriate medium and seed them onto the cell culture inserts.
- Induce Migration: Place the insert into a well containing a chemoattractant. For a stable gradient, consider embedding the chemoattractant in a hydrogel [58].
- Quantify Migration: After the desired incubation period, quantify the migrated cells from the bottom of the microporous membrane using a fluorescence microscope.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents for Bioactivity Assays
| Item | Function/Application | Key Consideration |
|---|---|---|
| PKH26 Dye | Stable fluorescent labeling of cell membranes for migration and uptake studies [58]. | More reliable than calcein AM for assays requiring prolonged signal stability [58]. |
| Trehalose | A stabilizer (cryoprotectant) added to exosome preparations before freezing [3]. | Helps maintain exosome integrity and reduces aggregation during freeze-thaw cycles [3]. |
| Dimethyl Sulfoxide (DMSO) | A common cryoprotectant for cells and biological samples. | Can be cytotoxic and may inhibit specific downstream processes for exosomes; use with caution [3]. |
| Hydrogel | Creates a sustained-release system for chemoattractants in migration assays [58]. | Maintains a stable chemical gradient, which is crucial for a successful and reproducible migration assay [58]. |
| MigraR Software | Open-source R-based platform for quantifying cell migration parameters (speed, straightness, direction) [59]. | Requires coordinate data (X, Y) from tracking software like ImageJ as input [59]. |
Signaling Pathways and Experimental Workflows
Exosome-Mediated Proliferation Signaling Pathway
The following diagram illustrates a key signaling mechanism identified for Y201 MSC-derived exosomes, which stimulate chondrocyte proliferation, a relevant pathway for proliferative function assays [61].
Workflow for Pre-Assay Exosome Handling and Quality Control
This workflow outlines the critical steps for preparing and validating exosomes prior to their use in functional bioactivity assays, integrating best practices for preventing aggregation.
The Scientist's Toolkit: Essential Research Reagents
The following table details key reagents and materials essential for experiments focused on EV storage and stability.
| Reagent/Material | Function & Explanation |
|---|---|
| Phosphate Buffered Saline (PBS) | A standard isotonic buffer for maintaining pH and osmotic pressure; commonly used as a control storage medium despite its limitations with EVs [62] [63]. |
| Trehalose | A non-permeable cryoprotectant that helps preserve EV integrity by stabilizing the lipid bilayer and preventing aggregation during freezing [3] [63]. |
| Dimethyl Sulfoxide (DMSO) | A permeable cryoprotectant that penetrates membranes to reduce ice crystal formation; requires careful evaluation for potential functional impacts [3] [63]. |
| Glycerol | A permeable cryoprotectant used to protect intracellular structures and improve viability in cryopreservation [63]. |
Troubleshooting Guides and FAQs
What are the most critical parameters to monitor when benchmarking a new EV storage protocol?
When evaluating a new storage protocol, you must measure a core set of physicochemical and functional parameters against the standard of -80°C PBS. The following table summarizes the key quantitative metrics for comparison.
| Evaluation Parameter | Experimental Method | Benchmarking Insight |
|---|---|---|
| Particle Concentration | Nanoparticle Tracking Analysis (NTA) | A significant drop indicates particle loss or massive aggregation. PBS storage often shows notable decreases [7] [63]. |
| Particle Size & Aggregation | NTA, Dynamic Light Scattering (DLS) | An increase in mean/median size and the proportion of particles >400nm signals aggregation. Common in PBS and after freeze-thaw cycles [3] [7]. |
| Morphology | Transmission Electron Microscopy (TEM) | Visualizes membrane integrity, vesicle rounding, and fusion. PBS-stored EVs often show deformation and aggregation [3] [63]. |
| Surface Charge (Zeta Potential) | Dynamic Light Scattering (DLS) | Indicates colloidal stability. A shift towards neutral values (e.g., from -30 mV to -10 mV) suggests decreased stability and higher aggregation propensity [63]. |
| Cargo Preservation | Protein (BCA assay), RNA (Spectrophotometer) | Quantifies the retention of core biomolecules. Multiple freeze-thaw cycles in PBS can significantly reduce RNA content [3] [63]. |
| Functional Bioactivity | In vitro cellular uptake or cytotoxicity assay | The ultimate test of therapeutic potential. Protocols that preserve functionality will show efficient cellular uptake and expected biological effects [7] [63]. |
The standard -80°C PBS storage causes high EV aggregation. How can I disperse these aggregates?
Water-bath sonication is a validated and effective method for dispersing aggregates formed during frozen storage in PBS [7].
Experimental Protocol: Dispersing Aggregated EVs via Sonication
- Sample Preparation: Thaw the frozen EV sample and vortex gently for a few seconds to mix.
- Sonication Setup: Place the EV sample in a water-bath sonicator. Ensure the water level is sufficient to immerse the sample tube adequately.
- Optimized Sonication Parameters:
- Post-Treatment Handling: Gently invert the tube after sonication. Avoid pipetting after sonication, as this can cause re-aggregation [7].
- Validation: Use NTA immediately after sonication to confirm the reduction in aggregate count and the increase in total particle concentration.
Besides PBS, what alternative storage buffers can I benchmark, and how do I test them?
Benchmarking should include cryoprotectants known to stabilize lipid bilayers and biological particles. A robust testing protocol goes beyond simple particle counts to assess drug delivery potential [63].
Experimental Protocol: Benchmarking Storage Buffers for EV Drug Delivery
- Buffer Preparation: Prepare the following test buffers:
- Control: 1x PBS, pH 7.4.
- Trehalose: PBS supplemented with 25 mM trehalose.
- DMSO: PBS supplemented with 6% DMSO.
- Glycerol: PBS supplemented with 30% glycerol.
- EV Storage: Resuspend purified EVs in the different buffers and store them at -80°C. A 10-week storage period is a suitable benchmark duration [63].
- Stability Assessment: After storage and thawing, characterize the EVs as outlined in the first table (using NTA, DLS, TEM, BCA, etc.) [63].
- Drug Delivery Efficiency Assessment:
- Drug Loading: Load a model drug (e.g., Doxorubicin) into the EVs after storage.
- Loading Capacity (LC): Calculate the LC using the formula:
LC (%) = (Weight of loaded drug / Weight of EVs) × 100[63]. - In vitro Efficacy: Treat target cells with the drug-loaded EVs and measure cytotoxicity compared to a control (e.g., freshly isolated, drug-loaded EVs). This directly tests the functional preservation of the EVs [63].
Our benchmarking study shows that adding trehalose to PBS best preserves EV concentration and function. How can I visualize the experimental workflow?
The following diagram illustrates the logical workflow for the comparative benchmarking experiment.
How do freeze-thaw cycles affect EVs stored in PBS, and what is a key preventative strategy?
Multiple freeze-thaw cycles are highly detrimental to EVs and should be avoided. Data indicates that subjecting EVs to multiple cycles decreases particle concentrations, impairs RNA content, and increases EV size and aggregation [3].
Best Practice Protocol:
- Aliquot for Single Use: Divide the EV stock into single-use aliquots of the volume required for a typical experiment.
- Thaw Once: Thaw one aliquot at a time at room temperature or in a 37°C water bath, and use it immediately. Do not re-freeze any leftover material.
- Use Cryoprotectants: The use of stabilizers like trehalose in the storage buffer has been shown to help EVs maintain their integrity across temperature stress [3].
Troubleshooting Guide: FAQs on Exosome Stability
FAQ 1: What is the optimal temperature for long-term storage of purified exosomes?
Answer: For long-term storage, -80°C is widely recommended. Data indicates that rapid freezing procedures and constant storage at this temperature best preserve EV quantity and cargo integrity [2]. Storage at -20°C or higher temperatures (4°C or 25°C) leads to a significant decrease in particle concentration and a rapid loss of protein activity [2]. For clinical applications where -80°C is impractical, lyophilization (freeze-drying) with cryoprotectants offers a promising alternative for room temperature storage [8].
FAQ 2: How do freeze-thaw cycles damage my exosome samples, and how can I prevent this?
Answer: Multiple freeze-thaw cycles are highly detrimental, causing:
- Decreased particle concentrations and increased particle size [2].
- Aggregation and membrane deformation, as observed via electron microscopy [2].
- Loss of RNA content and impaired bioactivity [2]. Prevention Strategy: Aliquot exosome samples into single-use volumes to avoid repeated freezing and thawing. The addition of stabilizers like trehalose (25 mM) to the storage buffer has been shown to maintain particle dispersion and integrity across multiple freeze-thaw cycles [1].
FAQ 3: My exosomes are aggregating. What can I do to improve colloidal stability?
Answer: Aggregation is a common issue, often caused by attractive forces between particles when electrostatic repulsion is low.
- Buffer Additives: Adding trehalose (25 mM) to PBS significantly narrows the particle size distribution and increases the number of individual particles, indicating reduced aggregation [1]. Trehalose acts by physically shielding exosomes and preventing fusion.
- Storage Matrix: Consider storing exosomes in their native biofluid or a specialized matrix. One study showed that encapsulating EVs in hyaluronic acid-based microneedles (EV@MN) effectively preserved their integrity and bioactivity for up to six months [2].
- Avoid Lyophilization without Protectants: Lyophilizing exosomes without cryoprotectants like trehalose causes severe aggregation [8]. Always use a protectant during freeze-drying.
FAQ 4: Are there any approved cryoprotectants for exosomes intended for therapeutic use?
Answer: While no universal protocol exists, trehalose is a leading candidate due to its strong track record. It is a natural, non-toxic disaccharide widely used as a stabilizer in the food and drug industry [1]. Studies confirm that trehalose protects exosomes from cryodamage during freezing and lyophilization, helping to maintain their physical properties and biological activity, which is crucial for therapeutics [1] [8]. Other cryoprotectants like DMSO are used in cell cryopreservation but may introduce cytotoxicity or inhibit specific downstream processes for exosomes [2].
FAQ 5: How does storage condition impact the therapeutic efficacy of my exosomes?
Answer: Suboptimal storage directly compromises therapeutic efficacy by damaging key exosome components.
- Structural Integrity: Membrane deformation and vesicle fusion from poor storage impair the exosome's ability to be taken up by target cells [2].
- Cargo Preservation: Damage can lead to the loss or degradation of functional proteins and RNA, which are essential for the exosome's biological effect [2] [8].
- Functional Assays: Macrophage immune assays demonstrated that extracellular vesicles stored in trehalose consistently stimulated higher TNF-α secretion compared to those stored in PBS, indicating better preservation of biological activity [1].
Quantitative Data on Storage Conditions
Table 1: Impact of Storage Conditions on Exosome Physicochemical Properties
| Storage Condition | Particle Concentration | Mean Size & Distribution | Morphology (EM) | Key Findings |
|---|---|---|---|---|
| -80°C (Long-term) | Minimal decrease [2] | Stable size distribution [2] | Intact vesicles [2] | Optimal for preserving quantity and cargo [2] |
| Multiple Freeze-Thaw Cycles (in PBS) | Decreased [2] [1] | Increased size & wider distribution [2] [1] | Vesicle enlargement, fusion, membrane deformation [2] | Highly detrimental; induces aggregation and cargo loss [2] |
| With Trehalose Stabilizer | Higher yield and maintained concentration [1] | Narrower size distribution, reduced mean size [1] | Reduced aggregation, intact membranes [1] | Prevents aggregation and cryodamage from freeze-thaw cycles [1] |
| Lyophilization with Trehalose | Maintained post-rehydration [8] | Unchanged polydispersity index [8] | No aggregation; morphology similar to fresh [8] | Enables room temperature storage without damage [8] |
Table 2: Impact of Storage Conditions on Exosome Cargo and Functional Activity
| Storage Condition | Protein Content/Activity | RNA Content/Integrity | In Vitro / Functional Bioactivity |
|---|---|---|---|
| -80°C (Long-term) | Well-preserved [2] | Well-preserved in short-term; degradation possible over years [2] [8] | Maintained, but may decrease over 28 days [8] |
| Multiple Freeze-Thaw Cycles (in PBS) | Not reported | Decreased RNA content [2] | Impaired bioactivity [2] |
| With Trehalose Stabilizer | Improved preservation [1] | Not reported | Consistently higher TNF-α stimulation in macrophages [1] |
| Lyophilization with Trehalose | Similar to -80°C stored samples [8] | Well-preserved, similar to -80°C stored samples [8] | Maintained function of cargo proteins and DNA [8] |
Detailed Experimental Protocols
Protocol 1: Assessing Stability Using Trehalose as a Stabilizer
This protocol is adapted from studies demonstrating that trehalose prevents aggregation and cryodamage [1].
1. Materials and Reagents:
- Purified exosome sample
- Phosphate-Buffered Saline (PBS)
- Trehalose
- Nanoparticle Tracking Analysis (NTA) system
- Transmission Electron Microscope (TEM)
- Ultracentrifuge and tubes
2. Procedure:
- Buffer Preparation: Prepare a 25 mM solution of trehalose in PBS (TRE-PBS). Use pure PBS as a control.
- Sample Preparation: After isolation, resuspend the exosome pellet in either TRE-PBS or PBS. Split the sample into single-use aliquots.
- Stability Challenge:
- Freeze-Thaw Cycles: Subject aliquots to multiple freeze-thaw cycles (e.g., 3-5 cycles). For each cycle, freeze at -80°C for 1 hour and thaw at room temperature.
- Long-Term Storage: Store other aliquots at -80°C for a defined period (e.g., 1, 3, or 6 months).
- Analysis:
- NTA: After storage/thawing, dilute samples appropriately and analyze with NTA to determine particle concentration and size distribution. A stable sample will show minimal changes in these parameters.
- TEM: Visualize the samples using TEM to assess morphology and the presence of aggregates.
- Functional Assay: Perform a relevant bioassay (e.g., cell uptake, cytokine secretion) to confirm preserved biological activity.
Protocol 2: Lyophilization of Exosomes for Room Temperature Storage
This protocol is based on methods developed to preserve exosomes at room temperature using lyophilization [8].
1. Materials and Reagents:
- Purified exosome sample
- Trehalose
- Lyophilizer
- Cryovials
2. Procedure:
- Sample Preparation: Mix the purified exosomes with trehalose as a cryoprotectant. The study used trehalose effectively to prevent damage [8].
- Freezing: Transfer the exosome-trehalose mixture into cryovials and freeze them at -80°C or in a freezer at -30°C to -50°C for several hours.
- Primary Drying: Place the frozen samples in the lyophilizer. Start the primary drying phase under a vacuum to sublimate the ice. This step typically takes 24-48 hours.
- Secondary Drying: After primary drying, a secondary drying phase may be applied at a slightly higher temperature to remove bound water.
- Storage and Reconstitution: Seal the vials under an inert gas if possible. Store the lyophilized powder at room temperature, protected from light. To reconstitute, add sterile water or PBS to the vial and gently vortex or pipette to resuspend.
Diagram 1: Recommended workflow for stable exosome storage.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Exosome Stabilization
| Reagent / Material | Function / Purpose | Key Consideration |
|---|---|---|
| Trehalose | Non-reducing disaccharide that acts as a cryoprotectant and lyoprotectant; prevents aggregation and stabilizes membranes during freezing and drying [1] [8]. | Preferred for its non-toxic nature and efficacy at concentrations around 25 mM. |
| Hyaluronic Acid (HA) | Forms a stabilizing matrix (e.g., in microneedles) that encapsulates exosomes, shielding them from environmental stresses during storage [2]. | Offers a novel approach for integrating stable exosomes into biomedical devices. |
| Dimethyl Sulfoxide (DMSO) | A common cryoprotectant for cells. | May cause cytotoxicity or interfere with downstream exosome function; use with caution [2]. |
| Size-Exclusion Chromatography (SEC) Columns | For purifying exosomes away of contaminants and transferring them into a desired storage buffer (e.g., PBS with trehalose). | Provides a gentler alternative to ultracentrifugation for buffer exchange, potentially improving yield. |
| Lyophilizer | Equipment for freeze-drying samples, enabling long-term storage of exosomes at room temperature. | Must be used with a cryoprotectant like trehalose to prevent massive aggregation and damage [8]. |
Conclusion
Preventing exosome aggregation is not merely a technical detail but a fundamental prerequisite for reproducible research and successful clinical translation. A holistic strategy—integrating purpose-driven buffer selection, stringent temperature control, judicious use of cryoprotectants like trehalose, and minimization of freeze-thaw cycles—is essential for maintaining exosome integrity from benchtop to bedside. Future progress hinges on developing universally accepted standardized protocols, advancing lyophilization techniques with improved lyoprotectants, and establishing robust, predictive quality control metrics that correlate physical stability with in vivo therapeutic efficacy. By adopting these evidence-based practices, the scientific community can overcome a major bottleneck and fully harness the potential of exosomes in precision medicine and regenerative therapy.
References
- 1. Trehalose prevents aggregation of exosomes and ... [https://www.nature.com/articles/srep36162]
- 2. Different storage and freezing protocols for extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11600612/]
- 3. Different storage and freezing protocols for extracellular vesicles [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-04005-7]
- 4. The impact of storage on extracellular vesicles - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC8804350/]
- 5. Preservation of small extracellular vesicles for functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7808382/]
- 6. Effect of pH, temperature and freezing-thawing on quantity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6418301/]
- 7. Efficient dispersion of aggregated extracellular vesicles [https://pmc.ncbi.nlm.nih.gov/articles/PMC12255699/]
- 8. Preservation of exosomes at room temperature using ... [https://www.sciencedirect.com/science/article/abs/pii/S0378517318307671]
- 9. Impact of buffer composition on biochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9764254/]
- 10. A Step-by-Step Guide to (Not) Failing at Extracellular ... [https://www.izon.com/news/a-step-by-step-guide-to-not-failing-at-extracellular-vesicle-western-blotting]
- 11. Tips & Tricks | Immunoassay Protocols & Troubleshooting [https://www.sigmaaldrich.com/US/en/technical-documents/product-supporting/milliplex/tips-tricks-immunoassay-protocol-troubleshooting-advice?srsltid=AfmBOoofB5mEiTm50kMiJ-yezHyYLjTN82zUwu0h8pOMv_-Pxl1xUkAJ]
- 12. Sample Preparation for Biofluids [https://www.biotage.com/biofluids-sample-prep]
- 13. Sucrose-based cryoprotective storage of extracellular vesicles [https://pubmed.ncbi.nlm.nih.gov/38665624/]
- 14. Cryopreservation of mesenchymal stromal cell-derived ... [https://www.sciencedirect.com/science/article/abs/pii/S0011224020306532]
- 15. Sucrose-based cryoprotective storage of extracellular ... [https://www.sciencedirect.com/science/article/pii/S2773041722000117]
- 16. Storage stability of exosomes in different buffers with/ ... [https://www.sciencedirect.com/science/article/abs/pii/S1773224725006811]
- 17. Preservation and Storage Stability of Extracellular Vesicles ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6582961/]
- 18. Navigating the Challenges of Exosome Stability in Long-Term ... [https://www.cd-bioparticles.net/resources/navigating-the-challenges-of-exosome-stability-in-long-term-therapeutic-storage.html?srsltid=AfmBOorZ7uofp2TPhD-YhyLFUutYk7dQmYk4sTRpA6odqiziLMOv3iTY]
- 19. Dissolving microneedles for long-term storage and ... [https://www.sciencedirect.com/science/article/abs/pii/S0142961222002848]
- 20. Microneedles Based on a Biodegradable Polymer— ... [https://www.mdpi.com/2073-4360/16/10/1396]
- 21. A review of sucrose, trehalose, cyclodextrins and dextrans [https://www.sciencedirect.com/science/article/pii/S0928098723002555]
- 22. Freeze-drying of mammalian cells using trehalose [https://www.nature.com/articles/s41598-017-06542-z]
- 23. Identification of storage conditions stabilizing extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9206228/]
- 24. Storage Stability of Exosomes in Different Buffers With/ ... [https://papers.ssrn.com/sol3/papers.cfm?abstract_id=5239658]
- 25. Evaluation of EV Storage Buffer for Efficient Preservation ... [https://www.mdpi.com/1422-0067/24/16/12841]
- 26. Frequently Asked Questions (FAQs) [https://www.creative-biolabs.com/exosome/frequently-asked-questions-faqs.htm]
- 27. Aliquoting FBS: Best Practices, Common Risks & Alternatives [https://www.capricorn-scientific.com/knowledge-center/aliquot-fbs]
- 28. Protocols and Best Practices for Freezing Cells [https://www.stemcell.com/cryopreservation-basics-protocols-and-best-practices-for-freezing-cells]
- 29. Cell Freezing Protocols | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/cell-culture-protocols/freezing-cells.html]
- 30. Automated Cryogenic Storage Systems (-190°C) [https://www.azenta.com/automated-sample-storage-systems/automated-cryogenic-storage-systems-190c]
- 31. Overcoming Obstacles in Automating Cryogenic Storage [https://www.technologynetworks.com/cell-science/articles/overcoming-obstacles-in-automating-cryogenic-storage-386685]
- 32. Evaluating stability and bioactivity of Rehmannia-derived ... [https://www.nature.com/articles/s41598-024-70334-5]
- 33. Exosome Isolation Protocols and Quantification Guidelines [https://www.sepscience.com/exosome-isolation-protocols-and-quantification-guidelines-12144]
- 34. Bio fluid exosomes: promises, challenges, and future ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12459069/]
- 35. Basic Guide for Approaching Drug Delivery with Extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11476574/]
- 36. FAQ: iPSC-derived Exosomes [https://www.reprocell.com/clinical-stem-cell-services/reprocell-japan-gmp-manufacturing-facility/faq-ipsc-derived-exosomes]
- 37. Understanding Lyophilization: What Makes it Special? [https://trimleaf.com/blogs/articles/understanding-lyophilization-what-makes-it-special?srsltid=AfmBOorQe2yPddl7hKM57oJvEnhTLNQ86rkEsb-2XhqRSxmAzherpASi]
- 38. Freeze-drying: what is it and what processes are involved? [https://www.barnalab.com/en/blog/freeze-drying-process-and-stages/]
- 39. Lyophilization: Guide to Freeze Drying in Pharmaceuticals [https://adragos-pharma.com/lyophilization-guide-to-freeze-drying-in-pharmaceuticals/]
- 40. Lyophilization vs. Traditional Drying Methods: Why Choose ... [https://www.specialtysolutions.co/blog/lyophilization-vs-traditional-drying-methods/?srsltid=AfmBOorNhfU6GCDGajP5EJ7eTbi4Pw_E9CyIr7cvXAdEnNeKEvPW94bp]
- 41. Impact of Storage Conditions on EV Integrity/Surface ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9146501/]
- 42. What's hot in freeze drying? Your lyophilization questions ... [https://www.patheon.com/us/en/insights-resources/blog/your-lyophilization-questions-answered.html]
- 43. Troubleshooting During the Manufacture of Lyophilized ... [https://www.americanpharmaceuticalreview.com/Featured-Articles/126958-Troubleshooting-During-the-Manufacture-of-Lyophilized-Drug-Product-Being-Prepared-for-the-Unexpected/]
- 44. Recent developments in isolating methods for exosomes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9879965/]
- 45. Exosome isolation and detection: Methods and applications [https://www.abcam.com/en-us/knowledge-center/cell-biology/exosome-isolation-and-detection]
- 46. Unveiling Exosome Isolation and Purification Protocols [https://www.thermofisher.com/blog/life-in-the-lab/a-deep-dive-into-ultracentrifugation-techniques/]
- 47. Progress in Exosome Isolation Techniques - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5327650/]
- 48. Exosomes Frequently Asked Questions (FAQs) [https://www.thermofisher.com/us/en/home/life-science/cell-analysis/exosomes/exosomes-webinar-q-and-a.html]
- 49. Nanoparticle Tracking Analysis (NTA) [https://www.protocols.io/view/nanoparticle-tracking-analysis-nta-d4298yh6.html]
- 50. Nanoparticle Tracking Analysis for the Quantification and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8243380/]
- 51. Dynamic Light Scattering Problems and Solutions [https://particletechlabs.com/ptl-press/dynamic-light-scattering-unexpected-results-and-false-positives/]
- 52. Overcoming technical challenges in extracellular vesicle ... [https://www.takarabio.com/about/bioview-blog/customer-stories/overcoming-technical-challenges-in-extracellular-vesicle-research?srsltid=AfmBOop3b_2wMFbnKBUM83FwQEcJaGhmizcFHNglsDm8ydFBf_8ee8kP]
- 53. Best practices for RNA storage and sample handling [https://www.qiagen.com/us/knowledge-and-support/knowledge-hub/bench-guide/rna/introduction/general-remarks-on-rna-handling-storage-and-stabilization]
- 54. Preserving the True Immune Landscape: Eliminating ... [https://blog.cellsonics.com/blog/preserving-immune-markers]
- 55. Impact of RNA integrity and blood sample storage conditions ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6016255/]
- 56. An efficient method for long-term room temperature storage ... [https://www.nature.com/articles/ejhg2013145]
- 57. Efficient dispersion of aggregated extracellular vesicles [https://www.nature.com/articles/s41598-025-10050-w]
- 58. Optimizing cell migration assays: Critical roles of ... [https://www.sciencedirect.com/science/article/pii/S0006291X24015341]
- 59. Advancing in vitro cell migration studies: a review of open- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12006941/]
- 60. Troubleshooting [https://bioassaysys.com/troubleshooting/]
- 61. Extracellular vesicle bioactivity and potential for clinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12532472/]
- 62. PBS Phosphate Buffered Saline for Cell Culture [https://biochemazone.com/pbs-phosphate-buffered-saline-for-cell-culture/?srsltid=AfmBOop7bsRw5yND5079TCZkFgU2tNx4Nz3OZ2C26FpQvWp-kKciCiuZ]
- 63. Preservation of extracellular vesicles for drug delivery [https://www.sciencedirect.com/science/article/abs/pii/S1773224725002539]