Ultracentrifugation Protocol for MSC Exosomes: A Complete Guide from Isolation to Characterization
This article provides a comprehensive guide for researchers and drug development professionals on isolating exosomes from Mesenchymal Stem Cell (MSC) conditioned media using ultracentrifugation.
Ultracentrifugation Protocol for MSC Exosomes: A Complete Guide from Isolation to Characterization
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on isolating exosomes from Mesenchymal Stem Cell (MSC) conditioned media using ultracentrifugation. It covers fundamental principles of exosome biogenesis and MSC sources, detailed step-by-step protocols for both standard and advanced ultracentrifugation methods, common troubleshooting and optimization strategies, and essential validation techniques. The content also includes comparative analysis of alternative isolation methods and discusses how isolation choices impact downstream therapeutic applications, offering a complete framework for obtaining high-purity, functionally intact MSC-derived exosomes for research and clinical translation.
Understanding MSC Exosomes: Biogenesis, Significance, and Pre-isolation Fundamentals
Exosome Definition and Core Characteristics
Mesenchymal stem cell-derived exosomes (MSC-Exos) are nanoscale extracellular vesicles (EVs) released by mesenchymal stem cells that function as crucial mediators of intercellular communication by transferring bioactive molecules between cells [1] [2]. These vesicles are defined by several core characteristics that distinguish them from other types of extracellular vesicles, as summarized in Table 1 below.
Table 1: Defining Characteristics of MSC-Derived Exosomes
| Characteristic | Description |
|---|---|
| Size Range | 30-150 nm in diameter [3] [4] |
| Density | 1.10-1.21 g/mL in sucrose gradient [2] [5] |
| Origin/Biogenesis | Endosomal pathway; formed as intraluminal vesicles (ILVs) within multivesicular bodies (MVBs) [6] [3] |
| General Morphology | Cup-shaped appearance under electron microscopy (an artifact of processing); true structure is a round, lipid-bilayer vesicle [6] [4] |
| Key Marker Proteins | Tetraspanins (CD9, CD63, CD81), ESCRT-related proteins (Alix, TSG101), Heat shock proteins (Hsp70, Hsp90) [2] [4] [5] |
Exosomes are one of several types of extracellular vesicles. The biogenesis pathway is the primary feature distinguishing exosomes from microvesicles (which bud directly from the plasma membrane) and apoptotic bodies (released during programmed cell death) [6] [2].
Endosomal Origin and Biogenesis Pathway
The formation of exosomes is a tightly regulated process originating within the endosomal system of the cell. The following diagram illustrates the key stages of exosome biogenesis and secretion.
The biogenesis process involves several key molecular players. The ESCRT (Endosomal Sorting Complex Required for Transport) protein complex, along with accessory proteins like Alix and VPS4, is centrally involved in sorting ubiquitinated cargo and facilitating the inward budding of the endosomal membrane to form ILVs [5]. This process can also occur via ESCRT-independent pathways involving lipids like ceramide [5]. Finally, RAB GTPase proteins (e.g., Rab27a, Rab27b) and SNARE complexes regulate the transport and fusion of MVBs with the plasma membrane, leading to the release of exosomes into the extracellular space [5].
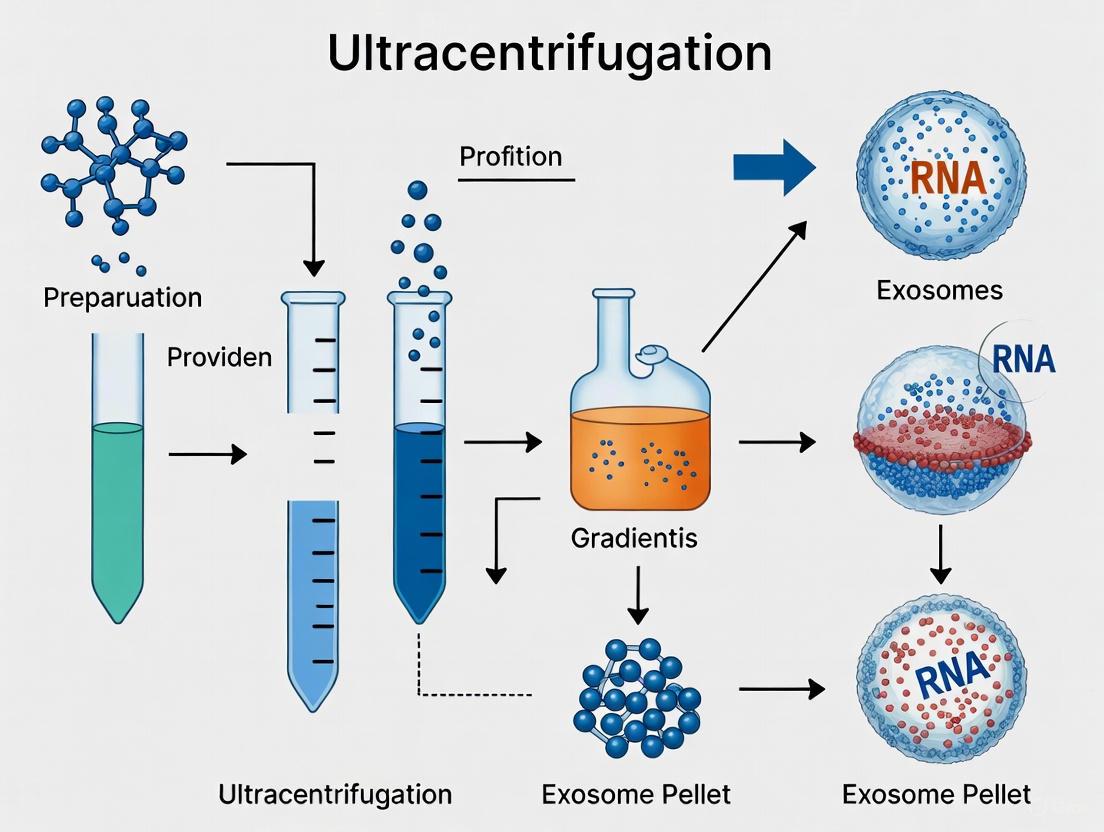
Standard Ultracentrifugation Isolation Protocol
Ultracentrifugation is the most commonly used method for isolating exosomes, accounting for approximately 56% of all isolation techniques used by researchers [3]. The differential ultracentrifugation protocol provides a robust foundation for research-scale exosome preparation. The workflow is visualized in the following diagram.
Detailed Protocol Notes:
- Starting Material: The protocol begins with conditioned medium from MSC cultures. It is critical to use fetal bovine serum (FBS) that has been ultracentrifuged (e.g., at 100,000 × g overnight) to remove bovine exosomes prior to cell culture, or to use serum-free media to avoid contamination [3].
- Centrifugation Steps: The sequential spins are designed to separate components based on size and density. The initial low-speed spins remove cells and large debris, while the 10,000 × g step pellets larger vesicles and organelles. The final ultracentrifugation at high force pellets the small exosomes [7] [3].
- Post-Isolation Handling: The final exosome pellet is typically resuspended in phosphate-buffered saline (PBS) or a specific buffer compatible with downstream applications. Isolated exosomes are stable for up to 6 months when stored at -20 °C or lower. Repeated freeze-thaw cycles should be avoided as they may degrade exosome integrity [2].
The Scientist's Toolkit: Essential Reagents and Equipment
Successful isolation and characterization of MSC-derived exosomes require specific reagents, equipment, and characterization techniques, as outlined in the table below.
Table 2: Research Reagent Solutions and Essential Materials for MSC Exosome Research
| Category | Item | Function/Application |
|---|---|---|
| Cell Culture | Alpha MEM / DMEM | Culture medium for expanding Mesenchymal Stem Cells [7]. |
| Exosome-Depleted FBS | Fetal Bovine Serum processed via ultracentrifugation to remove bovine vesicles, preventing contamination of the isolate [3]. | |
| Trypsin-EDTA | For detaching adherent MSCs during cell culture passaging [7]. | |
| Isolation | Ultracentrifuge & Rotor (e.g., SW32 Ti) | Essential equipment for achieving the high gravitational forces (>100,000 × g) required to pellet exosomes [7] [3]. |
| Ultra-Clear Tubes (e.g., Open-Top Thinwall, 38.5 ml) | Specialized centrifuge tubes designed to withstand the extreme pressures of ultracentrifugation [7]. | |
| Dulbecco's Phosphate Buffered Saline (PBS) | Used for washing cell pellets and, most importantly, for resuspending the final exosome pellet [7] [3]. | |
| Characterization | Nanoparticle Tracking Analyzer (e.g., NanoSight) | Instrument for determining the size distribution and concentration of particles in the exosome preparation [7]. |
| Transmission Electron Microscope (TEM) | Used to visualize the morphology and confirm the cup-shaped structure of isolated exosomes [7] [4]. | |
| Antibodies (CD63, CD81, CD9, TSG101, Alix) | Key reagents for Western Blot analysis to confirm the presence of canonical exosome marker proteins [2] [4]. | |
| Flow Cytometer | Can be used for the analysis of exosomes bound to beads, allowing for immunophenotyping of surface markers [7]. |
Therapeutic Mechanisms and Applications
MSC-derived exosomes exert their therapeutic effects primarily through their molecular cargo, which they transfer to recipient cells to alter cell function and promote repair. The key biochemical components and their therapeutic roles are summarized below.
Table 3: Therapeutic Cargo of MSC-Derived Exosomes and Applications
| Cargo Type | Key Components | Documented Therapeutic Effects / Mechanisms |
|---|---|---|
| Proteins | • Tetraspanins (CD9, CD63, CD81)• Heat Shock Proteins (Hsp70, Hsp90)• Growth Factors & Cytokines• Membrane Transport Proteins (RAB GTPases, Annexins) [6] [5] | • Immunomodulation: Inhibit T-cell proliferation, induce regulatory T-cells, promote M2 macrophage polarization [1] [4].• Tissue Repair: Promote angiogenesis, reduce apoptosis, and stimulate proliferation of tissue-specific cells [1] [2]. |
| Nucleic Acids | • mRNAs• microRNAs (miRNAs) [6] [2] | • Genetic Reprogramming: Transferred mRNAs and miRNAs can alter gene expression in recipient cells. For example, exosomal miRNAs can suppress pro-inflammatory pathways or inhibit fibrosis, supporting tissue regeneration [6] [8]. |
| Lipids | • Cholesterol• Ceramide• Phosphoglycerides [6] [2] | • Structural Integrity: Form the lipid bilayer structure of the exosome.• Bioactive Signaling: Ceramide is involved in the biogenesis of exosomes and can also influence signaling pathways in target cells [6] [5]. |
The therapeutic potential of MSC-derived exosomes has been demonstrated in preclinical models for a wide range of conditions, including:
- Cardiovascular Diseases: Reducing myocardial ischemia/reperfusion injury and promoting cardiac repair [2] [4].
- Kidney and Liver Diseases: Attenuating kidney injury in ischemia-reperfusion models and reducing liver fibrosis [1] [8].
- Neurological Disorders: Promoting neurogenesis and providing neuroprotective effects in models of stroke, spinal cord injury, and neurodegenerative diseases [1].
- Wound Healing: Enhancing cutaneous wound healing through immunomodulation and promotion of angiogenesis [1] [9].
This application note provides a foundational introduction to MSC-derived exosomes, detailing their defining characteristics, a standard isolation protocol, and their therapeutic relevance, thereby setting the stage for advanced research and protocol optimization within a thesis focused on ultracentrifugation methodologies.
Therapeutic Significance of MSC Exosomes in Regenerative Medicine and Drug Development
Mesenchymal stem cell-derived exosomes (MSC-Exos) represent a transformative advancement in regenerative medicine, offering a cell-free therapeutic alternative that addresses critical limitations associated with whole-cell therapies. These nanoscale extracellular vesicles (30-150 nm in diameter) encapsulate a diverse cargo of bioactive molecules—including proteins, lipids, and nucleic acids—that mediate intercellular communication and exert profound therapeutic effects [10]. Unlike their parent cells, MSC-Exos demonstrate lower immunogenicity, enhanced stability, reduced tumorigenicity risk, and an innate ability to cross biological barriers, positioning them as promising next-generation therapeutics [11] [10]. The integration of ultracentrifugation protocols as a foundational isolation methodology has been instrumental in standardizing MSC-Exos research and accelerating their translation from bench to bedside.
The therapeutic potential of MSC-Exos spans a remarkable spectrum of medical applications, encompassing neurodegenerative disorders, cardiovascular diseases, autoimmune conditions, orthopedic injuries, and aging-related pathologies [11] [10]. Their mechanisms of action include cargo delivery to recipient cells, potent immunomodulation through T-cell and macrophage polarization, and activation of endogenous repair pathways that collectively promote tissue regeneration, reduce inflammation, and restore homeostasis [10]. This application note delineates the standardized methodologies, mechanistic underpinnings, and clinical translation frameworks that establish MSC-Exos as powerful tools in regenerative medicine and drug development, with particular emphasis on ultracentrifugation-based isolation protocols.
Ultracentrifugation Protocol for MSC Exosome Isolation
Comprehensive Materials and Equipment
Table 1: Essential Reagents and Equipment for Ultracentrifugation Protocol
| Category | Specific Items | Specifications/Application |
|---|---|---|
| Cell Culture | Human umbilical cord MSCs (huMSCs) | Passages 6-8 recommended [12] |
| Culture medium | Alpha MEM supplemented with 10% FBS [7] | |
| Supplements | L-glutamine, penicillin-streptomycin [7] | |
| Isolation | Ultracentrifuge | Beckman Coulter Optima L100XP [7] |
| Rotor | SW32 Ti Swinging-Bucket Rotor [7] | |
| Centrifuge tubes | Open-Top Thinwall Ultra-Clear, 38.5 mL capacity [7] | |
| Filtration | 0.22 μm and 0.45 μm sterile syringe filters [7] | |
| Buffers | Phosphate-buffered saline (PBS) | Without Ca++ and Mg++ [7] |
| EDTA solution | For cell harvesting and processing [7] |
Stepwise Isolation Procedure
Cell Culture and Supernatant Collection: Culture human umbilical cord-derived MSCs in T75 or T150 flasks using complete alpha-MEM medium. When cells reach 60-80% confluency, replace medium with exosome-depleted serum medium. Collect conditioned supernatant after 48-72 hours of culture [7] [12].
Initial Clarification Centrifugation: Centrifuge the collected supernatant at 2,000 × g for 10 minutes at 4°C to remove cells and large debris. Transfer supernatant to new tubes without disturbing the pellet [7] [12].
Intermediate Filtration: Filter the supernatant through 0.45 μm sterile syringe filters to eliminate remaining particulates and microvesicles. For higher purity, sequential filtration through 0.22 μm filters is recommended [12].
Ultracentrifugation: Transfer the filtered supernatant to ultracentrifuge tubes. Balance tubes precisely and centrifuge at 100,000 × g for 90 minutes at 4°C using a swinging bucket rotor [7] [13].
Washing and Second Ultracentrifugation: Carefully discard supernatant and resuscentrifuge the exosome pellet in 10 mL PBS. Perform a second ultracentrifugation at 100,000 × g for 90 minutes at 4°C to enhance purity [7].
Final Resuspension and Storage: Resuspend the final exosome pellet in an appropriate volume of PBS (typically 100-200 μL). Aliquot and store at -80°C for downstream applications [7].
Quality Assessment and Characterization
Post-isolation characterization is critical for verifying exosome quality and functionality. The following analytical methods should be employed:
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration using instruments such as ZetaView PMX-430-Z QUATT or NanoSight NS300. Expected size range: 30-150 nm [7] [13].
- Transmission Electron Microscopy (TEM): Visualize exosome morphology and membrane integrity. Sample preparation involves negative staining with uranyl acetate [13].
- Western Blot Analysis: Confirm presence of exosomal markers (CD63, CD81, ALIX, TSG101) and absence of negative markers (calnexin) [14] [13].
- Zeta Potential Measurement: Assess surface charge and colloidal stability using ZetaView instrument [12].
Comparative Analysis of Exosome Isolation Methodologies
Technical Performance Metrics
Table 2: Comprehensive Comparison of Exosome Isolation Techniques
| Method | Principle | Particle Size (nm) | Purity | Cell Viability Improvement | Scalability | Time | Cost |
|---|---|---|---|---|---|---|---|
| Ultracentrifugation | Density/sedimentation | 60 [15] | Moderate | 22% [15] | High | 4-6 hours | $$$ [12] |
| Size-Exclusion Chromatography | Size separation | 50-200 [14] | High | N/A | Medium | ~20 min [14] | $$ [14] |
| Ion-Exchange Chromatography | Charge interaction | ~100 [12] | High | Strong clonogenic effect [12] | High | Moderate | $$ [12] |
| Ultrafiltration | Size exclusion | 122 [15] | Low-Moderate | 11% [15] | Medium | <2 hours | $ [15] |
| Precipitation | Solubility shift | 89 [15] | Low | 15% [15] | Medium | <2 hours | $ [15] |
Ultracentrifugation remains the gold standard for research applications due to its reliability and ability to process large sample volumes, though emerging techniques like ion-exchange chromatography demonstrate superior purity and functional efficacy in specific applications [15] [12]. The choice of isolation method significantly influences exosome characteristics, with ultracentrifugation yielding exosomes with smaller average size (60 nm) and narrow size distribution compared to ultrafiltration (122 nm) and precipitation (89 nm) methods [15].
Therapeutic Mechanisms and Signaling Pathways
Molecular Mechanisms of Action
MSC-Exos exert their therapeutic effects through multiple interconnected mechanisms:
Bioactive Cargo Delivery: MSC-Exos transfer proteins, mRNAs, miRNAs, and lipids to recipient cells, modifying their phenotype and function. This horizontal transfer of genetic material enables reprogramming of target cells without direct cell-cell contact [11] [10].
Immunomodulation: MSC-Exos polarize macrophages toward the anti-inflammatory M2 phenotype, suppress T-cell proliferation, and regulate dendritic cell maturation, creating an immunomodulatory microenvironment conducive to tissue repair [10].
Anti-apoptotic Effects: Through delivery of anti-apoptotic miRNAs and proteins, MSC-Exos inhibit programmed cell death in damaged tissues, enhancing cell survival under stress conditions [11].
Angiogenesis Promotion: MSC-Exos contain pro-angiogenic factors (VEGF, FGF, miR-126) that stimulate endothelial cell proliferation and new blood vessel formation, improving tissue perfusion and regeneration [10].
Pathway Activation in Specific Disease Contexts
The therapeutic efficacy of MSC-Exos is mediated through specific signaling pathways in different disease contexts:
Premature Ovarian Failure: MSC-Exos activate AMPK/NR4A1, TGF-β1/Smad3, Wnt/β-catenin, and Hippo signaling pathways, reducing granulosa cell apoptosis and promoting follicular development [11].
Neurodegenerative Disorders: Through miRNA-mediated regulation, MSC-Exos modulate neuroinflammatory responses and promote neuronal survival, potentially benefiting conditions like Alzheimer's disease and Parkinson's disease [10].
Cardiovascular Diseases: MSC-Exos enhance angiogenesis and cardiomyocyte survival through delivery of pro-angiogenic miRNAs and activation of survival pathways, improving cardiac function post-myocardial infarction [10].
Aging-Related Conditions: MSC-Exos mitigate hallmarks of aging including cellular senescence, mitochondrial dysfunction, and stem cell exhaustion through complex signaling network modulation [11].
Clinical Translation and Therapeutic Applications
Clinical Trial Landscape and Dosing Strategies
The clinical translation of MSC-Exos has accelerated significantly, with 66 registered clinical trials completed between 2014-2024 [16]. These trials span diverse therapeutic areas including respiratory diseases, neurological disorders, and autoimmune conditions.
Table 3: Clinical Administration Routes and Dosing Strategies for MSC-Exos
| Administration Route | Therapeutic Area | Dose Range | Efficacy Notes | Clinical Trial Phase |
|---|---|---|---|---|
| Aerosolized Inhalation | Respiratory diseases (COVID-19, ARDS) | ~10⁸ particles | Therapeutic effects at lower doses vs. IV [16] | Phase I/II [16] |
| Intravenous Infusion | Systemic diseases, GVHD | 10⁸-10¹¹ particles | Higher doses required vs. inhalation [16] | Phase I-III [16] |
| Local Injection | Orthopedic injuries, osteoarthritis | 10⁸-10¹⁰ particles | Direct targeting to affected tissue [11] | Phase I/II [11] |
| Intra-ovarian Injection | Premature ovarian failure | Species-dependent | Improves follicle count and hormone levels [11] | Preclinical/Phase I [11] |
Clinical evidence indicates that administration route significantly influences the effective dose window, with aerosolized inhalation achieving therapeutic effects at approximately 10⁸ particles—significantly lower than doses required for intravenous administration [16]. This route-dependent efficacy underscores the importance of optimizing delivery strategies for specific clinical indications.
Regulatory Advances and Approved Therapies
The regulatory landscape for MSC-based therapies has evolved substantially, with several landmark approvals:
Ryoncil (remestemcel-L): Received FDA approval in December 2024 as the first MSC therapy for pediatric steroid-refractory acute graft versus host disease (SR-aGVHD) [17]. This approval establishes a regulatory precedent for future MSC-derived products.
Omisirge (omidubicel-onlv): Approved in April 2023 for hematologic malignancies, representing advancement in cord blood-derived cellular therapies [17].
While no MSC-Exos have received full FDA approval to date, the growing clinical trial portfolio and established regulatory pathways for parent cell products signal imminent translation of exosome-based therapeutics into clinical practice.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Critical Reagents and Research Tools for MSC Exosome Research
| Reagent Category | Specific Product/Kit | Research Application | Functional Role |
|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Reagent (Thermo Fisher) | Rapid exosome precipitation | Pre-analysis concentration [14] |
| Characterization | ZetaView PMX-430-Z QUATT (Particle Metrix) | Size/concentration analysis | NTA measurements [12] |
| Chromatography | qEV Gen 2-35 nm columns (Izon Science) | Size-exclusion chromatography | High-purity isolation [14] |
| Cell Culture | Clin-SFM-Human MSC medium (Clin-Biotech) | MSC culture expansion | Serum-free formulation [12] |
| Antibodies | ALIX (Cell Signaling), CD63 (ABclonal) | Western blot validation | Exosome marker detection [14] |
| Microscopy | Transmission Electron Microscope | Morphological analysis | Ultrastructural visualization [13] |
Challenges and Future Perspectives
Despite considerable progress, several challenges remain in the widespread clinical implementation of MSC-Exos therapies. Biological variability stemming from different MSC sources (bone marrow, adipose tissue, umbilical cord), isolation methods, and characterization protocols continues to hamper standardization efforts [16] [10]. The absence of harmonized dosing frameworks and potency assays further complicates clinical translation and comparability between studies [16].
Future innovations will likely focus on bioengineering approaches to enhance targeting specificity and therapeutic potency. Genetic modification of parent MSCs to enrich exosomes with specific therapeutic molecules, surface engineering to improve tissue-specific targeting, and development of scalable manufacturing processes represent promising avenues for advancement [10]. Additionally, the emergence of iPSC-derived MSCs (iMSCs) offers opportunities for enhanced consistency and scalability compared to primary MSCs [17].
The continued refinement of ultracentrifugation protocols, coupled with orthogonal purification methods and rigorous characterization standards, will be essential for establishing MSC-Exos as mainstream therapeutic modalities. As regulatory frameworks evolve and manufacturing capabilities advance, MSC-Exos are poised to become indispensable tools in the regenerative medicine arsenal, potentially transforming treatment paradigms for numerous debilitating conditions.
The therapeutic potential of mesenchymal stem cell-derived exosomes (MSC-Exos) is increasingly recognized in regenerative medicine, diagnostic development, and drug delivery systems [18] [19]. These nanoscale extracellular vesicles (EVs), typically 40-160 nm in diameter, mediate intercellular communication by transferring proteins, lipids, and nucleic acids from parent MSCs to recipient cells [18] [20]. However, the biological cargo and consequent functional properties of MSC-Exos are not uniform; they are profoundly influenced by the tissue-specific origin of the parent MSCs [4]. This application note examines critical pre-isolation considerations regarding MSC sources—specifically bone marrow (BM), adipose tissue (AD), and umbilical cord (UC)—and their impact on exosome cargo composition, providing detailed methodologies for researchers working within an ultracentrifugation-focused framework.
MSCs can be isolated from various tissues, with bone marrow, adipose tissue, and umbilical cord representing the most common sources. Each source imparts distinct biological characteristics to the cells, which are subsequently reflected in the molecular cargo of the exosomes they produce [4]. This variation stems from differences in the native microenvironment and physiological role of the tissue of origin.
Bone Marrow-derived MSCs (BM-MSCs) were the first to be discovered and represent a gold standard in the field; however, their isolation is invasive, and their proliferative capacity decreases with donor age [4]. Adipose Tissue-derived MSCs (AD-MSCs) are obtained from lipoaspirate, offering an abundant and accessible source with strong proliferative potential [4]. Umbilical Cord-derived MSCs (UC-MSCs), harvested from Wharton's jelly, are characterized by rapid self-renewal, high doubling capacity, and minimal ethical concerns, making them a promising source for scalable production [21] [4].
The protein and nucleic acid cargo of exosomes directly determines their functional specificity upon delivery to recipient cells [20]. The proteomic profile of UC-MSC-derived exosomes, for instance, is enriched with proteins involved in extracellular matrix organization and vesicle-mediated transport [21]. The following table summarizes key comparative characteristics of MSCs from these primary sources.
Table 1: Comparative Analysis of Primary Mesenchymal Stem Cell (MSC) Sources
| Characteristic | Bone Marrow (BM) | Adipose Tissue (AD) | Umbilical Cord (UC) |
|---|---|---|---|
| Isolation Accessibility | Invasive, low yield [4] | Minimally invasive, high yield [4] | Non-invasive, high yield [21] |
| Proliferative Capacity | Moderate, age-dependent [4] | High [4] | Very high, stable doubling time [21] |
| Defining Surface Markers | CD73+, CD90+, CD105+, HLA-DR- [4] | CD73+, CD90+, CD105+, HLA-DR- [4] | CD73+, CD90+, CD105+, HLA-DR- [21] |
| Key Advantages | Considered the biological "gold standard" | Abundant tissue source, easy access | Young cell phenotype, low immunogenicity, no ethical issues [21] |
| Key Documented Cargo/Functional biases | Well-studied for immunomodulation | Promising for angiogenesis and wound healing | Enriched in ECM organization proteins; potent tissue repair [21] |
The functional potency of MSC-Exos is directly dictated by their biomolecular cargo. Proteomic analyses reveal that UC-MSC exosomes are uniquely enriched with proteins governing extracellular matrix (ECM) organization and structural integrity, making them particularly potent for wound healing applications [21]. In contrast, AD-MSC exosomes may carry cargo that promotes angiogenesis. Beyond proteins, the miRNA profile is equally critical; exosomal miRNAs (e.g., miR-21, miR-146a) can regulate recipient cell gene expression, influencing processes like immunomodulation and metabolic reprogramming [18].
Table 2: Quantitative and Functional Cargo Differences in MSC-Derived Exosomes
| Cargo Component | Bone Marrow (BM) | Adipose Tissue (AD) | Umbilical Cord (UC) |
|---|---|---|---|
| Proteomic Highlights | Alix, TSG101, CD63, CD81 [20] | Alix, TSG101, CD63, CD81 [20] | Enriched in ECM proteins (e.g., Collagens, Fibronectin) [21] |
| Distinct Protein Functions | Immunomodulation, vesicle biogenesis | Immunomodulation, vesicle biogenesis | Tissue scaffolding, cell adhesion, wound repair [21] |
| Key miRNA Examples | miR-21, miR-146a (Immunomodulation) [18] | Angiogenesis-related miRNAs (e.g., miR-31) | Pro-regenerative miRNAs (e.g., miR-21, let-7 family) [18] [22] |
| Functional Evidence from Studies | Attenuates inflammatory responses [18] | Promotes blood vessel formation | Superior acceleration of wound closure and epithelial regeneration in models [21] |
Experimental Protocol: Source-Specific Exosome Isolation via Ultracentrifugation
This protocol is designed for the isolation of exosomes from conditioned media of BM-MSCs, AD-MSCs, and UC-MSCs, emphasizing critical steps that account for source-specific variations.
Pre-isolation: Cell Culture and Conditioned Media Collection
- Cell Source Validation: Verify MSC identity via flow cytometry for CD73, CD90, CD105 (≥95% positive) and CD45, CD34, HLA-DR (≤2% positive) [4]. Confirm trilineage differentiation potential (osteogenic, chondrogenic, adipogenic) [4].
- Culture Conditions: Maintain MSCs in DMEM/F12 or α-MEM supplemented with 10% exosome-depleted FBS. Exosome depletion from FBS is achieved via ultracentrifugation at 100,000 × g overnight at 4°C, followed by 0.22 µm filtration [21].
- Collection of Conditioned Media (CM): Harvest CM from MSCs at 80-90% confluence. To remove cells and debris, perform sequential centrifugation: 300 × g for 10 min, 2,000 × g for 20 min, and 10,000 × g for 30 min at 4°C. Collect the supernatant promptly for isolation or store at -80°C [21].
Ultracentrifugation Isolation Workflow
The following diagram outlines the core ultracentrifugation protocol for exosome isolation.
Post-isolation: Characterization and Quantification
Consistent characterization is vital for correlating exosome cargo with MSC source. Adhere to MISEV (Minimal Information for Studies of Extracellular Vesicles) guidelines [18] [23].
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration. MSC-Exos should peak within 40-160 nm [21] [23].
- Transmission Electron Microscopy (TEM): Confirm spherical, cup-shaped morphology and bilayer membrane structure [21].
- Western Blotting: Detect positive markers (e.g., CD63, CD81, CD9, TSG101, Alix) and negative markers (e.g., Calnexin) to ensure purity [20].
- Downstream Cargo Analysis: For proteomics, use liquid chromatography with tandem mass spectrometry (LC-MS/MS). For miRNA profiling, employ next-generation sequencing (NGS) or RT-qPCR [20] [21].
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for MSC Exosome Isolation & Characterization
| Reagent / Material | Function / Application | Example & Notes |
|---|---|---|
| Exosome-Depleted FBS | Cell culture supplement prevents bovine exosome contamination in conditioned media. | Ultracentrifuged or commercial exosome-depleted FBS is essential for clean background [21]. |
| Dulbecco's Phosphate Buffered Saline (PBS) | Washing buffer; used for resuspending and washing the exosome pellet. | Plain PBS should be avoided for long-term storage; use stabilizers like BSA or trehalose [18]. |
| Protease & Phosphatase Inhibitors | Preserves protein integrity and phosphorylation states in exosomal cargo during isolation. | Add to conditioned media and lysis buffers for proteomic studies [21]. |
| RiboNuclease (RNase) Inhibitors | Protects RNA cargo (miRNA, mRNA) from degradation throughout the isolation process. | Critical for obtaining high-quality RNA for sequencing or PCR analysis [20]. |
| Antibodies for Characterization | Detection of exosomal surface and internal markers via Western Blot or flow cytometry. | Anti-tetraspanins (CD63, CD81, CD9); anti-biogenesis markers (Alix, TSG101) [20] [23]. |
| Sucrose Solution | Forms a density gradient for high-purity isolation as an alternative to differential ultracentrifugation. | Used in density gradient centrifugation to separate exosomes from contaminants [24]. |
The selection of an MSC source—bone marrow, adipose tissue, or umbilical cord—is a critical pre-isolation variable that directly and measurably impacts the protein and miRNA cargo of resultant exosomes. This cargo diversity underpins distinct therapeutic efficacies. A rigorous and standardized ultracentrifugation protocol, coupled with comprehensive characterization, is fundamental for ensuring the reproducibility of research and the accurate interpretation of data in the rapidly advancing field of MSC-exosome therapeutics. Researchers must carefully select their MSC source based on the intended biological outcome and account for this variable in their experimental design.
The isolation of exosomes from Mesenchymal Stem Cells (MSCs) is a critical step in harnessing their therapeutic potential for regenerative medicine, drug delivery, and disease treatment [25] [26]. Ultracentrifugation remains a cornerstone technique for exosome isolation, prized for its high yield, versatility, and reliability [27]. This application note provides a comprehensive overview of essential laboratory equipment and stringent safety protocols for implementing ultracentrifugation methods in MSC exosome research. The content is framed within a broader thesis on standardizing ultracentrifugation protocols to ensure the isolation of exosomes with high purity, integrity, and biological activity, which is imperative for reproducible downstream analysis and therapeutic applications [25] [28].
The Scientist's Toolkit: Essential Equipment and Reagents
Establishing a robust workflow for MSC exosome isolation requires specific instrumentation, consumables, and reagents. The following table details the core components of the laboratory setup.
Table 1: Essential Research Reagent Solutions and Equipment for MSC Exosome Isolation via Ultracentrifugation
| Item | Function/Application | Specific Examples & Notes |
|---|---|---|
| Ultracentrifuge & Rotors | Generates high centrifugal force (≥100,000 × g) to pellet exosomes [25] [27]. | Floor-model ultracentrifuge (e.g., Sorvall WX 90+, Beckman Optima XPN 90); Swinging bucket rotors (e.g., SW 28 Ti, SW 40 Ti) [25] [28]. |
| Ultracentrifuge Tubes | Holds samples during high-speed spins. | Must be compatible with the rotor and capable of withstanding ultracentrifugation forces. |
| Cell Culture Media | For expanding MSCs and conditioning for exosome production. | Serum-free media (e.g., STEMPRO MSC SFM CTS) or exosome-depleted FBS to avoid contaminating vesicles [25] [27]. |
| Density Gradient Medium | Separates exosomes based on buoyant density, enhancing purity by removing contaminants [25] [28]. | Sucrose (30%) or Iodixanol (e.g., OptiPrep) solutions [25] [28]. |
| Buffers | For resuspending, washing, and storing exosome pellets. | Phosphate-Buffered Saline (PBS) is commonly used [25] [27]. |
| Characterization Instruments | For validating the size, concentration, and identity of isolated exosomes. | Nanoparticle Tracking Analyzer (NTA), Transmission Electron Microscope (TEM), Western Blot apparatus [25] [29]. |
Core Ultracentrifugation Methods for MSC Exosome Isolation
Several ultracentrifugation-based methods have been developed, each with distinct advantages. The choice of method depends on the experimental requirements for yield, purity, and scalability.
Direct Ultracentrifugation (UC)
This traditional method involves pelleting exosomes directly through high-speed centrifugation. It is widely used but can subject exosomes to high shear forces and potential damage [25] [28].
Protocol:
- Pre-clearing: Centrifuge the conditioned cell culture media at 300 × g for 10 minutes at 4°C to remove cells. Transfer the supernatant [25] [27].
- Microvesicle Removal: Centrifuge the supernatant at 10,000 × g for 30 minutes at 4°C to pellet larger microvesicles and apoptotic bodies. Transfer the supernatant [25].
- Exosome Pelletting: Transfer the pre-cleared supernatant to ultracentrifuge tubes. Centrifuge at 100,000 - 120,000 × g for 70-120 minutes at 4°C [25] [27].
- Washing & Resuspension: Carefully discard the supernatant and resuspend the often invisible exosome pellet in a suitable volume of PBS. A second ultracentrifugation step can be performed for washing. The final pellet is resuspended in PBS and stored at -80°C [27].
Sucrose Cushion Ultracentrifugation (SUC)
This refined method uses a density cushion to protect exosomes from the pelletting forces, thereby improving vesicle integrity and yield [25].
Protocol:
- Pre-clearing: Perform steps 1 and 2 as in the Direct UC protocol [25].
- Cushion Layering: Carefully layer the pre-cleared media on top of a 4 mL cushion of 30% sucrose solution in an ultracentrifuge tube [25].
- Ultracentrifugation: Centrifuge at 100,000 × g for 90 minutes at 4°C. Exosomes will collect at the sample-sucrose interface rather than forming a hard pellet [25].
- Collection & Washing: Collect the sucrose layer containing the exosomes, dilute with PBS, and perform a second ultracentrifugation to pellet the exosomes. Resuspend the final pellet in PBS [25].
Cushioned–Density Gradient Ultracentrifugation (C-DGUC)
This high-performance method combines the protective cushion with a density gradient for superior purity, effectively separating exosomes from protein contaminants [28].
Protocol:
- Pre-clearing & Cushion: Pre-clear the conditioned media and layer it over a 60% iodixanol cushion. Centrifuge at 100,000 × g for 2-4 hours [28].
- Gradient Formation: Collect the concentrated exosomes from the cushion and layer them onto a pre-formed discontinuous iodixanol density gradient (e.g., 5%, 10%, 20%) [28].
- Equilibrium Centrifugation: Centrifuge at 100,000 × g for 12-18 hours (overnight) to allow exosomes to migrate to their equilibrium density (typically 1.10-1.14 g/mL in iodixanol) [28].
- Fraction Collection: After centrifugation, collect the gradient from the top in fractions. Analyze fractions for exosome presence and pool those containing pure exosomes [28].
Table 2: Comparison of Ultracentrifugation Methods for MSC Exosome Isolation
| Method | Key Principle | Relative Yield | Relative Purity | Impact on Exosome Integrity | Best For |
|---|---|---|---|---|---|
| Direct UC | Direct pelletting by high g-force [27]. | Moderate | Moderate; can have protein contamination [25]. | Can cause damage or aggregation due to pelletting forces [28]. | Standard, non-scaling applications. |
| Sucrose Cushion (SUC) | Cushion prevents direct pelletting, protects integrity [25]. | High [25] | Higher than UC; reduces protein contamination [25]. | Preserves cup-shaped morphology and integrity [25]. | Applications requiring high yield and biological activity. |
| C-DGUC | Combines cushioning with density-based separation [28]. | Good (from cushion) | Very High; effectively removes contaminants [28]. | Best preservation of integrity and function [28]. | High-purity applications and detailed functional studies. |
Workflow Diagram
The following diagram illustrates the key decision points and steps in the MSC exosome isolation workflow.
Essential Ultracentrifuge Safety Protocols
Operating an ultracentrifuge requires strict adherence to safety protocols to prevent catastrophic equipment failure, personal injury, and loss of valuable samples [30].
Routine Operational Safety
- Balance Loads Meticulously: Tubes must be balanced by mass (not volume) within the manufacturer's specified tolerance (typically 0.1 g) [30]. Imbalanced loads create violent vibrations that can damage the rotor and instrument.
- Inspect Rotors and Tubes: Visually inspect rotors for signs of corrosion, cracks, or stress before each use. Check ultracentrifuge tubes for scratches, cloudiness, or cracks, and do not reuse them if they are compromised [30].
- Use Proper Containment: Always close the centrifuge lid and ensure safety interlocks are engaged before starting a run. In the event of a failure, this contains debris within the chamber.
- Handle Hazards Appropriately: If spinning hazardous materials, ensure tubes are sealed properly and decontaminate the rotor and chamber after use.
Centrifuge Safety Checklist
- Rotor & Tubes: Confirm compatibility and absence of physical damage.
- Tube Content: Securely sealed and matched by mass across the rotor.
- Load Balance: Balanced within 0.1 g for all opposing positions.
- Lid & Locks: Lid is securely closed and locked.
- Run Parameters: Speed, time, and temperature are correctly set and within safe limits for the rotor.
- Clear Chamber: No foreign objects are in the centrifuge chamber.
Quality Control and Characterization of Isolated MSC Exosomes
Post-isolation, exosomes must be characterized to confirm their identity, purity, and integrity [25] [29]. Key techniques include:
- Nanoparticle Tracking Analysis (NTA): Determines the particle size distribution and concentration [25] [29].
- Transmission Electron Microscopy (TEM): Visualizes the cup-shaped morphology of intact exosomes [25].
- Western Blotting: Detects the presence of exosome-specific marker proteins (e.g., CD63, CD81, Alix, TSG101) and the absence of negative markers (e.g., calnexin) [25] [29].
A properly configured laboratory with a focus on both advanced methodology and rigorous safety is fundamental for successful MSC exosome research. The one-step sucrose cushion and C-DGUC methods provide refined approaches to isolate high-quality exosomes, overcoming some limitations of traditional ultracentrifugation. By integrating the equipment overview, detailed protocols, and stringent safety measures outlined in this document, researchers can enhance the reproducibility, reliability, and safety of their work, thereby accelerating the translational potential of MSC-derived exosomes in therapeutic applications.
The therapeutic potential of mesenchymal stem cell (MSCs) is increasingly attributed to their paracrine activity, particularly through the release of extracellular vesicles such as exosomes [31] [3]. These nanovesicles (30-150 nm in diameter) transfer functional cargoes including proteins, miRNAs, and mRNAs from MSCs to recipient cells, facilitating intercellular communication and tissue repair [3]. However, the isolation of high-purity exosomes for research and therapeutic applications faces a significant challenge: conventional fetal bovine serum (FBS) used in cell culture contains abundant bovine extracellular vesicles that contaminate the final exosome preparation [32]. These contaminating vesicles co-isolate with MSC-derived exosomes during ultracentrifugation, compromising downstream analyses and experimental results [32]. Therefore, the preparation of high-quality starting material through proper culture conditions and conditioned media collection represents a foundational step in MSC exosome research, directly influencing the validity and reproducibility of experimental outcomes.
Essential Research Reagent Solutions
Table 1: Key research reagents for exosome-depleted culture systems
| Reagent Category | Specific Examples | Function & Importance |
|---|---|---|
| Basal Media | Dulbecco's Modified Eagle Medium (DMEM), Serum-Free Media (SFM) | Provides nutritional foundation for MSC culture without introducing exogenous vesicle contaminants [33] [25]. |
| Exosome-Depleted FBS | Ultracentrifugation-prepared dFBS, Commercial kits (e.g., Exo-FBS from System Biosciences) | Supplements essential growth factors while dramatically reducing contaminating bovine vesicles [32] [34]. |
| Cell Culture Supplements | L-glutamine, Penicillin-Streptomycin | Maintains cell health and prevents microbial contamination during extended culture periods [35] [25]. |
| Processing Reagents | Phosphate-Buffered Saline (PBS), Protease Inhibitors | Used for cell washing and protecting exosomal proteins from degradation during processing [33] [3]. |
Protocol: Preparation of Exosome-Depleted Fetal Bovine Serum
The use of exosome-depleted FBS is critical for ensuring that isolated exosomes originate from MSCs rather than culture medium supplements. Multiple methods exist for preparing exosome-depleted FBS, with ultrafiltration and ultracentrifugation being the most common.
Ultrafiltration Protocol
Ultrafiltration provides an efficient, time-saving alternative to traditional ultracentrifugation for depleting EVs from FBS [32].
- Equipment Setup: Prepare Amicon Ultra-15 centrifugal filter devices (100 kDa molecular weight cutoff, Merck Millipore UFC910024) and a benchtop centrifuge [32].
- Processing: Load regular FBS into the centrifugal filter device according to manufacturer's capacity guidelines.
- Centrifugation: Centrifuge at 3,000 × g for 55 minutes at 4°C [32].
- Collection: Collect the flow-through, which constitutes the exosome-depleted FBS (UF-dFBS).
- Validation: Confirm depletion efficiency using nanoparticle tracking analysis (NTA) to quantify remaining particle concentrations [32].
Ultracentrifugation Protocol
Ultracentrifugation remains the historical gold standard for EV depletion, though it is more time-consuming [32].
- Tube Preparation: Aliquot regular FBS into ultracentrifugation tubes suitable for an SW28 rotor (or equivalent).
- Centrifugation: Ultracentrifuge at 100,000–121,896 × g for 16–19 hours at 4°C [32] [25].
- Collection: Carefully collect the top 9/10 of the supernatant without disturbing the pellet, which contains bovine EVs and aggregates.
- Filtration: Sterilize the collected supernatant by passing it through a 0.22 µm pore filter [32].
- Storage: Aliquot and store at -20°C or -80°C until use.
Comparative Performance of Depletion Methods
Table 2: Quantitative comparison of FBS exosome depletion methods
| Method | Hands-On Time | Total Processing Time | Particle Reduction Efficiency | Relative Cost | Key Advantages |
|---|---|---|---|---|---|
| Ultrafiltration | 10-15 minutes | ~1 hour | Highly efficient depletion [32] | Medium (48 euros/50 mL) [32] | Rapid, easy standardization, maintains cell growth [32] |
| Ultracentrifugation | ~2 hours | 16-19 hours | Only partially depletes EVs [32] | Low (32 euros/50 mL) [32] | Widely recognized, requires no specialized filters |
| Commercial dFBS | None | None | Variable between batches | High (224 euros/50 mL) [32] | Maximum convenience, ready-to-use |
Protocol: MSC Culture and Conditioned Media Collection
Proper MSC culture and conditioned media collection are crucial for maximizing exosome yield while maintaining cell viability and function.
Cell Culture Setup
- Cell Source: Use human bone marrow-derived MSCs (hMSCs) at passages 3-6 for optimal growth and exosome production [33].
- Culture Medium: Prepare complete culture medium using DMEM supplemented with 10% exosome-depleted FBS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin [33] [25].
- Cell Seeding: Seed MSCs at appropriate density (e.g., 5,000-10,000 cells/cm²) in standard tissue culture flasks and culture at 37°C in a humidified 5% CO2 incubator until they reach 70-80% confluence [35] [25].
Conditioning Phase
- Serum Deprivation: When MSCs reach 70-80% confluence, carefully aspirate the growth medium and wash cells twice with PBS to remove residual serum components [33] [25].
- Serum-Free Conditioning: Add serum-free DMEM or specialized serum-free media (e.g., STEMPRO MSC SFM CTS) to the cells [25]. Serum-free conditioning minimizes contaminating proteins in subsequent exosome isolates.
- Incubation: Incubate cells for 24-48 hours in the conditioning medium. The optimal conditioning time balances exosome yield against potential cellular stress from nutrient depletion [35] [25].
Conditioned Media Collection and Preliminary Processing
- Collection: Collect the conditioned media from MSC cultures into sterile centrifuge tubes [33].
- Cell Removal: Centrifuge the collected media at 300 × g for 10 minutes at 4°C to remove detached cells [33] [25].
- Debris Clearance: Transfer the supernatant to new tubes and centrifuge at 10,000-20,000 × g for 30 minutes at 4°C to eliminate apoptotic bodies, microvesicles, and cellular debris [33] [25] [34].
- Filtration: Filter the supernatant through 0.22 µm pore filters to remove remaining particulates [35].
- Storage: Either process immediately for exosome isolation or store at 4°C for short-term storage (up to 48 hours) or at -80°C for long-term preservation [33].
Diagram 1: Experimental workflow for preparing high-quality starting material from MSC culture to conditioned media collection.
Quality Assessment and Technical Considerations
Validation of Exosome Depletion
After preparing exosome-depleted FBS, validate the depletion efficiency using:
- Nanoparticle Tracking Analysis (NTA): Quantifies residual particle concentration and size distribution [32] [25].
- Transmission Electron Microscopy (TEM): Visualizes presence or absence of vesicles in the depleted FBS [32].
- Western Blotting: Detects exosomal markers (CD9, CD63, CD81) to confirm removal of bovine exosomes [32].
Critical Factors for Success
- Cell Health Monitoring: Regularly assess MSC morphology and viability throughout the conditioning phase. Serum deprivation can induce stress; therefore, conditioning duration should be optimized to balance yield against cell viability [36].
- Consistent Processing: Maintain consistent centrifugation speeds, times, and temperatures during media processing to ensure reproducible results between experiments.
- Contamination Prevention: Use sterile techniques throughout the protocol to prevent microbial contamination, which can significantly alter exosome content and function.
- Documentation: Meticulously record cell passage numbers, confluence levels, conditioning durations, and processing parameters to maintain experimental reproducibility.
The preparation of high-quality starting materials through proper MSC culture in exosome-depleted FBS and careful conditioned media collection represents the critical first step in obtaining pure, biologically relevant MSC-derived exosomes. The protocols outlined herein, when implemented consistently, significantly reduce contaminating bovine vesicles that would otherwise compromise downstream analyses and experimental validity. As research continues to elucidate the therapeutic potential of MSC-derived exosomes, standardization of these foundational protocols becomes increasingly important for generating comparable, reproducible data across the scientific community.
Step-by-Step Ultracentrifugation Protocols: From Basic to Advanced Techniques
Within the broader scope of developing a robust ultracentrifugation protocol for mesenchymal stem cell (MSC) exosome research, the initial steps of sample preparation are critical. The sequential centrifugation at low speeds (300g) and intermediate speeds (10,000g) serves as the foundational purification stage, aiming to remove cellular debris and microvesicles from the conditioned media. This preparatory phase is essential for ensuring the subsequent isolation of a pure exosome population, as contamination from larger particles can significantly impact the yield, characterization, and downstream experimental results [37] [38]. This application note details a standardized and optimized protocol for this crucial sample preparation step.
Theoretical Basis and Rationale
Differential centrifugation separates particles based on their size, density, and shape through the application of sequentially increasing centrifugal forces [37]. In a homogeneous starting solution, larger and denser particles sediment faster and are pelleted at lower centrifugal forces.
- Low-Speed Centrifugation (300g): This initial step is designed to pellet intact cells, large apoptotic bodies, and substantial cellular debris. The objective is to remove the largest contaminants while keeping the smaller vesicles, including microvesicles and exosomes, in suspension [25] [7].
- Intermediate-Speed Centrifugation (10,000g): The subsequent centrifugation at a higher force targets microvesicles (MVs), which are large extracellular vesicles (100–1000 nm in diameter) generated by the outward budding of the plasma membrane [39] [38]. Pelleting these vesicles prevents their co-isolation with the smaller exosomes (40–150 nm) in the final ultracentrifugation step [40] [25].
Theoretical analysis indicates that the efficiency of pelleting is dependent not only on the centrifugal force (g-force) and time but also on the rotor type (swinging-bucket vs. fixed-angle) and the sedimentation path length [37]. Furthermore, studies highlight that the selective loss of specific MV subpopulations can occur during the initial low-speed spin if parameters are not optimized, underscoring the need for a carefully considered protocol [38].
Materials and Equipment
Research Reagent Solutions
Table 1: Essential reagents and materials for sequential centrifugation.
| Item | Function/Description |
|---|---|
| Conditioned Cell Culture Media | Serum-free media collected from MSC cultures, the source of extracellular vesicles. |
| Dulbecco's Phosphate Buffered Saline (PBS) | Used for washing cell pellets and resuspending/diluting vesicle samples. |
| Serum-Free Media | Used during the cell conditioning phase to enrich for exosomes and avoid fetal bovine serum-derived vesicle contamination. |
| Protease Inhibitor Cocktails | Added to conditioned media to prevent protein degradation during processing. |
Laboratory Equipment
Table 2: Essential equipment for the protocol.
| Equipment | Specification |
|---|---|
| Refrigerated Benchtop Centrifuge | Capable of maintaining 4°C, with rotors for 15 mL and 50 mL conical tubes. |
| Centrifuge Rotors | Fixed-angle or swinging-bucket rotors. The rotor type influences pelleting efficiency [37] [38]. |
| Conical Tubes | 15 mL and 50 mL, capable of withstanding the required g-forces. |
| Pipettes and Serological Pipettes | For accurate and aseptic handling of media and supernatants. |
Step-by-Step Protocol
Pre-Conditioned Media Collection
- Culture MSCs from your desired source (e.g., umbilical cord, bone marrow, adipose tissue) to 70-80% confluency [40] [7].
- Wash the adherent cells gently with pre-warmed PBS to remove residual serum and dead cells.
- Add serum-free culture medium and incubate for 24-48 hours to condition the media. Serum-free conditions are recommended to avoid contamination with bovine exosomes from FBS [25] [7].
- Collect the conditioned medium into 50 mL conical tubes.
Sequential Centrifugation Steps
The following workflow outlines the sequential centrifugation process for clarifying conditioned media prior to exosome isolation.
Detailed Procedure:
First Spin: Removal of Cells and Large Debris
- Centrifuge the collected conditioned media at 300g for 10 minutes at 4°C [25] [38] [7].
- Carefully transfer the supernatant (S1) to new 50 mL conical tubes using a serological pipette. Avoid disturbing the soft pellet (P1), which contains cells and large cellular debris.
- The pellet (P1) can be discarded.
Second Spin: Removal of Microvesicles and Smaller Debris
- Centrifuge the supernatant (S1) at 10,000g for 30 minutes at 4°C [25] [38].
- Following centrifugation, carefully collect the supernatant (S2). This supernatant now contains the exosomes and soluble proteins and is ready for the final exosome isolation step, typically ultracentrifugation at 100,000g or higher.
- The resulting pellet (P2), containing microvesicles and smaller cellular debris, should be discarded [38].
Critical Parameters and Optimization
- Rotor Type Selection: The choice between a fixed-angle and a swinging-bucket rotor is significant. Fixed-angle rotors generally have shorter sedimentation path lengths, reducing run times, but can lead to pellet striation and potential resuspension issues. Swinging-bucket rotors provide a pellet at the bottom of the tube, which is easier to work with, but often require longer run times [37] [38]. For the initial 300g spin, using a swinging-bucket rotor or implementing a washing step for the P1 pellet in a fixed-angle rotor can mitigate the selective loss of microvesicles that may sediment with cells and debris [38].
- Temperature Control: All centrifugation steps must be performed at 4°C to minimize protease activity and preserve the integrity of vesicles.
- Handling of Supernatant: Always aspirate supernatants carefully without disturbing the pellet. Leaving a small volume of liquid above the pellet is recommended to avoid accidental collection of pelleted material.
Expected Results and Quality Control
Upon successful completion of the protocol, the final supernatant (S2) should be clear and devoid of visible particulates. The pellet from the 10,000g spin (P2) may appear as a small, translucent or white spot at the bottom of the tube.
Table 3: Troubleshooting common issues during sequential centrifugation.
| Problem | Potential Cause | Suggested Remedy |
|---|---|---|
| Low final exosome yield | Microvesicle loss in P1 pellet | Use a swinging-bucket rotor for the 300g spin or add a wash step to the P1 pellet [38]. |
| Cloudy supernatant after 10,000g spin | Incomplete pelleting of microvesicles | Ensure correct g-force and time calculations. Verify rotor calibration. Increase centrifugation time slightly. |
| Protein contamination in final exosome prep | Incomplete removal of soluble proteins | The 10,000g spin does not remove soluble proteins. Ensure subsequent ultracentrifugation steps are performed and consider using a sucrose cushion for higher purity [25]. |
Applications in Downstream Processing
The clarified supernatant (S2) obtained from this protocol is the direct input for downstream exosome isolation. The primary method for isolating exosomes from this pre-cleared media is ultracentrifugation, typically at forces of 100,000g to 200,000g for 70-120 minutes [40] [25] [7]. Alternative methods such as tangential flow filtration (TFF) can also be applied, which has been shown to significantly improve exosome yield and biological activity when combined with 3D MSC cultures [40]. Furthermore, density gradient ultracentrifugation or size-exclusion chromatography can be used following the 100,000g pellet to further purify exosomes from protein aggregates or other co-isolated contaminants [39] [25].
The following diagram summarizes the position of this sample preparation protocol within the complete workflow of MSC exosome research, from cell culture to characterization.
Within the rapidly advancing field of mesenchymal stem cell (MSC) research, exosomes have emerged as critical mediators of therapeutic effects, offering a promising cell-free alternative for regenerative medicine and drug delivery [16] [41]. These nano-sized extracellular vesicles (30-150 nm in diameter) facilitate intercellular communication by transferring bioactive molecules—including proteins, lipids, and nucleic acids—from parent MSCs to recipient cells [42] [43]. The isolation of high-purity exosomes is therefore paramount for both research and clinical translation.
Among various isolation techniques, differential ultracentrifugation remains the most widely adopted "gold standard" method due to its cost-effectiveness, reproducibility, and absence of requirement for specialized reagents [42]. This protocol details the standardized application of differential ultracentrifugation for isolating MSC-derived exosomes, with specific parameters for high-speed pelleting (100,000-120,000g for 70-90 minutes) and subsequent pellet collection. The procedures outlined herein are designed to ensure the isolation of exosomes with optimal yield, purity, and biological integrity for downstream applications.
Principles of Ultracentrifugation
Ultracentrifugation separates nanoparticles based on their size, density, and shape through the application of high centrifugal forces. The fundamental principle is described by the relative centrifugal force (RCF) equation, which guides the sedimentation of particles [42]:
RCF = (1.118 × 10⁻⁵) × (RPM)² × r
Where RCF is the relative centrifugal force (expressed as × g), RPM is revolutions per minute, and r is the rotor radius in centimeters. Differential ultracentrifugation employs a series of progressively increasing centrifugal forces to sequentially eliminate larger particles, ultimately resulting in the pelleting of exosomes at ultrahigh speeds [42]. This step-wise approach ensures the effective removal of cellular debris and larger extracellular vesicles before exosome collection, thereby enhancing the purity of the final isolate.
Materials and Equipment
Research Reagent Solutions
Table 1: Essential materials and reagents for exosome isolation via ultracentrifugation.
| Category | Item | Specification/Function |
|---|---|---|
| Starting Material | MSC Conditioned Medium | Serum-free medium collected from MSC cultures (48-72 hour collection) [43] |
| Buffers | Phosphate-Buffered Saline (PBS) | Sterile, cold (4°C); for dilution and resuspension of exosome pellets |
| Protease Inhibitors | Protease Inhibitor Cocktail | Added to PBS to prevent protein degradation during isolation |
| Centrifugation Equipment | Ultracentrifuge | Capable of reaching 100,000-120,000g |
| Fixed-Angle Rotor | Typically Type 50.2 Ti or similar; suitable for high-speed pelleting | |
| Polycarbonate Bottles/Tubes | Compatible with ultracentrifuge and rotor | |
| Post-Isolation Analysis | Nanoparticle Tracking Analyzer | For determining exosome size distribution and concentration [44] [29] |
| Transmission Electron Microscope | For morphological characterization [44] [29] | |
| Western Blot Equipment | For detection of exosomal markers (CD9, CD63, CD81, ALIX, TSG101) [44] [29] |
Step-by-Step Protocol
Sample Preparation
Begin with conditioned medium collected from MSC cultures. Critical note: To eliminate bovine exosomes from fetal bovine serum (FBS), culture MSCs in serum-free medium for 24-48 hours prior to collection, or use FBS that has been ultracentrifuged (100,000g overnight) to deplete exogenous vesicles [45].
Differential Centrifugation Steps
Table 2: Step-wise centrifugation parameters for exosome isolation.
| Step | Centrifugation Force | Duration | Temperature | Purpose | Pellet Content |
|---|---|---|---|---|---|
| 1 | 300 × g | 10 min | 4°C | Pellet and remove cells | Cells |
| 2 | 2,000 × g | 20 min | 4°C | Remove dead cells and large debris | Cellular debris |
| 3 | 10,000 × g | 30-45 min | 4°C | Pellet larger extracellular vesicles (microvesicles) | Microvesicles, organelles |
| 4 | 100,000-120,000 × g | 70-90 min | 4°C | Pellet exosomes | Exosomes, small vesicles |
Between each step, carefully decant or pipette the supernatant without disturbing the pellet. The supernatant from step 3 serves as the input for the final ultracentrifugation step.
Pellet Collection and Washing
Following the final ultracentrifugation step, promptly decant the supernatant. To purify the exosomes from co-precipitated proteins, resuspend the pellet in a generous volume of cold, sterile PBS (e.g., 10-30 mL). Subsequently, subject the resuspended solution to a second round of ultracentrifugation under the same conditions (100,000-120,000g for 70-90 minutes) [42]. The final exosome pellet should be resuspended in a small volume of PBS or a specific buffer suitable for downstream applications (e.g., 50-200 µL). Gently pipette to avoid mechanical shearing and damaging the exosomes. Aliquot to prevent repeated freeze-thaw cycles and store at -80°C.
Yield and Purity Considerations
Ultracentrifugation, while standard, faces challenges including low exosome recovery rates (potentially as low as 30% due to pellet resuspension difficulties) and potential co-precipitation of non-exosomal materials like protein aggregates [42]. The yield of exosomes is highly dependent on the MSC source, culture conditions, and passage number. For instance, bone marrow MSC-derived small extracellular vesicles (sEVs) have reported average yields of approximately 3,751-4,319 particles per cell [29]. Implementing a three-dimensional (3D) culture system for MSCs, as opposed to traditional 2D flasks, has been shown to increase total exosome production by up to 19.4-fold [43].
Characterization and Validation
Post-isolation characterization is critical to confirm the identity, purity, and quality of the isolated exosomes. The following table summarizes the key validation methods and expected outcomes.
Table 3: Essential characterization techniques for validating isolated MSC exosomes.
| Method | Key Metrics | Expected Outcome for MSC Exosomes |
|---|---|---|
| Nanoparticle Tracking Analysis (NTA) | Size distribution, mode, concentration [44] [29] | Peak particle size: 30-150 nm [16] [43] |
| Transmission Electron Microscopy (TEM) | Morphology and structure [44] [29] | Cup-shaped, spherical morphology with intact lipid bilayer |
| Western Blotting | Detection of marker proteins [44] [29] | Positive for CD9, CD63, CD81, ALIX, TSG101 |
| Negative Control | Assessment of purity | Negative for Calnexin (endoplasmic reticulum marker) |
Troubleshooting and Technical Notes
- Low Yield: Ensure the ultracentrifuge rotor is properly calibrated. Consider increasing the starting volume of conditioned medium. Optimize MSC culture conditions (e.g., using 3D culture or specific media like α-MEM) to enhance sEV secretion [29] [43].
- Protein Contamination: Include a washing step with PBS and repeat ultracentrifugation. Ensure the supernatant is completely removed after the 10,000g spin.
- Poorly Formed Pellet: Avoid overfilling centrifuge tubes, which can lead to pellet dispersion. Always use balanced tubes to prevent rotor imbalance and failed runs.
- Exosome Damage: Resuspend the final pellet gently by pipetting; avoid vortexing. The use of a blunt-ended pipette tip can help minimize shear forces.
Alternative and Advanced Methods
While ultracentrifugation is foundational, several alternative methods offer different advantages. Tangential Flow Filtration (TFF) has demonstrated statistically higher particle yields compared to ultracentrifugation and is more scalable for clinical-grade production [29]. Density gradient centrifugation, a variant of ultracentrifugation, can achieve higher purity by separating particles based on buoyant density in a sucrose or iodixanol gradient, effectively reducing protein contamination [42]. Emerging technologies like microfluidic-based isolation (e.g., Biologically intact Exosome Separation Technology, BEST) show great promise for high-purity, low-damage isolation with potential for diagnostic applications [42].
Within the rapidly advancing field of mesenchymal stem cell (MSC) research, exosomes have emerged as pivotal mediators of therapeutic effects, offering a promising cell-free alternative for regenerative medicine and drug development [25] [11]. These nanoscale extracellular vesicles (30-150 nm) shuttle bioactive molecules—including proteins, lipids, and nucleic acids—from MSCs to recipient cells, modulating immune responses, promoting tissue repair, and influencing regenerative processes [11] [46]. The translational potential of MSC-derived exosomes, however, is critically dependent on the isolation method, which must ensure high yield, purity, and biological integrity [25] [47].
Differential ultracentrifugation (UC) has long been the gold standard for exosome isolation. Despite its widespread use, this method faces significant limitations, including low yield, prolonged processing time, potential disruption of exosome membrane integrity, and co-precipitation of protein contaminants [25] [48]. These impurities, such as aggregated proteins and lipoproteins, can confound downstream analytical results and functional assays [47].
The one-step sucrose cushion ultracentrifugation (SUC) method has been developed as a refined approach to overcome these challenges. By leveraging a density barrier, this technique effectively separates exosomes from contaminating proteins, thereby enhancing the purity and yield of isolates essential for both research and clinical applications [25]. This protocol details the application of the SUC method for isolating exosomes from MSC-conditioned media.
Quantitative Method Comparison
The selection of an exosome isolation method involves trade-offs between yield, purity, time, and cost. The table below summarizes a comparative analysis of common techniques, highlighting the performance of the one-step sucrose cushion method.
Table 1: Comparison of Exosome Isolation Methods for MSC Research
| Isolation Method | Estimated Yield | Relative Purity | Key Advantages | Key Limitations | Suitability for MSC Exosomes |
|---|---|---|---|---|---|
| One-Step Sucrose Cushion UC | High [25] | High [25] | Preserves exosome integrity and morphology; reduces protein contaminants [25] | Requires ultracentrifuge; slightly more complex than UC [25] | Excellent for high-quality, functional exosomes [25] |
| Differential UC (Gold Standard) | Low to Moderate [25] [47] | Moderate | Widely accepted; no specialized reagents needed [49] | Time-consuming; can damage exosomes; high protein contamination [25] [47] | Good, but risk of impaired biological activity [25] |
| Tangential Flow Filtration (TFF) | Very High (92.5x UC) [47] | High (when combined with dFBS) [47] | Scalable for large volumes; fast processing [47] [48] | Requires specialized TFF system [47] | Excellent for large-scale production [47] |
| Polyethylene Glycol (PEG) Precipitation | Moderate [50] | Low to Moderate | Simple protocol; no special equipment [49] [50] | Co-precipitates contaminants; difficult to re-suspend pellet [50] [48] | Moderate, purity is a concern for therapeutic use [50] |
| Commercial Kits | Moderate [49] | Moderate | User-friendly and time-saving [49] | Can be costly; may include kit-specific contaminants [49] | Moderate for quick, small-scale studies [49] |
Experimental Protocols
One-Step Sucrose Cushion Ultracentrifugation Protocol
This protocol is adapted from established methodologies for isolating exosomes from human MSC-conditioned media [25] [51].
A. Pre-processing of Conditioned Media
- Culture MSCs in serum-free media for 48 hours to avoid contaminating bovine exosomes from fetal bovine serum (FBS). Using exosome-depleted FBS is an alternative [25] [47].
- Harvest conditioned media and subject it to sequential centrifugation steps:
- Filter the supernatant through a 0.22 µm pore filter to remove larger particles [25] [51].
B. Sucrose Cushion Preparation
- Prepare a 30% (w/w) sucrose solution in Dulbecco's Phosphate Buffuffered Saline (PBS). For enhanced stability, the solution can be prepared in deuterium oxide (D2O) [25] [51].
- Carefully layer 4 mL of the 30% sucrose solution into the bottom of an ultracentrifuge tube (e.g., Beckman Coulter Polycarbonate bottle) [25].
C. Ultracentrifugation and Exosome Recovery
- Gently layer ~22.5 mL of the pre-cleared conditioned media on top of the sucrose cushion, avoiding mixing of the two phases. A glass pipette can be used to facilitate layering [51].
- Ultracentrifuge the prepared tubes at 100,000 × g for 90 minutes at 4°C using a swinging bucket rotor (e.g., Beckman Coulter SW 45 Ti) [25].
- After centrifugation, carefully aspirate and discard the upper media layer. The exosomes are located at the sucrose-media interface and within the sucrose layer.
- Collect the ~5 mL of the sucrose layer and transfer it to a new ultracentrifuge bottle.
- Add ~20 mL of PBS to dilute the sucrose and fill the bottle for a washing step.
- Ultracentrifuge the diluted exosome suspension at 100,000 × g for 90 minutes at 4°C to pellet the exosomes [25].
- Discard the supernatant and resuspend the final exosome pellet in 100-500 µL of PBS [25]. Aliquot and store at -80°C for long-term preservation.
Figure 1: Workflow for One-Step Sucrose Cushion Ultracentrifugation
Downstream Characterization of Isolated Exosomes
Rigorous characterization is essential to confirm the identity, purity, and quantity of isolated exosomes.
Table 2: Key Characterization Techniques for MSC Exosomes
| Technique | Parameter Measured | Expected Outcome for MSC Exosomes |
|---|---|---|
| Nanoparticle Tracking Analysis (NTA) | Particle size distribution and concentration [25] [47] | A peak particle size between 30-150 nm [25] [44] |
| Transmission Electron Microscopy (TEM) | Morphology and ultrastructure [25] [49] | Cup-shaped, spherical vesicles with double-membrane structure [25] [49] |
| Western Blotting | Presence of exosomal marker proteins [25] [44] | Positive for CD63, CD81, CD9, Alix, TSG101 [25] [44] [48]. Negative for Calnexin (a negative control marker) [44] |
| Flow Cytometry | Surface marker profiling [25] | Positive for MSC-related markers (e.g., CD73, CD90, CD105) and tetraspanins [25] [11] |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials and Reagents for Sucrose Cushion Ultracentrifugation
| Item | Function / Role | Example & Notes |
|---|---|---|
| Ultracentrifuge | High-speed centrifugation to pellet exosomes | Beckman Coulter Optima series with swinging bucket rotor (e.g., SW 45 Ti, SW 41 Ti) [25] [44] |
| Sucrose | Forms density cushion to separate exosomes from contaminants | Prepare 30% (w/w) solution in PBS or D2O; density matches exosomes, sparing them from pellet-induced damage [25] [51] |
| Exosome-depleted FBS | Cell culture supplement free of contaminating bovine exosomes | Prepared by ultracentrifuging standard FBS (100,000 × g, 18 hours) or purchased commercially [47] |
| Serum-free Media | For conditioning to collect MSC exosomes | STEMPRO MSC SFM CTS or phenol red-free DMEM [25] [47] |
| Specific Antibodies | Characterizing exosomal markers via Western Blot/Flow Cytometry | Anti-CD63, Anti-CD81, Anti-CD9, Anti-Alix [25] [44] |
| PBS (pH 7.4) | Washing, resuspension, and buffer preparation | Used for diluting sucrose and final exosome resuspension [25] |
The one-step sucrose cushion ultracentrifugation method represents a significant advancement in the isolation of MSC-derived exosomes. By mitigating the primary drawbacks of conventional ultracentrifugation—specifically low yield and poor purity—this technique provides researchers with exosome preparations of superior quality. The enhanced integrity and reduced contaminant profile of SUC-isolated exosomes make them particularly suitable for demanding downstream applications, including functional studies in disease models, drug delivery vector development, and ultimately, clinical translation in regenerative medicine.
Within a broader research thesis on the ultracentrifugation protocol for mesenchymal stem cell (MSC) exosomes, the steps following initial isolation are critical. Post-isolation processing—comprising washing, resuspension, and storage—is not merely a concluding procedure but a fundamental phase that determines the integrity, purity, and functional viability of exosome preparations for downstream applications and drug development. Proper execution ensures that the biological characteristics of these 30-150 nm extracellular vesicles are preserved, directly impacting the reproducibility and reliability of experimental data [52] [25].
This document provides detailed application notes and protocols for these crucial steps, framing them within the context of standardized MSC exosome research.
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table catalogues the essential materials required for the post-isolation processing of MSC-derived exosomes.
Table 1: Key Research Reagents and Materials for Post-Isolation Processing
| Item | Function & Application | Critical Notes |
|---|---|---|
| Phosphate-Buffered Saline (PBS) | Resuspension and washing buffer; provides a physiological, isotonic environment for exosomes. | Pre-cool to 4°C for optimal stability. Use sterile, particle-free PBS [25] [53]. |
| Ultra-Clear or Polyallomer Centrifuge Tubes | Specially designed tubes for ultracentrifugation; minimize tube wall adhesion and withstand extreme g-forces. | Compatible with swinging bucket rotors (e.g., Beckman Coulter SW series) [54]. |
| Sucrose (for Cushion Purification) | Forms a density barrier (e.g., 30% solution) to purify exosomes from protein contaminants during ultracentrifugation. | Preserves exosome integrity and significantly improves purity compared to direct pelleting [25]. |
| Cryogenic Vials | Long-term storage of exosome aliquots at -80°C. | Use low-protein-binding tubes to prevent adsorption losses. |
| Protease Inhibitor Cocktails | Added to PBS to prevent proteolytic degradation of exosomal cargo during processing and storage. | Crucial for downstream proteomic analyses [25]. |
| BCA or Bradford Protein Assay Kits | Standard method for quantifying exosome protein concentration after resuspension. | Ensures accurate dosing for functional experiments [53]. |
Data-driven decisions are paramount. The following table summarizes key quantitative findings related to exosome processing and storage.
Table 2: Comparative Data on Processing Methods and Storage Impact
| Parameter | Direct Ultracentrifugation (UC) | Sucrose Cushion Ultracentrifugation (SUC) | Notes & References |
|---|---|---|---|
| Relative Exosome Yield | Baseline | ~2.5-fold higher than UC | SUC method results in a significantly higher concentration of exosomes per mL [54]. |
| Exosome Integrity & Morphology | Cup-shaped morphology can be compromised; potential membrane disruption. | Better preservation of cup-shaped morphology; more homogenous population. | The cushioning effect protects vesicle structure [25]. |
| Protein Contaminants | Higher levels of non-exosomal protein contaminants (e.g., apoproteins). | Reduced protein contamination; higher purity. | Sucrose separates exosomes from higher-density contaminants [25]. |
| Optimal Storage Temperature | -80°C for long-term storage; -20°C is also suitable. | -80°C for long-term storage; -20°C is also suitable. | Size remains constant over long periods at -20°C; multiple freeze-thaw cycles should be avoided [52]. |
| Impact of Freeze-Thaw Cycles | Multiple cycles can affect integrity (aggregation, cargo leakage). | Multiple cycles can affect integrity (aggregation, cargo leakage). | Aliquot exosomes to avoid repeated freezing and thawing [52]. |
Detailed Experimental Protocols
Protocol 1: Washing and Resuspension of the Exosome Pellet
This protocol describes the steps immediately following the initial ultracentrifugation pelleting of exosomes from MSC-conditioned media.
Materials:
- Isolated exosome pellet from 100,000g ultracentrifugation
- Cold (4°C) sterile PBS
- Ultracentrifuge and appropriate rotor (e.g., SW60 Ti, Type 70.1)
- Polyallomer ultracentrifuge tubes
Method:
- Discard Supernatant: After the initial ultracentrifugation, carefully decant the supernatant without disturbing the often-invisible pellet at the bottom of the tube. A residual volume of 50-100 µL may be left to avoid losing the pellet.
- Primary Resuspension: Gently resuspend the exosome pellet in a large volume (e.g., 10-35 mL, depending on tube capacity) of cold, sterile PBS. Use pipetting with wide-bore tips or gentle vortexing to achieve a homogeneous suspension. Avoid generating foam.
- Washing Ultracentrifugation: Transfer the resuspended exosomes to a new ultracentrifuge tube. Centrifuge again at 100,000g for 60-90 minutes at 4°C to pellet the washed exosomes [25] [53].
- Final Resuspension: Carefully discard the PBS supernatant. Resuspend the final, washed exosome pellet in a small, defined volume of PBS (e.g., 50-200 µL) suitable for downstream applications. Keep samples on ice throughout.
Protocol 2: One-Step Sucrose Cushion Ultracentrifugation for High-Purity Isolation
This modified method integrates purification during isolation, yielding exosomes with higher purity and integrity, ideal for sensitive downstream assays.
Materials:
- Pre-processed MSC-conditioned media (cleared of cells and debris)
- 30% sucrose solution in D-PBS (density ~1.12-1.18 g/mL)
- Ultracentrifuge and swinging bucket rotor
Method:
- Prepare Sucrose Cushion: Pipette 4 mL of 30% sucrose solution into an ultracentrifuge tube.
- Layer Sample: Slowly and carefully layer the pre-cleared conditioned media on top of the sucrose cushion, avoiding mixing at the interface.
- Ultracentrifugation: Centrifuge the layered sample at 100,000g for 90 minutes at 4°C [25]. Exosomes will band at the sucrose-PBS interface, while denser contaminants will pellet.
- Collect Fraction: After centrifugation, discard the top media layer. Carefully collect the ~5 mL sucrose layer containing the exosomes using a pipette.
- Dilution and Washing: Transfer the sucrose-exosome mixture to a new ultracentrifuge tube, dilute with a large volume of PBS, and centrifuge again at 100,000g for 90 minutes at 4°C to pellet the purified exosomes away from the sucrose.
- Final Resuspension: Discard the supernatant and resuspend the pure exosome pellet in a desired volume of PBS for immediate use or storage [25].
Protocol 3: Concentration Measurement, Aliquoting, and Storage at -80°C
Standardized protocols for quantification and storage are vital for experimental consistency.
Materials:
- Resuspended exosomes in PBS
- Bicinchoninic Acid (BCA) Protein Assay Kit
- Cryogenic vials
- -80°C freezer
Method:
- Protein Quantification:
- Lyse an aliquot of the resuspended exosomes (e.g., 5-10 µL) using a RIPA buffer supplemented with protease inhibitors [25].
- Perform a BCA protein assay according to the manufacturer's instructions to determine the exosome protein concentration. This serves as a proxy for exosome quantity.
- Aliquoting:
- Based on the quantification, aliquot the exosome suspension into single-use cryogenic vials to avoid repeated freeze-thaw cycles. Typical aliquots might contain 10-50 µg of exosomal protein.
- Storage:
Workflow and Pathway Visualizations
Post-Isolation Processing Workflow
The following diagram illustrates the logical workflow and decision points for processing exosomes after initial isolation.
Impact of MSC-Exos on Bone Regeneration Signaling Pathways
MSC-derived exosomes (MSC-Exos) exert their therapeutic effects by modulating key signaling pathways in recipient cells. The following diagram summarizes their role in bone regeneration, a key therapeutic area.
The transition of mesenchymal stem cell (MSC) exosome research from fundamental science to clinical applications necessitates a critical evaluation of isolation methodologies. Ultracentrifugation (UC), long considered the gold standard in research settings, faces significant challenges when scaled for industrial or clinical production, including exosome damage, low yield, and high time investment [55] [42]. This application note delineates the core considerations for adapting MSC exosome ultracentrifugation protocols across the development pipeline, providing a structured comparison of scalable alternatives to guide researchers and development professionals in method selection for their specific production stage.
Quantitative Comparison of Exosome Isolation Methods
The selection of an isolation method profoundly impacts the yield, purity, and biological functionality of the resulting exosomes, with clear trade-offs emerging between small-scale research and large-scale production needs. The table below summarizes key performance metrics for common isolation techniques.
Table 1: Performance Metrics of Exosome Isolation Methods for Scaling Production
| Method | Typical Yield | Purity | Scalability | Processing Time | Relative Cost | Key Applications |
|---|---|---|---|---|---|---|
| Differential Ultracentrifugation (UC) | Medium | High | Medium | Long (>4 hours) | Medium | Small-scale research, biomarker discovery [23] [42] |
| Sucrose Cushion UC | High | Very High | Low-Medium | Very Long (>90 mins) | Medium | High-purity research, functional studies requiring intact exosomes [55] |
| Size-Exclusion Chromatography (SEC) | Medium | Medium-High | High | Medium (~1 hour) | Medium-High | Mid-to-large scale production, high-purity requirements [23] [56] |
| Tangential Flow Filtration (TFF) | High | Medium | Very High | Short-Medium | High | Large-scale and industrial production [23] [42] |
| Polymer-Based Precipitation | Very High | Low | High | Short | Low | Rapid concentration, diagnostic assays where purity is secondary [23] [56] |
| Ion-Exchange Chromatography (IEC) | High (48.5% recovery) | High | High | Varies | Medium-High | Large-scale therapeutic preparation [57] |
Detailed Experimental Protocols for Scalable Production
Small-Scale Research: One-Step Sucrose Cushion Ultracentrifugation
This protocol is optimized for bench-scale research where maximizing exosome integrity and purity from small volumes of conditioned media is paramount [55].
Step 1: Cell Culture and Supernatant Collection
- Culture MSCs in serum-free media for 48 hours to eliminate contaminating bovine exosomes.
- Collect the conditioned media and perform initial clarifications:
- Centrifuge at 300 × g for 10 minutes to pellet cells.
- Transfer supernatant and centrifuge at 10,000 × g for 30 minutes to remove cell debris and microvesicles.
Step 2: Sucrose Cushion Ultracentrifugation
- Carefully layer the pre-cleared supernatant on top of a 4 mL cushion of 30% sucrose solution in an ultracentrifuge tube.
- Ultracentrifuge at 100,000 × g for 90 minutes at 4°C. Exosomes will form a pellet at the bottom of the tube.
- Discard the supernatant and resuspend the exosome pellet in phosphate-buffered saline (PBS).
Step 3: Washing and Storage
- Resuspend the pelleted exosomes in a large volume of PBS and ultracentrifuge again at 100,000 × g for 90 minutes to wash.
- Finally, resuspend the purified exosome pellet in 100–500 µL of PBS and store at -80°C.
Pilot to Large-Scale Production: Ion-Exchange Chromatography (IEC)
For scaling up exosome production, Ion-Exchange Chromatography (IEC) offers a robust and efficient method, leveraging the negative surface charge of exosomes [57].
Step 1: Sample Preparation
- Clarify large volumes (e.g., 750+ mL) of MSC-conditioned media by sequential centrifugation (2000 × g for 10 minutes) and filtration through a 0.45 µm membrane.
Step 2: Chromatography Setup
- Equilibrate an anion-exchange column (e.g., Source-30Q) with wash buffer (e.g., 100 mM Tris-HCl, 10 mM EDTA, 0.4 M NaCl, pH 7.5).
- Load the clarified supernatant onto the column at a controlled flow rate (e.g., 30 mL/min). Negatively charged exosomes bind to the column resin.
Step 3: Washing and Elution
- Wash the column with 1.5 column volumes (CV) of wash buffer to remove unbound protein contaminants.
- Elute the purified exosomes using an elution buffer with a higher salt concentration (e.g., 100 mM Tris-HCl, 10 mM EDTA, 1 M NaCl, pH 7.5).
- Collect the eluent based on UV absorbance at 280 nm.
Step 4: Concentration and Storage
- The eluted exosomes can be concentrated using ultrafiltration if necessary.
- Aliquot and store at -80°C.
Integrated Hybrid Protocol: Precipitation-Ultrafiltration
A 2025 study introduced a hybrid method combining chemical precipitation with ultrafiltration (CPF), designed for efficiency and high purity across different biofluids, suitable for translational research [56].
Step 1: Precipitation
- Mix the conditioned media or biofluid with a polyethylene glycol (PEG)-based precipitation reagent.
- Incubate the mixture overnight at 4°C to precipitate the exosomes.
Step 2: Low-Speed Centrifugation
- Centrifuge the sample at ≥10,000 × g for 30-60 minutes at 4°C to pellet the precipitated exosomes.
- Resuspend the pellet in PBS.
Step 3: Two-Step Filtration
- Pass the resuspended pellet through a 0.22 µm syringe filter to remove large aggregates.
- Use ultrafiltration devices (e.g., with a 100 kDa molecular weight cut-off) to concentrate and further purify the exosomes, exchanging the buffer to PBS.
Step 4: Characterization
- The resulting exosomes are suitable for downstream multi-omics analyses and functional assays [56].
The Scientist's Toolkit: Essential Reagents and Materials
Successful scaling of exosome production relies on a core set of reagents and instruments. The following table details essential items and their functions in the isolation workflow.
Table 2: Key Research Reagent Solutions for Exosome Isolation
| Item | Function/Application | Example Specifications |
|---|---|---|
| Serum-Free MSC Media | Cell culture for exosome collection without bovine exosome contamination | e.g., STEMPRO MSC SFM CTS [55] |
| Sucrose Cushion Solution | Density barrier for high-purity exosome isolation via UC | 30% sucrose in PBS, density ~1.12-1.18 g/mL [55] |
| Chromatography Resin | Large-scale purification based on surface charge | Anion-exchange resin (e.g., Source-30Q) [57] |
| Ultrafiltration Devices | Concentration and buffer exchange of exosome preparations | 100-500 kDa MWCO, Tangential Flow Filtration systems [56] [42] |
| PEG-Based Precipitation Kit | Rapid, high-yield exosome precipitation from large volumes | Commercial kits (e.g., PEG 6000-8000) [56] |
| PBS Buffer | Washing, resuspension, and storage of isolated exosomes | 1X, sterile, pH 7.4 |
| Protease Inhibitors | Prevention of proteolytic degradation during isolation | Added to lysis buffers for downstream analysis [55] |
Transitioning MSC exosome isolation from small-scale research to large-scale production requires a strategic shift in methodology. While ultracentrifugation with a sucrose cushion remains a powerful tool for achieving high-purity exosomes for foundational research, its scalability is limited. For larger volumes, methods such as Ion-Exchange Chromatography and integrated approaches like precipitation-ultrafiltration offer superior yields, better preservation of biological activity, and greater process efficiency. The optimal protocol is determined by the specific balance of priorities among yield, purity, scalability, and intended application, whether for discovery research or clinical development.
Troubleshooting Common Ultracentrifugation Challenges and Yield Optimization Strategies
Exosomes derived from Mesenchymal Stem Cells (MSCs) are at the forefront of regenerative medicine and drug delivery research due to their immunomodulatory capabilities, tissue repair properties, and high biosafety profile [58] [59]. A critical bottleneck in harnessing this potential for preclinical and clinical applications is the consistent production of high-quality exosomes in sufficient quantities. The isolation process is a key determinant of final yield and purity. Among the various isolation techniques, differential ultracentrifugation (dUC) remains the most widely used "gold standard" method [54] [23] [59]. However, standard dUC protocols often result in low yields, which can impede research progress and translational applications.
This Application Note provides a detailed framework for optimizing ultracentrifugation parameters—specifically centrifugal force, run duration, and rotor selection—to maximize exosome yield from MSC cultures without compromising vesicle integrity. The protocols and data presented herein are designed to be integrated into a broader thesis on standardizing ultracentrifugation protocols for MSC exosome research, providing researchers with actionable strategies to overcome the yield challenge.
Core Principles and Challenges in Ultracentrifugation
The fundamental principle of differential ultracentrifugation is the sequential application of increasing centrifugal forces to pellet particles of decreasing size and density. The typical workflow for isolating MSC exosomes begins with low-speed spins to remove cells and apoptotic debris, followed by intermediate-speed spins to pellet larger microvesicles, and culminates in a high-speed ultracentrifugation step to pellet the exosomes themselves [54] [59].
The primary challenges leading to low yield in standard protocols include:
- Inefficient Pelleting: The use of insufficient g-force or duration can leave a significant portion of exosomes in the supernatant [59].
- Vesicle Damage: Excessive g-forces or prolonged centrifugation can cause exosome deformation, aggregation, and even rupture, rendering them non-functional and reducing the measurable yield [59].
- Rotor Geometry: The efficiency of pelleting is highly dependent on the k-factor (clearing factor) of the rotor, which describes its efficiency at a given maximum speed. Rotors with lower k-factors are more efficient, requiring shorter run times to achieve the same sedimentation [54] [60].
- Instrument Calibration: Inaccurate calibration of ultracentrifuge temperature, radial distance, and timing can lead to significant inconsistencies and suboptimal yields [60].
Optimization of Critical Ultracentrifugation Parameters
To address the challenge of low yield, a systematic optimization of key ultracentrifugation parameters is essential. The following section provides optimized protocols and quantitative guidance.
Optimized Differential Ultracentrifugation Protocol
The following protocol is optimized for isolating exosomes from conditioned media of MSC cultures.
Pre-Analytical Processing (Critical for Yield):
- Cell Culture: Culture MSCs to 70-80% confluence. Replace growth medium with a serum-free, exosome-depleted medium [54] [59].
- Conditioned Media Collection: Collect conditioned media after 24-48 hours. Process immediately or store at -80°C to prevent degradation.
- Initial Clarification:
- Centrifuge at 300 × g for 10 min at 4°C to pellet cells.
- Transfer supernatant to a new tube and centrifuge at 2,000 × g for 20 min at 4°C to remove dead cells.
- Transfer supernatant and centrifuge at 10,000 × g for 30 min at 4°C to pellet larger microvesicles and debris. The resulting supernatant contains exosomes and soluble proteins [54] [59].
Ultracentrifugation for Exosome Pelleting:
- Transfer: Transfer the 10,000 × g supernatant to ultracentrifuge tubes (e.g., Polyallomer conical tubes for a SW60 Ti rotor). Balance tubes meticulously with PBS.
- High-Speed Spin: Ultracentrifuge at ≥100,000 × g for 70-90 min at 4°C. Note: The optimal duration within this range depends on the rotor's k-factor (see Table 2) [54] [59].
- Wash (Optional but Recommended): Resuspend the crude exosome pellet in a large volume of sterile, cold PBS. Perform a second ultracentrifugation step under the same conditions to improve purity by removing co-pelleted protein contaminants.
- Resuspension: Carefully decant the supernatant. Resuspend the final, often invisible, exosome pellet in 50-200 µL of PBS or a suitable buffer. Gently pipette up and down and incubate on ice for 10-20 minutes to facilitate complete resuspension.
Table 1: Summary of Optimized Ultracentrifugation Protocol Steps
| Step | Centrifugal Force | Duration | Temperature | Target |
|---|---|---|---|---|
| Cell Pelletting | 300 × g | 10 min | 4°C | Intact cells |
| Debris Removal | 2,000 × g | 20 min | 4°C | Dead cells, debris |
| Microvesicle Removal | 10,000 × g | 30 min | 4°C | Microvesicles, debris |
| Exosome Isolation | ≥100,000 × g | 70-90 min | 4°C | Exosomes |
| Wash Step | ≥100,000 × g | 70-90 min | 4°C | Contaminating proteins |
Parameter Optimization: Force, Duration, and Rotor Selection
The force and duration of the final ultracentrifugation step are interdependent and must be optimized relative to the rotor used. The k-factor is a critical value for calculating the required run time.
Table 2: Ultracentrifugation Parameters for Different Rotors
| Rotor Type | Tube Volume | Max RCF (× g) | k-Factor | Recommended Duration for >90% Yield | Key Considerations |
|---|---|---|---|---|---|
| Fixed-Angle (e.g., Type 70.1 Ti) | 12-38 mL | 100,000+ | ~42 | ~180 min | High capacity, lower efficiency, pellet on side of tube. |
| Swinging-Bucket (e.g., SW 60 Ti) | 4-5 mL | 100,000+ | ~31 | ~90 min | Higher efficiency, pellet at bottom, ideal for small volumes [54]. |
| Swinging-Bucket (e.g., SW 32 Ti) | 38 mL | 100,000+ | ~250 | ~10 hours | High volume capacity, but very long run times. |
The required run time (t) for a specific rotor to pellet a particle of a certain size can be calculated using the k-factor and the sedimentation coefficient (s) of the particle of interest, using the formula: t (in hours) = k / s
For exosomes, with a typical sedimentation coefficient of approximately 50-200 Svedberg (S), a rotor with a lower k-factor will always achieve pelleting in a shorter time, thereby reducing the time exosomes are subjected to high g-forces and potentially increasing functional yield [54] [60].
Yield vs. Purity Trade-Off and Alternative Methods
It is crucial to recognize that ultracentrifugation optimization often involves a trade-off between yield and purity. While the parameters above aim to maximize yield, they may also co-pellet non-exosomal structures like protein aggregates and lipoproteins [54] [23]. For applications requiring very high purity, a density gradient centrifugation step can be incorporated following the initial dUC. This method separates vesicles based on their buoyant density, effectively isolating exosomes from most contaminants, though it often results in a lower final yield [59].
Table 3: Comparison of Ultracentrifugation with Other Common Isolation Methods
| Isolation Method | Purity | Yield | Scalability | Impact on Exosome Integrity |
|---|---|---|---|---|
| Differential Ultracentrifugation | High | Medium | Medium | Risk of damage/aggregation from high forces [59]. |
| Density Gradient Centrifugation | Very High | Low | Low | Maintains integrity; complex and time-consuming [59]. |
| Size-Exclusion Chromatography | Medium-High | Medium | High | Gentle process; maintains vesicle structure and function [23] [59]. |
| Precipitation | Low | High | High | Can co-precipitate contaminants; may affect downstream applications [54] [23]. |
| Ultrafiltration | Medium | High | High | Shear stress may damage exosomes [59]. |
The Scientist's Toolkit: Essential Reagents and Materials
The following table lists key reagents and materials required for the optimized ultracentrifugation protocol described in this note.
Table 4: Research Reagent Solutions for MSC Exosome Isolation
| Item | Function/Description | Example |
|---|---|---|
| Serum-Free, Exosome-Depleted Media | Cell culture medium for producing conditioned media free of exogenous vesicles. | Commercial exosome-depleted FBS or defined serum-free media. |
| Phosphate-Buffered Saline (PBS) | Washing cells, balancing ultracentrifuge tubes, and resuspending final exosome pellets. | Sterile, cold 1X PBS, pH 7.4. |
| Polyallomer Ultracentrifuge Tubes | Tubes designed to withstand extreme centrifugal forces without cracking. | Beckman Coulter Polyallomer tubes (e.g., for SW 60 Ti rotor). |
| Ultracentrifuge and Rotors | Instrumentation for achieving forces >100,000 × g. Swinging-bucket rotors are preferred for efficiency. | Beckman Coulter Optima XPN with SW 60 Ti or Type 70.1 Ti rotors. |
| Protease Inhibitor Cocktails | Added to PBS or conditioned media to prevent proteolytic degradation of exosomal cargo. | Commercial tablets or liquid cocktails. |
| Cryogenic Vials | For long-term storage of isolated exosomes at -80°C. | Sterile, internal-threaded vials. |
Workflow and Quality Control
A successful isolation strategy involves not just the run itself, but also careful pre-processing and post-isolation analysis. The following diagram and section outline the complete workflow and critical quality control checks.
MSC Exosome Isolation and QC Workflow
Post-Isolation Quality Control
Following isolation, it is imperative to characterize the exosome preparation to confirm yield, size, and purity.
- Nanoparticle Tracking Analysis (NTA): This is the preferred method for determining particle concentration (yield) and size distribution. It confirms the presence of vesicles in the expected 30-200 nm range [54] [23].
- Transmission Electron Microscopy (TEM): Provides visual confirmation of exosome morphology (cup-shaped morphology under vacuum) and membrane integrity [61].
- Western Blotting: Used to assess purity by detecting positive protein markers (e.g., CD63, CD81, TSG101, Alix) and the absence of negative markers (e.g., GM130, Calnexin) which indicate cellular contamination [61] [62].
Optimizing ultracentrifugation protocols is a critical step in overcoming the significant challenge of low exosome yield in MSC research. By moving beyond a one-size-fits-all approach and carefully considering the interplay between centrifugal force, run duration calculated via k-factor, and rotor geometry, researchers can significantly improve their isolation efficiency. The protocols and data summarized in this Application Note provide a robust foundation for enhancing the yield and quality of MSC-derived exosomes, thereby accelerating their path from basic research to clinical application.
The isolation of pure, functional exosomes from mesenchymal stem cell (MSC) conditioned media is a critical step in downstream therapeutic applications and mechanistic studies. Differential ultracentrifugation, while widely used, often co-precipitates significant protein contaminants that compromise exosomal integrity and biological activity. This application note details a refined one-step sucrose cushion ultracentrifugation protocol that effectively minimizes protein contamination. We provide a comprehensive comparison of this method against direct ultracentrifugation, supported by quantitative data on yield, purity, and integrity, along with a detailed workflow to guide researchers in achieving highly pure MSC-derived exosomes.
Mesenchymal stem cell (MSC)-derived exosomes are nanoscale extracellular vesicles (30-150 nm) that mediate the paracrine effects of their parent cells, holding immense promise for cell-free regenerative therapy, immunomodulation, and drug delivery [55] [7]. The isolation of these vesicles with high purity is paramount for accurately delineating their biological functions and for developing safe, effective therapeutics.
Differential ultracentrifugation (UC) remains the most common isolation technique due to its cost-effectiveness and handling of large volumes [55]. However, a significant drawback of UC is the co-isolation of non-exosomal proteins and aggregates, which sediment at high gravitational forces [55] [35]. These contaminants can skew proteomic analyses, impede functional characterization, and potentially trigger unintended immune responses in therapeutic contexts.
The one-step sucrose cushion ultracentrifugation (SUC) method addresses this fundamental issue. By leveraging the density differential between exosomes (1.15-1.19 g/mL) and contaminating proteins (density ~1.22 g/mL), the sucrose cushion acts as a barrier that allows exosomes to pellet while retaining lighter contaminants in the supernatant [55] [25]. This technical note directly compares the UC and SUC methods, providing a validated protocol to enhance the purity and quality of isolated MSC-exosomes.
Comparative Analysis of Isolation Methods
Quantitative Comparison of Yield and Purity
The following table summarizes key performance metrics for the direct ultracentrifugation (UC) and one-step sucrose cushion (SUC) methods, based on data from MSC-conditioned media [55] [25].
Table 1: Comparative Analysis of Exosome Isolation Methods
| Parameter | Direct Ultracentrifugation (UC) | One-Step Sucrose Cushion (SUC) |
|---|---|---|
| Principle | Sequential centrifugation based on size and density | Density barrier separation using 30% sucrose |
| Reported Particle Yield | Lower | Comparatively higher [55] |
| Protein Contamination | Higher levels of co-pelleted contaminants | Significantly reduced [55] |
| Exosome Integrity | Risk of damage and aggregation due to hard pellet | Better preservation of vesicle integrity and cup-shaped morphology [55] [25] |
| Operational Time | Standard protocol duration | Similar to UC, with minimal time added for cushion preparation |
| Best Suited For | Initial, rapid concentration of vesicles | Applications requiring high-purity exosomes (e.g., proteomics, functional studies, therapeutics) |
Key Findings from Experimental Data
- Enhanced Morphology and Yield: Nanoparticle tracking analysis (NTA) and electron microscopy confirm that the SUC method yields a greater number of exosomes with a homogenous population and preserved cup-shaped morphology compared to UC [55] [25].
- Reduced Contaminants: Western blot analysis demonstrates that exosomes isolated via SUC exhibit stronger signals for canonical exosome markers (CD63, Alix) and reduced presence of contaminating proteins like GAPDH, indicating superior purity [55].
- Mechanism of Purity: The 30% sucrose solution (density ~1.12-1.18 g/mL) creates an optimal density cushion. During ultracentrifugation, exosomes pass through and pellet, while less dense protein aggregates and other contaminants are retained above or within the sucrose layer, preventing them from reaching the pellet [55].
Experimental Protocol: One-Step Sucrose Cushion Ultracentrifugation
This protocol is optimized for the isolation of exosomes from human MSC-conditioned serum-free media.
Materials and Reagents
Table 2: Research Reagent Solutions for Sucrose Cushion Ultracentrifugation
| Reagent/Equipment | Specification/Function |
|---|---|
| Sucrose Solution | 30% (w/v) sucrose in 1x PBS. Creates the density cushion for separation. Must be filtered (0.22 µm) before use. |
| Phosphate-Buffered Saline (PBS) | 1x, cold (4°C). Used for preparing sucrose solution and washing exosome pellets. |
| Ultracentrifuge | Equipped with a swinging bucket rotor (e.g., Sorvall WX+ series, Beckman Coulter SW series). |
| Polyallomer Centrifuge Tubes | For swinging bucket rotors (e.g., Beckman Coulter, ref 335650). Compatible with high g-forces. |
| Serum-Free Cell Culture Media | For conditioning MSCs (e.g., STEMPRO MSC SFM CTS). Essential to avoid fetal bovine serum exosome contamination. |
| Filtration Devices | 0.22 µm filters for sterilizing solutions and clarifying conditioned media. |
Step-by-Step Procedure
Step 1: Preparation of Conditioned Media
- Culture MSCs to 70-80% confluence in standard growth medium.
- Replace the medium with serum-free media and culture for 48 hours.
- Collect the conditioned media and perform preliminary clarifications:
Step 2: One-Step Sucrose Cushion Ultracentrifugation
- Prepare the sucrose cushion by pipetting 4 mL of 30% sucrose solution into a polyallomer ultracentrifuge tube.
- Carefully layer the pre-cleared conditioned media on top of the sucrose cushion. Avoid mixing the two layers.
- Load the tubes into the rotor and centrifuge at 100,000 × g for 90 min at 4°C [55] [25].
- After centrifugation, carefully aspirate and discard the supernatant, which contains soluble proteins and contaminants.
- Collect the sucrose layer (~5 mL), which now contains the exosomes, and transfer it to a new ultracentrifuge tube.
Step 3: Washing and Final Pellet Recovery
- Add a generous volume (e.g., 35-40 mL) of cold 1x PBS to the collected sucrose layer and mix thoroughly. This step dilutes the sucrose for effective pelleting.
- Centrifuge the mixture at 100,000 × g for 90 min at 4°C to pellet the exosomes.
- Completely decant the supernatant. The exosome pellet may be loose and translucent; exercise caution to not dislodge it.
- Resuspend the final, purified exosome pellet in 50-500 µL of 1x PBS.
- Aliquot and store at -80°C for downstream applications.
The following workflow diagram illustrates the key procedural steps and their logical progression.
Diagram 1: SUC Method Workflow
Downstream Characterization and Quality Control
To validate the success of the isolation, employ the following characterization techniques:
- Nanoparticle Tracking Analysis (NTA): Determines the particle size distribution and concentration. Expect a primary peak in the 30-150 nm range [55] [44].
- Transmission Electron Microscopy (TEM): Visualizes the morphology of exosomes. Vesicles isolated via SUC typically exhibit a well-preserved cup-shaped morphology [55] [25].
- Western Blotting: Confirms the presence of exosome-enriched marker proteins (e.g., CD63, CD9, CD81, Alix, TSG101) and the absence of negative markers (e.g., Calnexin, GM130) to assess purity [55] [44].
The one-step sucrose cushion ultracentrifugation method provides a significant advancement over direct ultracentrifugation for isolating MSC-derived exosomes. By effectively minimizing protein contamination and preserving vesicle integrity, this protocol ensures the recovery of high-purity exosomes suitable for demanding downstream applications in research and drug development. Its reproducibility and cost-effectiveness make it an ideal candidate for establishing a standard operating procedure in the field of extracellular vesicle research.
Exosomes, small extracellular vesicles ranging from 30-150 nm in diameter, are indispensable mediators of intercellular communication, carrying a complex cargo of proteins, nucleic acids, and lipids that reflect the physiological state of their parent cells [23] [63]. In Mesenchymal Stem Cell (MSC) research, the diagnostic and therapeutic potential of exosomes hinges entirely on preserving their structural and functional integrity throughout isolation and processing. These nanovesicles mirror the molecular composition of MSCs, making them invaluable for biomarker discovery and regenerative medicine applications [23]. However, their integrity is notoriously fragile, susceptible to both disruptive forces that compromise membrane architecture and aggregation phenomena that reduce bioavailability and alter experimental outcomes.
The primary challenges in maintaining exosome integrity stem from the inherent tension between isolation efficiency and preservation of native vesicle properties. Vesicle disruption typically occurs through mechanical shear forces, osmotic stress, or improper handling, leading to loss of cargo and biological activity. Conversely, aggregation—often induced by freezing or chemical precipitants—diminishes effective concentration, hampers cellular uptake, and introduces artifacts in downstream analysis [64]. For MSC-derived exosomes intended for therapeutic delivery, both extremes fundamentally undermine their functional capacity, whether through compromised payload delivery or reduced target engagement. This Application Note establishes robust, standardized protocols within the context of ultracentrifugation-based workflows to navigate these challenges, ensuring the recovery of intact, biologically functional exosomes for research and development.
Quantitative Comparison of Exosome Isolation Methods
Selecting an appropriate isolation strategy requires careful consideration of how each method balances yield, purity, and most critically, the preservation of exosome integrity. The following table summarizes the performance characteristics of common techniques, with particular emphasis on factors affecting structural preservation.
Table 1: Performance Metrics of Common Exosome Isolation Methods
| Method | Purity | Yield | Impact on Integrity | Scalability | Processing Time |
|---|---|---|---|---|---|
| Differential Ultracentrifugation | High [23] | Medium [23] | High shear forces can cause deformation and damage; aggregation upon resuspension is common [63] [37]. | Medium [23] | High (3-6 hours) [14] [54] |
| Density Gradient UC | Very High [42] | Low to Medium | Superior purity and reduced shear stress minimize aggregation, better preserving morphology [63] [42]. | Low | Very High (>12 hours) [42] |
| Size-Exclusion Chromatography (SEC) | Medium–High [23] | Medium [23] | Gentle separation maintains structural integrity and biological activity; low co-precipitation of contaminants [14] [63]. | High [23] | Low (~20 minutes) [14] |
| Polymer-Based Precipitation | Low [23] | High [23] [54] | Induces significant aggregation and can co-precipitate non-vesicular contaminants, impacting downstream functionality [23] [65]. | High [23] | Low [54] |
| Tangential Flow Filtration (TFF) | Medium [23] | High [23] | Membrane fouling and shear stress at high flow rates can potentially damage vesicles [23] [42]. | High [23] | Medium |
| Immunoaffinity Capture | Very High [23] | Low [23] | Gentle binding preserves integrity but elution conditions (low pH) may compromise vesicle stability and function [23] [63]. | Low [23] | Medium |
As the prevailing "gold standard," differential ultracentrifugation can generate high-purity exosome preparations. However, the high g-forces (100,000–150,000 × g) and prolonged run times are major contributors to vesicle damage and aggregation. Studies demonstrate that the pelleting and subsequent resuspension of the final exosome pellet are critical points where integrity is lost [63] [37]. In contrast, techniques like SEC effectively separate exosomes from soluble proteins and lipoproteins with minimal applied force, resulting in superior structural preservation and functionality, albeit with a trade-off in moderate yield and potential dilution of the sample [14] [63]. Precipitation methods, while offering high yield and convenience, introduce significant aggregation and impurity issues, making them suboptimal for applications requiring pristine vesicle quality [23] [65].
Ultracentrifugation Protocol for MSC Exosomes with Integrity Focus
The following optimized protocol for differential ultracentrifugation incorporates specific steps to minimize disruption and aggregation during the isolation of exosomes from MSC-conditioned media.
Pre-Analytical Phase: MSC Culture and Sample Preparation
- MSC Culture and Conditioning: Culture MSCs to 70-80% confluence in standard media. Replace with exosome-depleted serum media for 48 hours. Collect conditioned media and process immediately or store at 4°C for a maximum of 48 hours to prevent degradation and pre-isolation vesicle aggregation.
- Initial Clarification: Centrifuge the conditioned media at 300 × g for 10 minutes at 4°C to sediment live cells. Transfer the supernatant to a new tube and centrifuge at 2,000 × g for 20 minutes at 4°C to remove dead cells and large debris [54] [66]. For enhanced purity, filter the supernatant sequentially through 0.45 µm and 0.22 µm syringe filters [14]. Critical Note: Avoid allowing samples to warm to room temperature, as this can promote proteolytic degradation.
Ultracentrifugation and Post-Processing
- Exosome Pelletization: Transfer the clarified supernatant to ultracentrifuge tubes, balancing them with high precision. Centrifuge at 100,000 × g for 70 minutes at 4°C using a fixed-angle rotor [54] [66]. Integrity-Preserving Tip: Use a slow acceleration ramp and, crucially, turn off the brake for deceleration to prevent disturbing the soft pellet.
- Pellet Resuspension and Washing: Following ultracentrifugation, carefully decant the supernatant. To mitigate aggregation, resuspend the pellet not in a large volume, but in a small, precise amount of a compatible buffer such as phosphate-buffered saline (PBS) or 0.25 M sucrose. Use a wide-bore pipette tip and gentle pipetting—avoiding vortexing—to resuspend. For a washing step, dilute the resuspended exosomes in a larger volume of PBS and repeat the 100,000 × g centrifugation for 70 minutes [63]. Finally, resuspend the final pellet in 50-100 µL of PBS or a specific storage buffer.
Table 2: The Scientist's Toolkit: Essential Reagents for Ultracentrifugation Protocol
| Item | Function/Application | Integrity Consideration |
|---|---|---|
| Ultracentrifuge with Swinging-Bucket or Fixed-Angle Rotor | High-speed pelleting of exosomes. | Swinging-bucket rotors offer a consistent sedimentation path, while fixed-angle rotors require protocol adjustments to avoid forced pelleting and damage [37]. |
| Polyallomer Centrifuge Tubes | Withstand high g-forces; chemically inert. | Preferred over polycarbonate for reduced vesicle adhesion to tube walls, thereby improving yield and minimizing shear during resuspension. |
| Wide-Bore Pipette Tips | Resuspending the final exosome pellet. | Critical for reducing shear forces that can disrupt vesicle membranes during pipetting. |
| PBS (pH 7.4) or Sucrose Buffer | Washing and resuspending exosomes. | Isotonic buffers prevent osmotic shock. Sucrose can provide a more stabilizing environment for long-term stability. |
| Protease/Phosphatase Inhibitors | Added to collection and resuspension buffers. | Preserves the protein and phosphoprotein cargo of exosomes by inhibiting degradation. |
| 0.22 µm PES Syringe Filters | Pre-filtration of clarified media. | Removes residual debris without excessive protein binding, which can reduce yield. |
Managing and Reversing Exosome Aggregation
Aggregation is a pervasive issue that compromises the accurate quantification, functionality, and dosing of exosome preparations. It commonly occurs during freeze-thaw cycles and in pellets from ultracentrifugation or precipitation.
- Prevention Strategies: For long-term storage, aliquot exosomes into single-use volumes to avoid repeated freeze-thaw cycles. While -80°C is standard, the addition of cryoprotectants (e.g., 1-5% trehalose) to the suspension buffer should be empirically tested for specific MSC-exosome preparations [64].
- Dispersion of Pre-Existing Aggregates: For aggregated samples, gentle water-bath sonication is an effective dispersion method. Apply sonication at a low power (e.g., 40 kHz, 100 W) for 15 minutes. This treatment has been shown to significantly increase measurable particle concentration and reduce the proportion of large aggregates without damaging vesicle structure, ultimately restoring cellular uptake efficiency [64]. Note: Routine pipetting is ineffective at dispersing aggregates and can even promote re-aggregation after sonication [64].
Workflow and Integrity Checkpoints
The following diagram illustrates the complete experimental workflow, highlighting the critical points where exosome integrity is most vulnerable and the corresponding preservation actions required.
The translational promise of MSC-derived exosomes in drug development and regenerative medicine is critically dependent on the quality of the isolated vesicles. Adherence to integrity-focused protocols during ultracentrifugation and subsequent handling is not merely a technical detail but a fundamental requirement for generating reliable, reproducible, and biologically meaningful data. By recognizing the vulnerabilities of exosomes to disruption and aggregation—and implementing the targeted strategies outlined in this Application Note—researchers can significantly enhance the validity of their downstream analyses and the efficacy of exosome-based therapeutic applications.
The study of mesenchymal stem cell-derived exosomes (MSC-Exos) represents a rapidly advancing frontier in regenerative medicine and drug development. A critical, yet often underestimated, prerequisite for this research is the use of culture media completely devoid of fetal bovine serum (FBS)-derived exosomes. Contaminating bovine exosomes co-isolate with MSC-Exos, obscuring proteomic, genomic, and functional analyses, and fundamentally compromising the validity of experimental data [67]. Within the broader context of optimizing ultracentrifugation protocols for MSC exosomes research, the initial and complete depletion of FBS exosomes is a non-negotiable step for ensuring reproducibility and accurate interpretation of results. This Application Note provides a detailed, evidence-based guide to achieving this critical objective, presenting and comparing validated protocols to equip researchers with the tools necessary for success.
The Critical Need for Exosome Depletion in MSC Research
FBS is a complex supplement containing essential growth factors, lipids, and a high concentration of bovine extracellular vesicles (EVs), including exosomes [32]. When standard FBS is used in MSC cultures, these bovine exosomes are co-isolated during the ultracentrifugation process designed to harvest MSC-Exos. This contamination presents several intractable problems:
- Data Ambiguity: It becomes impossible to distinguish whether observed biological effects or molecular cargo (proteins, RNA) originate from the MSCs or the bovine serum [67].
- Irreproducible Results: Variations in FBS batches can lead to inconsistent contaminant profiles, undermining experimental reproducibility across studies and laboratories.
- Compromised Therapeutic Development: For clinical translation, the presence of xenogenic components is unacceptable from both a regulatory and safety perspective.
Therefore, employing a robust FBS exosome depletion protocol is not merely a best practice but a foundational requirement for any serious investigation into the native biology or therapeutic application of MSC-derived exosomes.
Comparative Analysis of FBS Exosome Depletion Methods
Researchers have developed several strategies to deplete exosomes from FBS. The table below provides a quantitative comparison of the most common methods, highlighting their efficiency and practical considerations.
Table 1: Quantitative Comparison of FBS Exosome Depletion Methods
| Method | Reported Particle Depletion Efficiency | Reported Protein Depletion Efficiency | Hands-On Time | Relative Cost | Key Advantages | Key Limitations |
|---|---|---|---|---|---|---|
| Ultracentrifugation (UC) | 78.27% ± 4.58% [67] | 70.41% ± 6.68% [67] | ~2 hours [32] | Low [32] | Widely accessible, low consumable cost | Incomplete depletion; time-consuming; requires specialized equipment |
| Ultrafiltration (UF) | 89.77% ± 4.55% [67] | 99.11% ± 0.34% [67] | 10-15 minutes [32] | Medium [32] | High efficiency, especially for proteins; fast; easy to standardize | Requires specific centrifugal filters; potential for membrane clogging |
| Commercial EV-Depleted FBS | Variable (Vendor-dependent) | Variable (Vendor-dependent) | None [32] | High [32] | Maximum convenience; saves time and labor | Highest cost; potential for residual contamination [67] |
| Serum-Free Media | 100% (Theoretical) | 100% (Theoretical) | None | Variable | No bovine EV contamination | Can induce cellular stress, alter MSC phenotype and impair immunomodulatory function [67] |
Detailed Experimental Protocols
Below are step-by-step protocols for the two most common in-house methods: Ultracentrifugation and Ultrafiltration.
Ultracentrifugation-Based Depletion Protocol
This protocol is adapted from conventional methods and involves prolonged high-speed centrifugation to pellet FBS-derived exosomes [32] [67].
Materials:
- Ultracentrifuge (e.g., Beckman Coulter Optima series)
- Fixed-angle or swinging-bucket rotor (e.g., SW28, SW40Ti)
- Ultracentrifuge tubes (e.g., Polypropylene, Polycarbonate)
- Sterile 0.22 µm PES filter unit
Procedure:
- Preparation: Thaw FBS at 4°C overnight if frozen. Ensure it is completely thawed and mixed gently.
- Initial Clarification (Optional): Transfer the FBS to appropriate ultracentrifuge tubes. Centrifuge at 12,000 × g for 20 minutes at 4°C to remove large aggregates and debris.
- High-Speed Centrifugation: Carefully load the clarified supernatant into new ultracentrifuge tubes. Balance the tubes precisely.
- Centrifuge at 120,000 × g for 16-18 hours (overnight) at 4°C [67].
- Collection of Depleted FBS: Following centrifugation, the exosomes will form a tight pellet at the bottom of the tube. Gently aspirate the top ~9/10 of the supernatant without disturbing the pellet. This supernatant is the ultracentrifugation EV-depleted FBS (UC-dFBS).
- Sterile Filtration: Filter the collected UC-dFBS through a 0.22 µm PES filter to ensure sterility.
- Aliquoting and Storage: Aliquot the sterile UC-dFBS into convenient volumes and store at -20°C or -80°C.
Ultrafiltration-Based Depletion Protocol
This protocol utilizes size-exclusion membranes to efficiently remove exosomes and is notably faster than ultracentrifugation [32].
Materials:
- Benchtop centrifuge with a swinging-bucket rotor
- 100 kDa molecular weight cut-off (MWCO) centrifugal filters (e.g., Amicon Ultra-15, Merck Millipore UFC910024)
Procedure:
- Preparation: Thaw FBS at 4°C overnight. Assemble the centrifugal filter units according to the manufacturer's instructions.
- Loading: Pipette up to 15 mL of FBS into the sample reservoir of the Amicon Ultra-15 centrifugal filter.
- Centrifugation: Centrifuge at 3,000 × g for 55 minutes at 4°C [32].
- Collection: The flow-through that passes into the collection tube is the ultrafiltration EV-depleted FBS (UF-dFBS). The exosomes and other large molecules are retained in the concentrator.
- Sterile Filtration (Optional but Recommended): Filter the collected UF-dFBS through a 0.22 µm filter to ensure sterility.
- Aliquoting and Storage: Aliquot the sterile UF-dFBS and store at -20°C or -80°C.
Impact on MSC Phenotype and Experimental Considerations
The choice of depletion method can significantly impact MSC biology, which must be considered when designing experiments.
Table 2: Functional Impact of Depletion Methods on MSCs
| Cellular Attribute | UC-dFBS | UF-dFBS | Serum-Free |
|---|---|---|---|
| Morphology | Retains fibroblastic, multipolar morphology [67] | Becomes more bipolar, fibroblastic [67] | Becomes more bipolar, fibroblastic [67] |
| Proliferation Rate | Normal proliferation [67] | Slower proliferation [67] | Slower proliferation [67] |
| Cell Viability & Surface Markers | Unaffected [67] | Unaffected [67] | Unaffected [67] |
| Immunomodulatory Capacity (e.g., IDO production) | Retained [67] | Significantly reduced [67] | Significantly reduced [67] |
| Particle Yield in Subsequent EV Harvest | Moderate | High | Low [67] |
Key Considerations:
- Protocol Selection: For studies where preserving native MSC immunomodulatory function is critical, UC-dFBS may be the preferred option despite its lower depletion efficiency. For applications requiring the highest purity of exosome preparations, such as proteomic studies, UF-dFBS is superior.
- Donor Variability: Be aware that the magnitude of the impact (e.g., on proliferation) can vary between different MSC donors [67].
- Quality Control: Always validate the efficiency of your depletion protocol using Nanoparticle Tracking Analysis (NTA) and protein quantification (e.g., BCA assay) to confirm the reduction in particle count and soluble protein [67].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for FBS Exosome Depletion
| Reagent / Material | Function / Application | Example Product / Reference |
|---|---|---|
| Fetal Bovine Serum (FBS) | Standard cell culture supplement requiring exosome depletion. | Various suppliers; ensure consistent sourcing. |
| 100 kDa MWCO Ultrafilters | For size-based depletion of exosomes via ultrafiltration. | Amicon Ultra-15 Centrifugal Filters (UFC910024) [32] |
| Ultracentrifuge & Rotors | For high-speed centrifugation-based depletion and exosome isolation. | Beckman Coulter Optima series with SW28 or 50.4Ti rotors [68] [34] |
| Exosome-Depleted FBS (Commercial) | Pre-depleted serum for convenience, though cost and residual contamination are concerns. | Exo-FBS (System Biosciences) [32] [34] |
| 0.22 µm PES Filters | For sterilizing depleted FBS post-processing. | Millipore Stericup-GP [32] |
| Nanoparticle Tracking Analyzer | Essential instrument for quantifying depletion efficiency by measuring vesicle concentration and size. | NanoSight NS300 (Malvern Panalytical) [69] [32] |
Workflow and Decision Pathway
The following diagram illustrates the experimental workflow for preparing exosome-depleted media and the subsequent decision-making process for selecting the appropriate method based on research goals.
Workflow Diagram Title: FBS Exosome Depletion and MSC Culture Pathway
Complete depletion of FBS-derived exosomes is an essential, non-negotiable first step in the rigorous isolation and study of MSC-derived exosomes. While no method is perfect, the choice between ultracentrifugation, ultrafiltration, and commercial sources must be a deliberate one, informed by a clear understanding of the trade-offs between depletion efficiency, impact on MSC biology, and practical logistics. By adopting and rigorously validating these optimized protocols, researchers in both academic and drug development settings can significantly enhance the reliability, reproducibility, and translational potential of their MSC exosome research.
Mesenchymal stem cell-derived exosomes (MSC-exosomes) have emerged as a promising cell-free therapeutic strategy in regenerative medicine, offering advantages over whole-cell therapies including reduced immunogenicity, absence of tumorigenic potential, and enhanced stability [19]. Preconditioning of parent MSCs prior to exosome isolation represents a crucial strategy to enhance the therapeutic efficacy of these nanovesicles. By exposing MSCs to specific environmental cues such as hypoxia or biochemical stimuli like cytokines, researchers can manipulate the cargo and functionality of the resulting exosomes, tailoring them for enhanced performance in targeted applications [70] [58]. Within the framework of a broader thesis focused on ultracentrifugation protocols for MSC exosome research, this application note provides detailed methodologies and quantitative data supporting the implementation of preconditioning strategies to maximize the therapeutic potential of isolated exosomes.
Quantitative Analysis of Preconditioning Efficacy
Therapeutic Outcomes of Preconditioned MSC-Exosomes
Table 1: Efficacy of Hypoxia-Preconditioned MSC-Exosomes in Preclinical Models
| Therapeutic Area | Exosome Source | Key Findings | Mechanistic Insights | Citation |
|---|---|---|---|---|
| Angiogenesis | Olfactory Mucosa MSCs | Enhanced proliferation, migration & angiogenesis of HBMECs | Upregulation of exosomal miR-612, stimulating HIF-1α-VEGF signaling | [70] |
| Myocardial Repair | Bone Marrow MSCs (Primate) | Promoted angiogenesis and increased vascular density | Upregulation of miR-486-5p, inhibiting MMP19 and promoting VEGFA | [70] |
| Cardiac Function | hCVPCs (Human) | Reduced apoptosis, increased angiogenesis, reduced scar formation | Hypoxic preconditioning enhanced repair of infarcted myocardium | [70] |
| Angiogenesis | Adipose-Derived MSCs | Altered proteomic profile; enhanced angiogenic potential | Upregulated LOXL2, CXCR4, and SDF-1 | [71] |
Table 2: Efficacy of Cytokine-Preconditioned MSC-Exosomes
| Preconditioning Agent | Exosome Source | Disease Model | Key Outcomes | Citation |
|---|---|---|---|---|
| IFN-γ | Bone Marrow MSCs | Myocardial Infarction (Rat) | Suppressed apoptosis, enhanced neovascularization, improved cardiac function | [70] |
| IL-1β | Murine MSCs | Sepsis (Mouse) | Induced M2 macrophage polarization, alleviated symptoms, improved survival | [70] |
| MIF | Umbilical Cord MSCs | Myocardial Infarction (Rat) | Enhanced proliferation, migration, angiogenesis of HUVECs; improved cardiac function | [70] |
| TNF-α (10 ng/mL) | Umbilical Cord MSCs | In vitro immunomodulation | Increased exosomal miR-146a content | [58] |
Meta-Analysis of MSC-Exosome Performance
A 2025 meta-analysis incorporating seven studies on imiquimod-induced psoriasis murine models demonstrated that MSC-exosome treatment significantly reduced clinical severity scores (Standardized Mean Difference [SMD]: -1.886; 95% Confidence Interval [CI]: -3.047 to -0.724) and epidermal thickness (SMD: -3.258; 95% CI: -4.987 to -1.529) compared to controls [44]. Meta-regression within this analysis specifically revealed that exosomes derived from human umbilical cord MSCs (hUCMSCs) showed a statistically greater improvement in clinical scores compared to those from other MSC sources (p=0.030) [44].
Experimental Protocols
Core Protocol: Ultracentrifugation for MSC-Exosome Isolation
This protocol forms the foundational basis for exosome isolation in the preconditioning studies cited [44] [7].
- Step 1: Cell Culture and Preconditioning. Culture MSCs to 50-60% confluence. Replace standard growth medium with preconditioning medium (e.g., hypoxic or cytokine-supplemented) for the prescribed duration.
- Step 2: Conditioned Medium Collection. After the preconditioning period, collect the cell culture supernatant. Centrifuge at 800 × g for 20 minutes at 4°C to remove cells and large debris [72].
- Step 3: Ultracentrifugation. Transfer the supernatant to ultracentrifuge tubes. Using a swinging-bucket rotor (e.g., Beckman Coulter SW32 Ti), perform ultracentrifugation at 100,000 × g for 70 minutes at 4°C to pellet the exosomes [7].
- Step 4: Washing (Optional). Resuspend the exosome pellet in a large volume of sterile, cold PBS. Perform a second ultracentrifugation under the same conditions (100,000 × g, 70 minutes, 4°C) to wash the exosomes [7].
- Step 5: Resuspension and Storage. Finally, resuspend the purified exosome pellet in a small volume of PBS or 0.9% NaCl. Aliquot and store at -80°C [72]. Stability can be maintained for up to one year under these conditions [72].
Preconditioning Methodologies
Hypoxic Preconditioning
- Objective: To mimic the physiological niche of MSCs and enhance exosome secretion of pro-angiogenic and pro-survival factors [71].
- Procedure: Culture MSCs in a specialized tri-gas incubator set to maintain 1-5% O₂, 5% CO₂, and balance N₂ for 24-48 hours prior to conditioned medium collection [70] [71]. The specific oxygen concentration and duration should be optimized for the MSC source and intended application.
Cytokine Preconditioning
- Objective: To prime MSCs toward a specific immunomodulatory or reparative phenotype.
- Procedure: Supplement the standard culture medium with the cytokine of choice. Common protocols include:
- IFN-γ: Stimulate MSCs with IFN-γ to enhance immunomodulatory properties [70] [58].
- TNF-α: Treat MSCs with 10-20 ng/mL of TNF-α to upregulate anti-inflammatory miRNAs like miR-146a in exosomes [58].
- IL-1β: Precondition MSCs with IL-1β to generate exosomes that promote macrophage M2 polarization, beneficial for sepsis and inflammatory conditions [70].
Signaling Pathways in Preconditioned Exosomes
Hypoxia-Induced Signaling Pathways
Figure 1: Hypoxia-Preconditioned Exosome Pathway. This diagram illustrates the core molecular mechanism through which hypoxic preconditioning enhances the pro-angiogenic capacity of MSC-exosomes, primarily via HIF-1α stabilization and subsequent miRNA-mediated VEGF signaling activation [70] [71].
Cytokine-Induced Immunomodulatory Pathways
Figure 2: Cytokine-Preconditioned Exosome Immunomodulation. This figure outlines how cytokine priming shapes the immunomodulatory function of MSC-exosomes, particularly through the delivery of specific miRNAs that reprogram macrophage responses and suppress inflammation [70] [58].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Kits for Preconditioning and Exosome Research
| Reagent / Kit | Function / Application | Examples / Specifications | Citation |
|---|---|---|---|
| Tri-Gas Incubator | Creates a controlled hypoxic environment for cell preconditioning. | 1-5% O₂, 5% CO₂, balance N₂. | [70] [71] |
| Proinflammatory Cytokines | Biochemically preconditions MSCs to enhance exosome potency. | IFN-γ, TNF-α (10-20 ng/mL), IL-1β. Recombinant human proteins. | [70] [58] |
| Ultracentrifuge | Gold-standard instrument for exosome isolation via differential centrifugation. | Beckman Coulter Optima series with Type 50.2 Ti or SW32 Ti rotors. | [44] [7] |
| Nanoparticle Tracking Analyzer | Characterizes exosome concentration and size distribution. | Malvern NanoSight NS300 system with NTA software. | [44] [72] |
| Exosome Characterization Antibodies | Confirms exosome identity via Western Blot. | Anti-CD9, Anti-CD63, Anti-ALIX, Anti-TSG101; Calnexin for purity control. | [44] [29] |
| Human Platelet Lysate (hPL) | Xeno-free supplement for GMP-compliant MSC culture media. | Commercially available, e.g., MultiPL100, Stemulate. | [72] [29] |
| Density Gradient Medium | Enhances purity of exosome isolation. | Sucrose or iodixanol gradients. | [26] |
| Tangential Flow Filtration (TFF) System | Scalable, GMP-compliant alternative for large-scale sEV isolation. | Higher particle yields compared to ultracentrifugation. | [29] |
The integration of preconditioning strategies—notably hypoxia and cytokine stimulation—with robust ultracentrifugation isolation protocols significantly augments the therapeutic profile of MSC-derived exosomes. The quantitative data and detailed methodologies provided in this application note offer researchers a validated framework for implementing these techniques. As the field advances toward clinical translation, the standardization of these preconditioning parameters alongside GMP-compliant production and isolation methods will be paramount for realizing the full potential of MSC-exosomes as a consistent, safe, and effective biotherapeutic product.
Comprehensive Characterization and Comparative Analysis of Isolation Methodologies
The therapeutic potential of mesenchymal stem cell (MSC)-derived exosomes is vast, ranging from tissue regeneration and immunomodulation to targeted drug delivery. The efficacy and reproducibility of these applications, however, are contingent upon the consistent production and thorough characterization of exosome preparations. Following isolation via ultracentrifugation, a multi-parametric analytical approach is mandatory to confirm the identity, purity, and quality of the isolated vesicles. This document outlines a core suite of characterization techniques—Nanoparticle Tracking Analysis (NTA) for size and concentration, Transmission Electron Microscopy (TEM) for morphology, and Western Blot for specific protein markers—standardized within the context of an MSC exosome research workflow. Adherence to these protocols ensures that exosome samples are accurately defined, facilitating reliable cross-comparison of experimental data and advancing the field towards clinical translation [73] [74].
Nanoparticle Tracking Analysis (NTA) for Size and Concentration
Principle and Application
NTA is a widely used technique that characterizes particles in a liquid suspension by visualizing and tracking their Brownian motion. A laser beam illuminates the particles, and the scattered light is captured by a high-sensitivity camera. The software then calculates the hydrodynamic diameter of each particle based on its diffusion coefficient using the Stokes-Einstein equation. Furthermore, NTA provides a concentration measurement by estimating the number of particles in the illuminated volume [73] [75]. For MSC exosomes, NTA in light-scattering mode is indispensable for determining the mean particle size, the size distribution profile (which should predominantly fall within the 30-150 nm range for exosomes), and the absolute concentration of particles in a sample (e.g., particles per milliliter) [73]. This information is critical for standardizing dosing in functional experiments and for comparing the yield and size profiles of exosomes isolated from different MSC sources or under different culture conditions [76].
Detailed NTA Protocol for MSC Exosomes
Materials:
- Resuspended exosome pellet (after ultracentrifugation)
- Sterile, filtered phosphate-buffered saline (PBS)
- NTA instrument (e.g., NanoSight NS300)
- 1 mL syringes
Method:
- Sample Preparation: Thaw the exosome sample on ice if frozen. Dilute the sample in filtered PBS to achieve a concentration within the ideal detection range of the NTA instrument (typically 10^8 to 10^9 particles/mL). The required dilution factor must be determined empirically and recorded precisely [73] [77].
- Instrument Calibration: Calibrate the NTA instrument using latex beads of known size (e.g., 100 nm) according to the manufacturer's instructions. Verify camera level and laser alignment.
- Data Acquisition:
- Load the diluted sample into the sample chamber using a syringe.
- Ensure the camera level is optimized to visualize particles as sharp, distinct points against the background.
- Adjust the focus until the laser beam is clearly visible on the screen.
- Set the detection threshold to exclude background noise.
- Capture five videos of 60 seconds each per sample, with a 10-second delay between captures. Perform at least three technical replicates.
- Data Analysis:
- Process all videos using the same software settings (e.g., detection threshold, blur size).
- Report the mean, mode, and D10/D50/D90 values for particle size.
- Report the particle concentration, factoring in the dilution factor to back-calculate the original sample concentration.
- Visually inspect the trajectories overlay to ensure accurate particle tracking.
Critical Considerations:
- Sample Purity: The light-scattering mode cannot distinguish exosomes from other similarly sized particles like protein aggregates or lipoprotein contaminants. Results must be interpreted in conjunction with other characterization data [73].
- Instrument Settings: Consistent instrument settings (camera shutter, gain, threshold) and sample preparation protocols are crucial for intra- and inter-study reproducibility [73].
Table 1: Key NTA Parameters and Specifications for MSC Exosome Analysis
| Parameter | Specification / Typical Range for MSC Exosomes | Notes |
|---|---|---|
| Size Range | 30 - 150 nm | Mode size often reported between 80-120 nm [76] |
| Concentration | Varies by source & isolation | Critical for dosing standardization [73] |
| Laser Wavelength | 405 nm, 488 nm, 532 nm, or 642 nm | Standard on commercial systems [73] |
| Measurement Volume | ~0.3 mL | Instrument-dependent |
| Analysis Temperature | 25°C (or controlled) | Required for accurate Stokes-Einstein calculation |
Diagram 1: NTA workflow for exosome characterization.
Transmission Electron Microscopy (TEM) for Morphology
Principle and Application
TEM uses a beam of electrons transmitted through an ultra-thin specimen to reveal information about its morphology, composition, and structure. For exosomes, TEM provides the highest resolution imaging, allowing researchers to visually confirm the spherical or cup-shaped morphology characteristic of exosomes, which often appears cup-shaped due to dehydration during sample preparation [78]. This technique is crucial for validating that the isolated vesicles are intact, membrane-bound structures and for excluding large contaminants or precipitates. When combined with immunogold labeling, TEM can also provide spatial information on the location of specific surface markers [78].
Detailed TEM Protocol for MSC Exosomes (Negative Staining)
Materials:
- Formvar/carbon-coated copper grids (200-400 mesh)
- Glutaraldehyde (2%)
- Uranyl acetate solution (2%)
- Filter paper
Method:
- Sample Application: Glow-discharge the grids to make them hydrophilic. Pipette 5-10 µL of the purified exosome suspension onto a parafilm. Float the grid on the droplet for 1 minute.
- Fixation: Carefully transfer the grid to a drop of 2% glutaraldehyde for 2 minutes.
- Washing: Transfer the grid sequentially to drops of distilled water (5-7 drops) to remove salts and fixative residues.
- Negative Staining: Float the grid on a drop of 2% uranyl acetate for 1 minute. Avoid prolonged staining to prevent crystal formation.
- Drying: Briefly touch the edge of the grid to a piece of filter paper to wick away excess stain. Allow the grid to air-dry completely in a clean, dust-free environment.
- Imaging: Image the grid using a TEM operated at 80-100 kV. Capture images at various magnifications to assess morphology and heterogeneity.
Critical Considerations:
- Sample Purity: The sample must be free of salts and aggregates, which can obscure the view of exosomes.
- Artifacts: The cup-shaped appearance is a common artifact of chemical fixation and dehydration. Cryo-TEM is an advanced alternative that preserves the native, spherical structure of exosomes in a vitrified ice layer [78].
Table 2: Key TEM Staining Reagents for Exosome Visualization
| Reagent | Function | Application Note |
|---|---|---|
| Uranyl Acetate | Heavy metal salt; scatters electrons to create contrast around particles. | Common negative stain; handle with appropriate safety precautions. |
| Phosphotungstic Acid | Alternative negative stain. | Can be used at neutral pH. |
| Glutaraldehyde | Cross-linking fixative; stabilizes exosome structure. | Used for pre-fixation to preserve morphology. |
| Osmium Tetroxide | Fixative and stain; binds to lipids. | Provides additional membrane contrast; requires careful handling. |
Western Blot for Protein Marker Characterization
Principle and Application
Western blotting is a fundamental technique for detecting specific proteins in a complex sample. It involves separating proteins by gel electrophoresis, transferring them to a membrane, and probing with antibodies specific to target antigens. For MSC exosomes, Western blot is used to confirm the presence of positive markers associated with exosome biogenesis and identity (e.g., CD63, CD81, CD9, Alix, TSG101) and the absence of negative markers from non-exosomal cellular compartments (e.g., Calnexin from the endoplasmic reticulum, GM130 from the Golgi apparatus) [76] [74]. This confirmation is essential for verifying that the isolated vesicles are indeed exosomes and not other extracellular vesicles or cellular debris.
Detailed Western Blot Protocol for MSC Exosome Markers
Materials:
- Exosome lysate (prepared in RIPA buffer with protease inhibitors)
- 4-12% Bis-Tris protein gel
- Nitrocellulose or PVDF membrane
- Primary antibodies: Anti-CD63, Anti-CD81, Anti-CD9, Anti-Alix, Anti-Calnexin
- HRP-conjugated secondary antibodies
- Chemiluminescent substrate
Method:
- Sample Preparation: Lyse the exosome pellet in RIPA buffer. Determine the protein concentration using a compatible assay (e.g., BCA). Load an equal amount of protein (e.g., 10-20 µg) for both exosome and whole cell lysate (positive control) samples.
- Electrophoresis and Transfer: Separate the proteins by SDS-PAGE and transfer them to a PVDF membrane using standard protocols.
- Antibody Probing:
- Block the membrane with 5% non-fat milk in TBST for 1 hour.
- Incubate with primary antibodies diluted in blocking buffer overnight at 4°C.
- Typical antibodies: Anti-CD63 (1:1000), Anti-CD81 (1:1000), Anti-CD9 (1:1000), Anti-Alix (1:1000), Anti-Calnexin (1:1000).
- Wash the membrane 3 times for 5 minutes each with TBST.
- Incubate with the appropriate HRP-conjugated secondary antibody for 1 hour at room temperature.
- Wash the membrane 3 times for 5 minutes each with TBST.
- Detection: Develop the membrane with a chemiluminescent substrate and image using a chemiluminescence detection system.
Critical Considerations:
- Antibody Validation: Ensure antibodies are validated for exosome detection, as protein conformation and accessibility may differ from whole cell lysates.
- Loading Controls: Due to the lack of a universal "housekeeping" protein for exosomes, loading equal protein amounts and using a positive marker like TSG101 or a stain-free gel total protein normalization is recommended.
- Specificity: The use of whole cell lysate as a control helps confirm antibody specificity.
Table 3: Essential Protein Markers for MSC Exosome Characterization by Western Blot
| Marker Category | Specific Markers | Expected Result | Biological Rationale |
|---|---|---|---|
| Positive Markers | CD63, CD81, CD9 | Present (Enriched) | Tetraspanins enriched on exosome membranes [76] [74]. |
| Alix, TSG101 | Present | ESCRT-associated proteins involved in exosome biogenesis [74]. | |
| Negative Markers | Calnexin, GRP94 | Absent (or not enriched) | Endoplasmic reticulum markers; confirm absence of cell debris [76]. |
| Cytochrome C | Absent | Mitochondrial marker; confirms purity from organelles [74]. |
Diagram 2: Logical framework for Western Blot analysis.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for Exosome Characterization
| Reagent / Kit | Function / Application | Example Use Case |
|---|---|---|
| Fluorescent Exosome Standards | Pre-labeled exosomes for instrument calibration and protocol optimization. | Validating NTA and flow cytometry settings [75]. |
| Antibody Panels (CD9, CD63, CD81) | Multiplex kits for simultaneous detection of multiple exosome surface markers. | Streamlining Western Blot or flow cytometry confirmation of exosome identity [79]. |
| Magnetic Beads (CD63-conjugated) | Immunoaffinity capture for isolating specific exosome subpopulations. | Purifying exosomes directly from complex biofluids prior to characterization [79]. |
| Size Exclusion Columns | Gentle, size-based separation of exosomes from soluble proteins. | Post-ultracentrifugation polishing step to improve sample purity for downstream assays [79]. |
Within the context of mesenchymal stem cell (MSC) research for therapeutic applications, assessing the purity of exosome preparations isolated via ultracentrifugation is a critical prerequisite for reliable downstream analysis and interpretation. Pure exosome preparations ensure that observed biological effects are genuinely attributable to the exosomes themselves and not to co-isolated contaminants. The presence of non-vesicular materials, such as soluble proteins and lipoproteins like Low-Density Lipoprotein-cholesterol (LDL-c), can significantly confound functional studies, biomarker discovery, and therapeutic efficacy evaluations [80] [47]. This application note details two fundamental, quantitative approaches for assessing exosome purity: calculating the particle-to-protein ratio and detecting lipoprotein contaminants, specifically LDL-cholesterol. These methods provide researchers with essential tools for quality control, enabling standardization and improving reproducibility in MSC exosome research.
Quantitative Purity Benchmarks
The following tables summarize key quantitative benchmarks and the impact of various factors on the purity of MSC-derived exosomes.
Table 1: Expected Purity Metrics for MSC-Derived Exosomes from Different Isolation and Culture Conditions
| Parameter | Standard Ultracentrifugation (with FBS) | TFF Isolation (with UF-dFBS) | Sucrose Cushion UC | Reference |
|---|---|---|---|---|
| Particle-to-Protein Ratio (particles/μg) | ~1.0 x 10⁷ (Conditioned Media) | 9.25 x 10⁸ (Estimated yield increase) | ~6.0 x 10⁸ (Post-isolation) | [80] [47] |
| LDL-cholesterol Contamination | High (Detectable) | Negligible (Not detected) | Information Missing | [47] |
| CD73 Purity (MSC marker) | Low (Baseline) | ~15.6x enhancement | Information Missing | [47] |
| Key Advancement | Baseline | High yield & high purity | High purity from complex media | [80] [47] |
Table 2: Impact of Contaminants and Process Changes on Purity Metrics
| Factor | Impact on Particle-to-Protein Ratio | Impact on Lipoprotein Contamination | Experimental Evidence |
|---|---|---|---|
| Addition of BSA | Decreases ratio by ~50% with 40-50% exogenous protein | Not directly measured | [80] |
| FBS Exosome Depletion (UF-dFBS) | Increases effective ratio by enhancing MSC-exosome specificity | Significantly reduces LDL-c contamination | [47] |
| Wash Step in UC | Increases ratio by ~60-fold vs. unpurified media | Helps remove some soluble contaminants | [80] |
| Isolation Method (TFF vs UC) | TFF yield ~92.5x higher than UC; impacts functional ratio | TFF with UF-dFBS minimizes lipoprotein co-isolation | [47] |
Experimental Protocols
Protocol 1: Determining Particle-to-Protein Ratio
This protocol provides a straightforward method to estimate sample purity by comparing the number of nano-vesicles to the protein concentration [80].
Materials and Equipment
- NanoSight LM10/NS300 system (Malvern Technologies) or ZetaView QUATT (Particle Metrix) [80] [47]
- Micro-BCA Protein Assay Kit (Thermo Scientific Pierce)
- Optima MAX Ultracentrifuge with TLA-110 rotor (Beckman Coulter) or equivalent
- Phosphate-Buffered Saline (PBS), particle-free water
Step-by-Step Procedure
- Sample Preparation: Resuspend the final exosome pellet obtained from ultracentrifugation of MSC-conditioned media in particle-free PBS.
- Nanoparticle Tracking Analysis (NTA):
- Dilute the exosome sample with particle-free water to a concentration between 2x10⁸ and 9x10⁸ particles/mL.
- Inject the sample into the NTA system using a syringe pump for controlled fluid flow.
- Capture six videos of 30-60 seconds duration, with a 10-second delay between recordings.
- Set the camera level, detection threshold, and other parameters according to the manufacturer's guidelines and keep them consistent for all measurements.
- Use the NTA software to analyze the videos, generating a particle size distribution and concentration for each. Calculate the average particle concentration from the six replicates [80].
- Protein Quantification (BCA Assay):
- Dilute the same exosome preparation 1:8 to 1:20 in PBS.
- Perform the micro-BCA assay in triplicate, following the manufacturer's instructions.
- Compare the absorbance of the exosome samples against a serially diluted Bovine Serum Albumin (BSA) standard curve.
- Extrapolate the protein concentration from the standard curve [80].
- Ratio Calculation:
- Calculate the particle-to-protein ratio using the formula: Particle-to-Protein Ratio = (Total Particle Concentration) / (Total Protein Concentration)
- Report the ratio as particles per microgram (μg) of protein [80].
Data Interpretation
A higher particle-to-protein ratio indicates a purer vesicle preparation, as it signifies more vesicles relative to soluble protein contaminants. The addition of exogenous protein (like BSA) decreases this ratio, demonstrating its sensitivity to contamination [80]. The use of culture medium with ultrafiltration-derived exosome-depleted FBS (UF-dFBS) can dramatically improve the ratio by enhancing the specificity for MSC-derived exosomes [47].
Protocol 2: Detecting LDL-cholesterol Contamination
Lipoproteins, particularly LDL, are common contaminants in exosome preparations from serum-containing media and can be quantified using a clinical biochemistry analyzer [47].
Materials and Equipment
- Hitachi 7020 Automatic Biochemistry Analyzer or equivalent clinical analyzer
- Control Serum I and II (Wako Pure Chemical Industries) for calibration
- LDL-cholesterol assay reagents (as per analyzer manufacturer)
- Ultracentrifugation equipment
Step-by-Step Procedure
- Sample Preparation: Isolate exosomes from MSC-conditioned media using the standard ultracentrifugation protocol. The final exosome pellet should be dissolved in deionized water (DW) for analysis [47].
- System Calibration:
- Prior to analysis, calibrate the automatic biochemistry analyzer using the Control Serum I and II, following the manufacturer's instructions to establish a standard curve for LDL-cholesterol [47].
- Sample Analysis:
- Load the dissolved exosome sample onto the calibrated analyzer.
- Run the LDL-cholesterol detection protocol according to the manufacturer's specified instructions for the reagent kit in use [47].
- Data Collection:
- The analyzer will output the concentration of LDL-cholesterol present in the exosome sample.
Data Interpretation
The concentration of LDL-cholesterol directly indicates the degree of lipoprotein contamination. In highly purified MSC-exosome preparations, particularly those isolated from culture media using exosome-depleted FBS (e.g., UF-dFBS), LDL-cholesterol levels can be negligible [47]. This assay is crucial for confirming that observed biological activities or biomarker profiles are not influenced by lipoprotein contaminants.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Kits for Exosome Purity Assessment
| Item | Function / Application | Example Product / Source |
|---|---|---|
| Nanoparticle Tracking Analyzer | Measures particle size distribution and concentration in exosome preparations. | NanoSight NS300 (Malvern), ZetaView (Particle Metrix) [80] [81] |
| Colorimetric Protein Assay | Quantifies total protein concentration in the exosome sample. | Micro-BCA Protein Assay Kit (Thermo Scientific Pierce) [80] [47] |
| Clinical Biochemistry Analyzer | Quantifies specific contaminants like LDL-cholesterol in exosome preparations. | Hitachi 7020 Analyzer [47] |
| Exosome-Depleted FBS | Cell culture supplement that reduces serum-derived vesicle and lipoprotein contaminants. | Ultracentrifugation (UC-dFBS) or Ultrafiltration (UF-dFBS) processed FBS [47] |
| Ultracentrifuge | Core instrument for isolating exosomes from biological fluids and culture media. | Optima Series (Beckman Coulter) [80] [54] |
Workflow and Data Interpretation
The following diagram illustrates the logical workflow integrating the two purity assessment protocols into a typical MSC exosome research pipeline.
Integrating particle-to-protein ratio calculation and LDL-cholesterol detection provides a robust, quantitative framework for assessing the purity of MSC-derived exosomes. These quality control measures are indispensable for ensuring the reliability of data generated in functional studies, the accuracy of biomarker profiling, and the safety and efficacy of exosome-based therapeutics. As the field progresses toward clinical applications, adopting standardized purity assessments will be crucial for validating isolation protocols and comparing results across different studies and laboratories.
The transition of mesenchymal stem cell (MSC) exosome research from bench to bedside is critically dependent on the development of scalable, reproducible, and efficient isolation methodologies. For years, ultracentrifugation (UC) has been the undisputed gold standard in laboratory-scale exosome isolation, forming the backbone of foundational MSC research [82] [24]. However, its limitations in large-scale production—including low yield, prolonged processing times, and potential vesicle damage—have prompted the exploration of alternative technologies [82] [83]. Tangential Flow Filtration (TFF) has emerged as a powerful solution to these challenges, offering a scalable and gentle approach for processing large volumes of cell culture conditioned media [84].
This Application Note provides a comparative analysis of Ultracentrifugation and Tangential Flow Filtration for the isolation of MSC-derived small extracellular vesicles (sEVs), with a particular focus on their application in large-scale production workflows for therapeutic and diagnostic development. We present structured quantitative data, detailed experimental protocols, and visual workflows to guide researchers in selecting and optimizing their exosome isolation strategies.
Key Comparative Data at a Glance
Table 1: Direct Comparison of Ultracentrifugation and Tangential Flow Filtration for sEV Isolation
| Parameter | Ultracentrifugation (UC) | Tangential Flow Filtration (TFF) |
|---|---|---|
| Basic Principle | Separation based on size and density using high g-forces [24] | Size-based separation via cross-flow filtration parallel to membrane [82] [84] |
| Typical sEV Yield | Significantly lower; ~1.02E+10 - 1.74E+09 particles from plasma/saliva [83] | Significantly higher; "surpasses UC in yield" for large-scale applications [82] |
| Processing Time | Lengthy (often > 4 hours for multiple cycles) [24] | Rapid; "time efficiency" is a noted advantage [82] |
| Scalability | Limited by centrifuge rotor capacity [85] | Highly scalable from mL to 1000s of Liters [84] |
| sEV Integrity | Risk of damage/aggregation due to high g-forces [82] [24] | Better preservation of integrity and function; gentler process [82] |
| Purity | Moderate; often requires additional purification steps (e.g., SEC) [82] | High purity when coupled with SEC; reduced co-isolation of contaminants [82] |
| Cost & Infrastructure | Requires expensive ultracentrifuge; lower consumable cost [24] | Lower equipment cost; potential for higher membrane consumable cost [82] |
| Reproducibility | User-dependent; potential for pellet disruption [24] | High reproducibility and ease of standardization [82] [84] |
Table 2: Quantitative sEV Recovery from Different Biological Samples Using Various Methods [83]
| Isolation Method | sEV Concentration from Saliva (particles/mL) | sEV Concentration from Plasma (particles/mL) | sEV Concentration from Cell Culture Media (particles/mL) |
|---|---|---|---|
| PEG Precipitation (CP) | 2.43E + 11 | 1.76E + 11 | 1.46E + 10 |
| PEG + Ultrafiltration (CPF) | Lower than CP but higher than UC | Lower than CP but higher than UC | Lower than CP but higher than UC |
| Ultracentrifugation (UC) | 1.74E + 09 | 1.02E + 10 | 1.30E + 09 |
| Size Exclusion Chromatography (SEC) | Successively lower than CP but higher than UC | Successively lower than CP but higher than UC | Successively lower than CP but higher than UC |
Experimental Protocols
Detailed Protocol: Ultracentrifugation for MSC-sEVs
This protocol is adapted from standardized methods for isolating sEVs from serum-containing cell culture media, a common challenge in MSC research [82].
Pre-processing of MSC Conditioned Media:
- Culture MSCs in media supplemented with EV-depleted FBS for 48 hours [82].
- Clarify conditioned media by centrifugation at 500 × g for 10 minutes to remove detached cells and large debris.
- Filter the supernatant through a 0.22 µm filter to remove other large particle contaminants [82].
Ultracentrifugation Process:
- Transfer the clarified conditioned media into ultracentrifuge tubes compatible with a fixed-angle rotor (e.g., Type 70.1 Ti).
- Perform first ultracentrifugation at 100,000 × g at 4°C for 120 minutes.
- Carefully decant the supernatant. The crude sEV pellet may be difficult to see.
- Resuspend the pellet in a large volume (e.g., 35 mL) of sterile, ice-cold Phosphate Buffered Saline (PBS). This step helps to wash the pellets and reduce contaminating proteins.
- Perform second ultracentrifugation at 100,000 × g at 4°C for 120 minutes [82].
- Discard the supernatant and resuspend the final sEV pellet in a small volume (e.g., 100-200 µL) of PBS or a suitable buffer for downstream applications.
- Store the isolated MSC-sEVs at -80°C.
Detailed Protocol: Tangential Flow Filtration for MSC-sEVs
TFF isolates sEVs based on size using a cross-flow mechanism that minimizes membrane fouling, making it ideal for processing the large volumes of media generated in MSC biomanufacturing [82] [84].
Pre-processing of MSC Conditioned Media:
- Follow the identical clarification and filtration steps (Steps 1-3) from the UC protocol above.
TFF Setup and Process:
- Select a TFF System: For research and pilot scale, a benchtop system with a peristaltic pump, pressure gauges, and a holder for flat sheet cassettes is suitable.
- Choose a Membrane: A flat sheet cassette with a pore size of 300-500 kD or ~0.1 µm is appropriate for retaining sEVs while allowing smaller proteins and solutes to pass through [84].
- System Preparation: Flush and equilibrate the entire TFF system with PBS according to the manufacturer's instructions.
- Load and Concentrate: Pump the clarified conditioned media into the TFF system. The feed stream flows tangentially across the membrane, and sEVs are retained while the buffer and small molecules pass through as permeate. The process continues until the retentate (sEV-containing fraction) is concentrated to a desired volume (e.g., 10-50x concentration) [82].
- Diafiltration (Buffer Exchange): To enhance purity, continuously add PBS or the final formulation buffer to the retentate reservoir at the same rate as permeate is being removed. This step washes out remaining contaminating proteins. Typically, 5-10 diafiltration volumes are sufficient.
- Final Recovery: Once diafiltration is complete, recover the highly concentrated sEV retentate. The system may be flushed with a small volume of buffer to maximize recovery.
- Optional Post-Purification: For the highest purity, the TFF-retentate can be further purified using Size Exclusion Chromatography (SEC) to remove any remaining soluble proteins or aggregates [82].
- Store the final TFF-isolated MSC-sEVs at -80°C.
Workflow and Decision Pathway
The following diagram illustrates the key steps and logical flow for both isolation methods, highlighting the more streamlined nature of the TFF process.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Materials and Reagents for MSC-sEV Isolation and Characterization
| Item | Function / Application | Examples / Specifications |
|---|---|---|
| EV-Depleted FBS | Nutrient supplementation for MSC culture without introducing contaminating serum EVs. | Prepared by ultracentrifugation (e.g., 100,000 × g overnight) or commercially sourced. Essential for clean sEV preps from conditioned media [82]. |
| TFF System & Membranes | Scalable concentration and purification of sEVs from large volumes. | Benchtop TFF systems with peristaltic pumps. Flat sheet cassettes (300-500 kD MWCO) for high flux or Hollow Fiber Modules for shear-sensitive samples [84]. |
| Ultracentrifuge & Rotors | High-g-force separation for differential centrifugation protocols. | Fixed-angle or swinging bucket rotors (e.g., Type 70.1 Ti, 50.2 Ti). Requires high-speed ultracentrifuge [82]. |
| Size Exclusion Columns | High-resolution size-based purification to remove contaminating proteins after UC or TFF. | Sepharose-based columns (e.g., qEV original) for final polishing step to achieve high-purity sEVs [82] [83]. |
| Nanoparticle Tracking Analyzer | Quantification of sEV particle concentration and size distribution. | Instruments (e.g., NanoSight NS300) measure Brownian motion to determine particle size and count, critical for dose standardization [16] [72]. |
| Characterization Antibodies | Confirmation of sEV identity via detection of surface and intravesicular markers. | Antibodies against tetraspanins (CD63, CD81, CD9), TSG101, and Flotillin-1 for Western Blot analysis [83]. |
The choice between Ultracentrifugation and Tangential Flow Filtration for large-scale MSC exosome production is strategic, dictated by the specific goals of the research or development program. While ultracentrifugation remains a valuable benchmark for discovery-phase research requiring multiple, small, parallel samples, its technical and scalability limitations are evident in a manufacturing context.
Tangential Flow Filtration presents a superior and industrially viable alternative for processing large volumes of MSC-conditioned media, offering significant advantages in yield, processing time, scalability, and gentle handling to preserve sEV integrity. The integration of TFF with a final polishing step, such as Size Exclusion Chromatography, provides a robust and reproducible pipeline for generating high-quality MSC-sEVs suitable for demanding downstream therapeutic and diagnostic applications.
The isolation of pure and functional extracellular vesicles (EVs), particularly exosomes, from Mesenchymal Stromal Cell (MSC) conditioned media is a critical step in downstream therapeutic and diagnostic applications. The choice of isolation methodology significantly impacts the yield, purity, and biological functionality of the isolated vesicles. This application note provides a detailed, data-driven comparison of three predominant isolation techniques—Ultracentrifugation (UC), Commercial Precipitation Kits, and Size-Exclusion Chromatography (SEC)—framed within the context of MSC exosome research. We summarize key performance metrics and provide optimized protocols to guide researchers and drug development professionals in selecting the most appropriate method for their specific applications.
Table 1: Overall Method Comparison for MSC Exosome Isolation
| Feature | Ultracentrifugation (UC) | Sucrose Cushion UC | Size-Exclusion Chromatography (SEC) | Commercial Precipitation Kits |
|---|---|---|---|---|
| Mechanism | Size/Density, Gravitational force [7] | Density/Buoyancy [25] | Size/Shape, Molecular sieve [86] | Solubility, Surface Charge [86] |
| Relative Purity | Intermediate to High [86] | High [25] | High [86] [87] | Low [88] [86] |
| Relative Yield | Intermediate [86] | High [25] | High [86] | High [86] |
| Sample Volume | Large volumes (e.g., 4mL/tube) [7] | Large volumes [25] | Small to Moderate (e.g., 0.5-2mL) [86] [89] | Small to Moderate [88] |
| Functional Vesicles | Preserved [7] | Preserved, cup-shaped morphology [25] | Well-preserved, functional for RNomics [86] [87] | Variable, potential polymer contamination |
| Cost & Equipment | Requires ultracentrifuge [7] | Requires ultracentrifuge [25] | Low equipment cost, reusable columns [86] | Low equipment cost, kit expense |
| Key Advantage | Good reproducibility, handles large volumes [7] | High yield & purity, protects vesicle integrity [25] | High purity & preserved RNA function, rapid [86] [87] | Simple protocol, no specialized equipment [86] |
| Key Disadvantage | Time-consuming, potential for co-pelleted contaminants [7] [25] | Additional sucrose step required | Sample dilution, may require a concentration step [86] | High contaminant co-precipitation (proteins, lipoproteins) [88] [86] |
MSCs have a protective effect on the progression of various diseases, which is not only due to their transdifferentiation capacity but also their paracrine mechanisms, including the release of EVs [7]. MSC-derived exosomes exhibit immunomodulatory, anti-inflammatory, anti-apoptotic, and pro-angiogenic functions, making them a promising cell-free therapeutic [7] [25]. The efficacy of these exosomes in downstream applications, from in vivo experiments to potential use in organ perfusion machines, is profoundly influenced by the isolation method chosen [7]. Different protocols can isolate different subpopulations of EVs with varying compositions of proteins and nucleic acids, making the selection of an isolation technique a cornerstone of experimental design and reproducibility [7] [86].
The following workflow outlines the logical decision-making process for selecting an appropriate exosome isolation method based on key research requirements:
Quantitative Data Comparison
The following tables synthesize quantitative and qualitative data from comparative studies to inform method selection.
Table 2: Purity and Contaminant Analysis Across Methods
| Isolation Method | EVs:Protein Ratio | Lipoprotein Contamination (APOB/APOE) | Albumin Contamination | Key Purity Findings |
|---|---|---|---|---|
| Ultracentrifugation (UC) | Lower vs. SEC [89] | Present [81] | Present [90] | Five cycles of UC required to remove >95% serum proteins [89]. |
| Sucrose Cushion UC | Higher than UC [25] | Not Specifically Tested | Reduced vs. UC [25] | Sucrose density separates exosomes from higher-density protein contaminants [25]. |
| Size-Exclusion Chromatography (SEC) | High [89] [81] | Lower than UC and kits [81] | Low/None [90] | Effectively separates vesicles from soluble proteins and lipoproteins [86] [81] [90]. |
| Commercial Kits | Low [88] [86] | High [88] [81] | Present in all kits tested [88] | Purity varies significantly between kits; overall low purity due to co-precipitation [88] [86]. |
Table 3: Particle Yield, Size, and Functional Cargo Analysis
| Isolation Method | Particle Yield & Size | RNA / miRNA Analysis | Protein Cargo | Key Functional Findings |
|---|---|---|---|---|
| Ultracentrifugation (UC) | Good yield [86]; ~60 nm mean size [15] | Lower useful reads for RNA-Seq vs. SEC [87] | Standard UC isolates EV markers (CD63, TSG101) [81] | Vesicles are functional, but prolonged forces may damage integrity [25]. |
| Sucrose Cushion UC | Higher yield than UC; cup-shaped morphology [25] | Not Specifically Tested | Strong expression of EV markers (CD63, Alix) [25] | Isolated exosomes retain biological activity and membrane integrity [25]. |
| Size-Exclusion Chromatography (SEC) | High yield [86]; ~50-200 nm size range [86] | Highest EVs-specific RNA; best for miRNA/mRNA-Seq [87] | Lower AGO2 (non-vesicular miRNA carrier) vs. other methods [87] | Ideal for downstream RNomics; vesicles are functional and intact [86] [87]. |
| Commercial Kits | High yield but variable [88] [86]; ~89 nm mean size [15] | Cytokine detection varies deeply by kit; no correlation between kits [88] | EV markers present; high contaminant proteins [88] | Function may be impaired by polymer contaminants or aggregation. |
Detailed Experimental Protocols
This protocol is considered the historical gold standard and is applicable for processing large volumes of MSC-conditioned media.
Research Reagent Solutions & Equipment:
- Alpha MEM Complete Medium: Cell culture medium for MSC expansion.
- Dulbecco’s Phosphate Buffered Saline (PBS): For washing and dilution.
- Ultra-Clear Tubes (e.g., Beckman Coulter, 344058): Special tubes for high-force centrifugation.
- Preparative Ultracentrifuge with Swinging-Bucket Rotor (e.g., Beckman Optima L100XP, SW32 Ti): Essential equipment.
Procedure:
- Cell Culture & Conditioning: Culture MSCs to 70-80% confluence in complete Alpha MEM with 10% FBS. To produce conditioned medium for exosome isolation, replace the growth medium with serum-free medium (e.g., STEMPRO MSC SFM CTS) and culture for 48 hours [25].
- Pre-Clearing: Collect the conditioned medium and perform sequential centrifugation steps to remove cells and debris.
- Ultracentrifugation: Transfer the pre-cleared supernatant to Ultra-Clear tubes. Ultracentrifuge at 100,000 × g for 90-120 minutes at 4°C [7] [25].
- Washing & Final Pellet: Carefully discard the supernatant. Resuspend the pellet in a large volume of PBS (e.g., 4 mL). Ultracentrifuge again at 100,000 × g for 70-90 minutes at 4°C to wash the exosomes. Discard the supernatant and resuspend the final, purified exosome pellet in a small volume of PBS (e.g., 50-100 µL) [7] [89].
- Storage: Aliquot and store the exosome suspension at -80°C.
This modified UC method offers enhanced yield and purity for MSC exosomes by protecting vesicle integrity during pelleting.
Procedure:
- Pre-Clearing: Perform steps 1 and 2 as described in the Standard UC protocol.
- Sucrose Cushion Preparation: Prepare a 30% sucrose solution in PBS. Add 4 mL of this solution to an Ultra-Clear tube.
- Layer Sample: Gently layer the pre-cleared conditioned medium (e.g., up to 30 mL) on top of the sucrose cushion, forming a distinct interface.
- Ultracentrifugation: Ultracentrifuge at 100,000 × g for 90 minutes at 4°C. During this step, exosomes migrate to the sucrose interface, while higher-density contaminants pellet.
- Harvest Exosomes: After centrifugation, carefully aspirate and discard the supernatant above the sucrose layer. Collect the sucrose layer (~5 mL) containing the exosomes and transfer it to a new tube.
- Dilution and Pellet: Dilute the sucrose-exosome mixture with a large volume of PBS (e.g., 35 mL to dilute sucrose). Ultracentrifuge this mixture at 100,000 × g for 90 minutes at 4°C to pellet the exosomes.
- Final Resuspension: Discard the supernatant and resuspend the final pellet in PBS. Store at -80°C.
SEC is an excellent alternative for obtaining high-purity, functional exosomes, especially from smaller sample volumes.
Research Reagent Solutions & Equipment:
- SEC Columns (e.g., qEV original, Izon Science): Pre-packed columns with sepharose (e.g., CL-2B, CL-4B, CL-6B) resin [86] [91].
- Phosphate Buffered Saline (PBS): Elution buffer.
- Ultrafiltration Concentrator (e.g., 100K Amicon Ultra-15): Optional, for concentrating the eluted exosomes [87].
Procedure:
- Sample Preparation: Pre-clear the MSC-conditioned medium or biofluid as in UC steps 1 and 2. For viscous samples like plasma/serum, dilution with PBS is recommended [89].
- Column Equilibration: Rinse the SEC column with at least 15 mL of PBS to equilibrate it, following the manufacturer's instructions.
- Sample Loading & Elution: Load the pre-cleared sample onto the column (typical load volume is 0.5-2 mL). Continuously add PBS to the column and begin collecting sequential fractions (typically 0.5-1 mL each).
- Fraction Collection: Discard the initial void volume (first ~3.5 mL for a 10mL column). The subsequent fractions (e.g., fractions 7-10) contain the purified exosomes. Later fractions will contain soluble proteins and other small contaminants [86] [89].
- Concentration (Optional): If a concentrated exosome sample is required, the exosome-containing fractions can be concentrated using ultrafiltration concentrators (100K MWCO) by centrifuging at 4,000 × g until the desired volume is reached [87].
The Scientist's Toolkit: Essential Research Reagents & Equipment
Table 4: Key Materials for MSC Exosome Isolation and Characterization
| Item Category | Specific Examples | Critical Function in Workflow |
|---|---|---|
| Cell Culture | Alpha MEM, Serum-free Media (e.g., STEMPRO MSC SFM) | MSC expansion and production of exosome-containing conditioned media. |
| Centrifugation | Refrigerated Benchtop Centrifuge, Preparative Ultracentrifuge (e.g., Beckman Optima L100XP), SW32 Ti Rotor, Ultra-Clear Tubes | Removal of cells, debris, and microvesicles; pelleting of exosomes via UC. |
| Chromatography | SEC Columns (e.g., qEV, Izon Science; Sepharose CL-2B/4B/6B) | Size-based separation of exosomes from contaminating proteins and lipoproteins. |
| Buffers & Reagents | Dulbecco's PBS (1x), 30% Sucrose Solution (in PBS) | Washing, dilution, and creation of density cushion for high-purity isolation. |
| Characterization | NanoSight NS300 (NTA), Transmission Electron Microscope, Western Blot reagents, Antibodies (CD63, CD81, TSG101, Alix, APOB) | Determining particle size/concentration, visualizing morphology, and confirming presence of EV markers/absence of contaminants. |
The choice of an exosome isolation method is a strategic decision that balances yield, purity, and intended downstream application.
- For maximum purity and optimal RNA recovery, particularly for sequencing and biomarker discovery, SEC is the recommended method. It effectively separates exosomes from soluble proteins and lipoproteins, yielding functional vesicles with high-quality RNA [86] [87].
- For high yield and purity from large volumes of MSC-conditioned media, the Sucrose Cushion UC method is highly advantageous. It provides a superior yield and purity profile compared to standard UC while protecting vesicle integrity [25].
- Standard UC remains a reliable and reproducible workhorse, especially for laboratories with established ultracentrifugation capabilities and when processing large sample volumes. However, researchers should be aware of potential contaminants and may need to implement multiple washing cycles [7] [89].
- Commercial Precipitation Kits, while convenient and requiring no specialized equipment, generally result in lower purity with significant co-isolation of contaminants. They should be selected with caution, especially for sensitive downstream analytical applications like proteomics or functional studies [88] [86] [81].
For the specific context of an MSC exosome research thesis, combining an initial UC or Sucrose Cushion UC step for concentration from large volumes of media, followed by a final SEC polishing step for maximum purity, represents a powerful hybrid approach for generating the highest quality exosome preparations [89] [91].
The isolation of mesenchymal stem cell (MSC)-derived exosomes via ultracentrifugation represents a critical first step in harnessing their therapeutic potential. However, physical characterization through nanoparticle tracking analysis (NTA) and Western blotting, while essential for establishing identity and purity, remains insufficient for confirming biological functionality. Functional validation through uptake studies and in vitro efficacy assays constitutes an indispensable component of the quality control pipeline, bridging the gap between physical characterization and pre-clinical animal studies [92]. For researchers and drug development professionals, establishing robust, predictive functional assays is paramount for correlating exosome properties with biological activity, ensuring batch-to-batch consistency, and ultimately demonstrating therapeutic potency as defined by regulatory guidelines [92] [93].
This application note details standardized protocols for assessing the biological activity of ultracentrifugation-isolated MSC exosomes, framed within the broader context of a thesis on MSC exosome research. We provide detailed methodologies for visualizing exosome uptake by recipient cells and for quantifying functional outcomes in relevant in vitro disease models, with a focus on immunomodulation—a key therapeutic mechanism of MSC exosomes [44] [94].
Experimental Protocols
Protocol 1: Confocal Microscopy for Exosome Uptake and Localization
This protocol validates the fundamental prerequisite for bioactivity: the internalization of exosomes by target cells. The following workflow outlines the key stages of the experiment.
2.1.1 Materials and Reagents
- Purified MSC Exosomes: Isolated via ultracentrifugation (110,000× g) and resuspended in PBS [34].
- Lipophilic Tracers: PKH67 (green fluorescence) or PKH26 (red fluorescence) dyes.
- Ultracentrifugation Equipment: Beckman Coulter Optima series with Type 50.2 Ti rotor or equivalent.
- Target Cells: Immortalized cell lines (e.g., THP-1 monocytes, keratinocytes) or primary cells relevant to the disease model.
- Confocal Microscopy Dish: Glass-bottom dishes suitable for high-resolution imaging.
- Fixative: 4% Paraformaldehyde (PFA) in PBS.
- Counterstains: Phalloidin (for F-actin cytoskeleton), DAPI (for nuclei), and LysoTracker (for lysosomes).
2.1.2 Step-by-Step Procedure
- Exosome Labeling:
- Dilute the purified exosome preparation (typically 10-100 µg of protein) in 1 mL of Diluent C.
- Prepare a 2X dye solution by adding 4 µL of PKH67 or PKH26 ethanolic dye stock to 1 mL of Diluent C.
- Mix the exosome suspension with the dye solution immediately and incubate for 5 minutes at room temperature, protected from light.
- Stop the staining reaction by adding 2 mL of 1% Bovine Serum Albumin (BSA) in PBS.
- Remove unincorporated dye via ultracentrifugation at 110,000× g for 70 minutes at 4°C [72]. Carefully aspirate the supernatant and resuspend the labeled exosome pellet in an appropriate volume of sterile PBS.
Cell Seeding and Incubation:
- Seed target cells (e.g., human keratinocytes for psoriasis models [44]) onto glass-bottom confocal dishes at a density of 1-2 x 10^5 cells/dish and culture until 60-70% confluent.
- Gently add the labeled exosomes to the culture medium. A typical working concentration ranges from 10 to 50 µg exosome protein per mL of culture medium [94].
- Incubate the cells with exosomes for 4-24 hours at 37°C and 5% CO₂ to allow for uptake.
Cell Fixation and Staining:
- Aspirate the medium and wash the cells gently three times with PBS.
- Fix the cells with 4% PFA for 15 minutes at room temperature.
- Permeabilize and block with 0.1% Triton X-100 and 5% BSA in PBS for 30 minutes.
- Stain with Phalloidin (1:500) for 30 minutes to visualize the actin cytoskeleton and with DAPI (1 µg/mL) for 5 minutes to label nuclei. To track lysosomal trafficking, incubate live cells with LysoTracker Deep Red for 30 minutes prior to fixation.
Imaging and Analysis:
- Image the cells using a confocal laser scanning microscope with appropriate laser lines and filters for the fluorophores used.
- Acquire Z-stack images to confirm intracellular localization.
- Use image analysis software (e.g., ImageJ/Fiji) to perform co-localization analysis (e.g., Pearson's coefficient) between the exosome signal (PKH) and organelle markers (e.g., LysoTracker) to quantify trafficking and uptake efficiency.
Protocol 2: In Vitro Efficacy Assay for Immunomodulatory Activity
This protocol assesses a key therapeutic function of MSC exosomes: the suppression of inflammatory responses in immune cells, providing a direct measure of biological potency [44].
2.2.1 Materials and Reagents
- THP-1 Human Monocyte Cell Line.
- Phorbol 12-myristate 13-acetate (PMA).
- Lipopolysaccharide (LPS) and Interferon-gamma (IFN-γ).
- MSC Exosomes: Test articles isolated from different sources (e.g., umbilical cord, placenta, adipose tissue) can be compared [44] [94].
- ELISA Kits: For human TNF-α, IL-6, and IL-10.
- Flow Cytometry Antibodies: Anti-CD86 (M1 marker), Anti-CD206 (M2 marker), and appropriate isotype controls.
2.2.2 Step-by-Step Procedure
- Macrophage Differentiation and Polarization:
- Culture THP-1 monocytes in RPMI-1640 medium supplemented with 10% FBS.
- Differentiate THP-1 cells into macrophages by treating with 100 nM PMA for 48 hours in 12-well plates. Allow the differentiated macrophages to rest in fresh medium for 24 hours.
- Polarize the macrophages to a pro-inflammatory (M1) state by stimulating with 100 ng/mL LPS and 20 ng/mL IFN-γ for 24 hours.
Exosome Treatment:
- Following M1 polarization, treat the macrophages with MSC exosomes (10-50 µg/mL) for 24-48 hours. Include control wells with PBS (negative control) and known immunosuppressants (positive control).
- Use exosomes derived from different MSC sources (e.g., hUCMSC vs. hPMSC) to compare functional efficacy [44].
Sample Collection:
- After incubation, collect the cell culture supernatant and centrifuge at 300 × g for 10 minutes to remove any cellular debris. Aliquot and store at -80°C for cytokine analysis.
- For flow cytometry, harvest the cells by gentle scraping and wash with PBS.
Analysis of Immunomodulation:
- Cytokine Secretion Profile: Quantify the levels of TNF-α and IL-6 (pro-inflammatory) and IL-10 (anti-inflammatory) in the supernatant using commercial ELISA kits, following the manufacturer's protocols.
- Surface Marker Phenotype: Resuspend the harvested cells in flow cytometry buffer and stain with anti-CD86 (M1 marker) and anti-CD206 (M2 marker) antibodies for 30 minutes on ice, protected from light. Analyze the cells using a flow cytometer. Calculate the percentage of CD86+ and CD206+ cells and the mean fluorescence intensity (MFI).
Data Presentation and Analysis
The table below summarizes potential experimental outcomes, illustrating how data from these protocols can be synthesized to assess the potency of different MSC exosome preparations.
Table 1: Exemplary Data from In Vitro Functional Validation of MSC Exosomes
| Exosome Source | Uptake Efficiency (MFI) | TNF-α Secretion (pg/mL) | IL-10 Secretion (pg/mL) | M1/M2 Phenotype Ratio (CD86/CD206) | Inferred Potency |
|---|---|---|---|---|---|
| hUCMSC-Exos | 15,200 ± 1,100 | 350 ± 45 | 180 ± 20 | 1.5 ± 0.3 | High [44] |
| hPMSC-Exos | 14,800 ± 950 | 380 ± 50 | 165 ± 18 | 1.7 ± 0.4 | High |
| Ad-MSC-Exos | 12,500 ± 800 | 450 ± 60 | 140 ± 15 | 2.1 ± 0.5 | Moderate [94] |
| PBS Control | 250 ± 50 | 950 ± 110 | 45 ± 8 | 5.8 ± 0.9 | N/A |
The Scientist's Toolkit: Key Research Reagent Solutions
A curated list of essential materials and their critical functions for establishing these functional assays is provided below.
Table 2: Essential Research Reagents for Functional Validation Assays
| Reagent / Kit | Primary Function | Key Application Note |
|---|---|---|
| PKH67/PKH26 Dyes | Lipophilic membrane labeling for tracking. | Validates exosome uptake and intracellular trafficking; critical for Protocol 1. |
| Ultracentrifuge & Rotors | High-speed isolation and purification of exosomes. | Essential for post-labeling cleanup to remove dye artifacts [34]. |
| Confocal Microscope | High-resolution 3D imaging of cellular uptake. | Enables Z-stack imaging and co-localization analysis for Protocol 1. |
| THP-1 Cell Line | A model human monocyte line for immunology. | Can be differentiated into macrophages for potency assays in Protocol 2. |
| LPS & IFN-γ | Potent inducers of pro-inflammatory M1 macrophage polarization. | Creates a robust in vitro inflammation model for testing exosome efficacy. |
| TNF-α/IL-6/IL-10 ELISA Kits | Quantitative measurement of cytokine profiles. | Provides a key quantitative readout for immunomodulatory potency in Protocol 2. |
| CD86/CD206 Antibodies | Surface marker staining for macrophage phenotyping via flow cytometry. | Confirms functional phenotypic shift from M1 to M2, indicating anti-inflammatory activity. |
The integration of robust uptake studies and disease-relevant in vitro efficacy assays forms the cornerstone of a comprehensive functional validation strategy for MSC-derived exosomes. The protocols detailed herein provide a framework for researchers to quantitatively assess the biological activity of their ultracentrifugation-isolated exosomes, moving beyond mere physical characterization. By implementing these standardized assays, scientists can directly link critical quality attributes (CQAs) to specific biological functions, thereby de-risking the development path for exosome-based therapeutics and providing crucial evidence of potency required for regulatory approval [92] [93]. This systematic approach to functional validation is indispensable for realizing the full clinical potential of MSC exosomes in regenerative medicine and immunotherapy.
Conclusion
Ultracentrifugation remains a fundamental method for MSC exosome isolation, particularly when enhanced with techniques like sucrose cushion centrifugation to improve purity and yield. The choice of isolation methodology directly impacts exosome characteristics, purity, and biological functionality, making rigorous validation through multiple characterization techniques essential. As the field advances, future directions should focus on standardizing protocols for clinical translation, developing integrated systems that combine ultracentrifugation with other techniques like TFF for scalable GMP production, and further exploring preconditioning strategies to enhance exosome therapeutic potency. The continued optimization of ultracentrifugation protocols will play a crucial role in advancing MSC exosome research from bench to bedside, enabling their full potential as next-generation cell-free therapeutics in regenerative medicine and drug development.
References
- 1. Mesenchymal stem cell-derived exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC7477653/]
- 2. Mesenchymal Stem Cell-Derived Exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC5071792/]
- 3. Mesenchymal stem cell derived-exosomes: a modern ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-020-02622-3]
- 4. Mesenchymal stem cell-derived extracellular vesicles for ... [https://www.nature.com/articles/s41419-022-05034-x]
- 5. Mesenchymal stem cell-derived exosomes in cancer therapy ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-022-01650-5]
- 6. Mesenchymal stem cell-derived extracellular vesicles - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8327470/]
- 7. Isolation of Extracellular Vesicles Derived from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8032486/]
- 8. Engineered mesenchymal stem cell-derived exosomes [https://www.sciencedirect.com/science/article/pii/S2405580825004005]
- 9. Current understanding of the mesenchymal stem cell ... [https://www.sciencedirect.com/science/article/pii/S2215017X21000746]
- 10. Mesenchymal stem cell-derived exosomes: A paradigm ... [https://www.sciencedirect.com/science/article/abs/pii/S0014482725002125]
- 11. Advances in mesenchymal stem cell and exosome-based ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04318-1]
- 12. Large-scale separation and purification of exosomes using ... [https://bmrat.biomedpress.org/index.php/BMRAT/article/view/881]
- 13. Protocol for isolating and identifying small extracellular ... [https://www.sciencedirect.com/science/article/pii/S2666166724003629]
- 14. Comparative Proteomics of Seminal Exosomes Reveals ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561774/]
- 15. Comparison of the efficiency of ultrafiltration, precipitation ... [https://www.sciencedirect.com/science/article/pii/S2405580824000323]
- 16. Trends in mesenchymal stem cell-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488731/]
- 17. Current Landscape of FDA Stem Cell Approvals and Trials ... [https://www.reprocell.com/blog/current-landscape-of-fda-stem-cell-approvals-and-trials-2023-2025]
- 18. Mesenchymal stem cell-derived exosomes as cell-free ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12632147/]
- 19. Clinical applications of stem cell-derived exosomes [https://www.nature.com/articles/s41392-023-01704-0]
- 20. Regulation of cargo selection in exosome biogenesis and ... [https://www.nature.com/articles/s12276-024-01209-y]
- 21. Comprehensive proteomic analysis of exosome mimetic ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-022-03008-6]
- 22. Compounding engineered mesenchymal stem cell-derived ... [https://www.sciencedirect.com/science/article/pii/S0753332224003081]
- 23. Exosome Isolation Protocols and Quantification Guidelines [https://www.sepscience.com/exosome-isolation-protocols-and-quantification-guidelines-12144]
- 24. Efficient methods of isolation and purification of extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12463815/]
- 25. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells | Stem Cell Research & Therapy | Full Text [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-018-0923-0]
- 26. Mesenchymal stem cell-derived extracellular vesicles [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1626996/full]
- 27. Exosome Ultracentrifugation Protocol - Creative Proteomics [https://www.creative-proteomics.com/resource/exosome-ultracentrifugation-protocol.htm]
- 28. Cushioned–Density Gradient Ultracentrifugation (C–DGUC): a Refined and High Performance Method for the Isolation, Characterization & Use of Exosomes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6476194/]
- 29. Comparison of production methods for mesenchymal stem ... [https://www.nature.com/articles/s41598-025-21218-9]
- 30. 6 Safety Tips for Operating a Centrifuge [https://www.labmanager.com/6-safety-tips-for-operating-a-centrifuge-19835]
- 31. Mesenchymal stromal/stem cell (MSC)-derived exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10079493/]
- 32. Efficient ultrafiltration-based protocol to deplete extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5795649/]
- 33. Exosomes from conditioned media of bone marrow-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6872157/]
- 34. Ad-MSC exosome extraction [https://bio-protocol.org/exchange/minidetail?id=9313092&type=30]
- 35. Optimized exosome isolation protocol for cell culture ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4507751/]
- 36. Updated protocols for cell lysate, secretome, and exosome ... [https://www.sciencedirect.com/science/article/abs/pii/S1046202325000568]
- 37. Isolation of exosomes by differential centrifugation [https://www.nature.com/articles/srep17319]
- 38. Selective loss of microvesicles is a major issue of the ... [https://www.nature.com/articles/s41598-021-83241-w]
- 39. Small extracellular vesicles isolation and separation [https://pmc.ncbi.nlm.nih.gov/articles/PMC9516236/]
- 40. Exosomes Produced from 3D Cultures of MSCs by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6277553/]
- 41. Bringing exosomes into the game: Current situation ... [https://www.sciencedirect.com/science/article/pii/S2590049825000682]
- 42. Efficient methods of isolation and purification of extracellular ... [https://nanoconvergencejournal.springeropen.com/articles/10.1186/s40580-025-00509-x]
- 43. Three-dimensional culture of MSCs produces exosomes with ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-020-01719-2]
- 44. A Murine Study and Meta-Analysis of Experimental Studies [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467663/]
- 45. The role of natural exosomes from SHED-MSC in ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1550280/full]
- 46. Stem cell derived exosome trilogy: an epic comparison of ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04440-0]
- 47. Defined MSC exosome with high yield and purity to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8060739/]
- 48. Recent developments in isolating methods for exosomes [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2022.1100892/full]
- 49. Characterization and Comparison of Mesenchymal Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9985774/]
- 50. Characterization of Stem Cell Exosomes Isolated by ... [https://link.springer.com/article/10.1134/S2635167625600166]
- 51. Exosome Isolation: A Density-based Ultracentrifugation ... [https://www.jove.com/t/30119/exosome-isolation-density-based-ultracentrifugation-technique-using]
- 52. Exosomes Derived from Mesenchymal Stem Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3975389/]
- 53. Exosome isolation and purification [https://bio-protocol.org/exchange/minidetail?id=6641117&type=30]
- 54. Exosome Isolation by Ultracentrifugation and Precipitation [https://pmc.ncbi.nlm.nih.gov/articles/PMC8088761/]
- 55. An improvised one-step sucrose cushion ultracentrifugation method... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6033286/]
- 56. A simplified and efficient method for isolating small ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12066714/]
- 57. - Large separation and purification of scale using... exosomes [https://bmrat.org/index.php/BMRAT/article/view/881]
- 58. preconditioning strategies for MSC-derived extracellular ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1509418/full]
- 59. Mesenchymal stem cell-derived extracellular vesicles [https://pmc.ncbi.nlm.nih.gov/articles/PMC12399395/]
- 60. Improving the Thermal, Radial and Temporal Accuracy of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3826449/]
- 61. An optimized exosome production strategy for enhanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9618217/]
- 62. Exploring Regulatory Frameworks for Exosome Therapy [https://pmc.ncbi.nlm.nih.gov/articles/PMC12371722/]
- 63. Principles and Problems of Exosome Isolation from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9202659/]
- 64. Efficient dispersion of aggregated extracellular vesicles [https://www.nature.com/articles/s41598-025-10050-w]
- 65. Comparison of exosomes purified via ultracentrifugation ... [https://www.sciencedirect.com/science/article/abs/pii/S0166093418304233#!]
- 66. Protocol for the separation of extracellular vesicles by ... [https://www.sciencedirect.com/science/article/pii/S2666166721000101]
- 67. Extracellular Vesicle Depletion Protocols of Foetal Bovine ... [https://www.mdpi.com/1422-0067/24/11/9242]
- 68. Optimization of ultracentrifugation‐based method to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11263976/]
- 69. Quantification of Exosomes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5039048/]
- 70. Exosomes from preconditioned mesenchymal stem cells [https://pmc.ncbi.nlm.nih.gov/articles/PMC10875222/]
- 71. Advances in application of hypoxia-preconditioned ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11375678/]
- 72. Extracellular vesicles isolated from adipose tissue-derived ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04435-x]
- 73. Nanoparticle Tracking Analysis: An Effective Tool to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11477862/]
- 74. Exosome Processing and Characterization Approaches for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9130923/]
- 75. Nanoparticle Tracking Analysis (NTA) for Exosome ... [https://www.creative-biostructure.com/nanoparticle-tracking-analysis-nta-for-exosome-characterization.htm?srsltid=AfmBOoodHacqcnGsFg6IjgWE0G04yNKo1GQn6a7K2dtuvRjQDwjJstum]
- 76. Extracellular Vesicles from Different Mesenchymal Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11989379/]
- 77. Overview of extracellular vesicle characterization ... [https://www.spiedigitallibrary.org/journals/neurophotonics/volume-9/issue-2/021903/Overview-of-extracellular-vesicle-characterization-techniques-and-introduction-to-combined/10.1117/1.NPh.9.2.021903.full]
- 78. Transmission Electron Microscopy (TEM) for Exosome ... [https://www.creative-biostructure.com/transmission-electron-microscopy-tem-for-exosome-characterization.htm?srsltid=AfmBOopg792rYQe9r_bow-Ku0VSgL-5j8KcX0jjMXCq1C-CjNC3SmYen]
- 79. Exosome isolation and detection: Methods and applications [https://www.abcam.com/en-us/knowledge-center/cell-biology/exosome-isolation-and-detection]
- 80. How pure are your vesicles? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3760653/]
- 81. A comparison of methods for the isolation and separation ... [https://www.nature.com/articles/s41598-020-57497-7]
- 82. Comparative analysis of tangential flow filtration and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9486818/]
- 83. A simplified and efficient method for isolating small ... [https://www.nature.com/articles/s41598-025-99822-y]
- 84. Tangential Flow Filtration: A Complete Guide for ... [https://www.repligen.com/unit-operations/uf-df/tangential-flow-filtration-guide]
- 85. A comparison of centrifugation and tangential flow filtration ... [https://www.sciencedirect.com/science/article/abs/pii/S1674200119301026]
- 86. A Review of Exosomal Isolation Methods: Is Size Exclusion ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7556044/]
- 87. Extracellular vesicles isolated by size-exclusion ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-021-02775-9]
- 88. Comparison of six commercial serum exosome isolation ... [https://pubmed.ncbi.nlm.nih.gov/30990781/]
- 89. Comparison of an Optimized Ultracentrifugation Method ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6292670/]
- 90. Isolation of Exosomes from Blood Plasma: Qualitative and ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0145686]
- 91. Investigate the efficacy of size exclusion chromatography ... [https://www.sciencedirect.com/science/article/abs/pii/S1570023223003926]
- 92. Functional assays to assess the therapeutic potential of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7890556/]
- 93. Overcoming challenges in MSC-sEV therapeutics [https://www.sciencedirect.com/science/article/pii/S1465324925005912]
- 94. Mesenchymal stromal/stem cell (MSC)-derived exosomes in ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-023-03287-7]