Unveiling the Mechanisms: How MSC Exosomes Enter and Reprogram Keratinocytes and Endothelial Cells for Regenerative Therapy
This article provides a comprehensive analysis of the cellular uptake mechanisms of Mesenchymal Stem Cell (MSC)-derived exosomes by keratinocytes and endothelial cells, two critical cell types in cutaneous wound healing...
Unveiling the Mechanisms: How MSC Exosomes Enter and Reprogram Keratinocytes and Endothelial Cells for Regenerative Therapy
Abstract
This article provides a comprehensive analysis of the cellular uptake mechanisms of Mesenchymal Stem Cell (MSC)-derived exosomes by keratinocytes and endothelial cells, two critical cell types in cutaneous wound healing and vascular repair. We explore the foundational biology of exosome internalization, methodological approaches for tracking and enhancing uptake, strategies to overcome experimental and therapeutic bottlenecks, and comparative validation of exosomes from different MSC sources. Tailored for researchers and drug development professionals, this review synthesizes current evidence and technological advances to guide the rational design of more effective exosome-based nanotherapeutics for regenerative medicine.
The Cellular Gateway: Foundational Principles of MSC Exosome Uptake
Exosomes are nanoscale extracellular vesicles that play a critical role in intercellular communication through their specialized cargo. This technical guide provides a comprehensive examination of exosome biogenesis pathways, cargo sorting mechanisms, and compositional analysis. Framed within research on mesenchymal stem cell (MSC) exosome uptake by keratinocytes and endothelial cells, this review synthesizes current understanding of these sophisticated biological entities and their potential therapeutic applications in regenerative medicine.
Exosome Biogenesis: Cellular Production Pathways
Exosome biogenesis involves a meticulously orchestrated intracellular process that begins with endocytosis and culminates in extracellular release. These vesicles are defined by their endosomal origin and characteristic size range of 30-200 nanometers in diameter [1] [2]. The biogenesis pathway can be categorized into four distinct phases, each regulated by specific molecular machinery.
Early Endosome Formation
The biogenesis pathway initiates with the inward budding of the plasma membrane, forming early sorting endosomes [3] [4]. This process is regulated by specific protein complexes including:
- Clathrin-mediated endocytosis: Facilitates concentration of cargo into clathrin-coated pits [3]
- Caveolin-1 dependent pathways: Particularly important in caveolae generation and membrane invagination [3]
- Rab GTPase proteins: Especially Rab5a, which when knocked down decreases exosome excretion [3]
The formation of early endosomes can be influenced by tubular carriers containing MICAL-like protein 1 (MICAL-L1) and syndapin 2, a Bin/amphiphysin/Rvs (BAR) domain protein that inserts into the endosomal bilayer structure and bends the membrane [3].
Multivesicular Body Formation and Maturation
Early endosomes undergo significant transformation into late endosomes and subsequently into multivesicular bodies (MVBs) through a second inward budding process that creates intraluminal vesicles (ILVs) within larger endosomal compartments [1] [3]. Two primary mechanisms regulate this critical step:
ESCRT-Dependent Pathway: The endosomal sorting complex required for transport (ESCRT) comprises approximately 30 proteins organized into four distinct complexes (ESCRT-0, -I, -II, and -III) along with associated proteins including VPS4 and Alix [1]. This machinery operates sequentially: ESCRT-0 recognizes and sorts ubiquitinated intracellular cargos; ESCRT-I and -II deform the membrane into buds with sequestered vesicles; and ESCRT-III facilitates vesicle scission [1].
ESCRT-Independent Pathways: Several alternative mechanisms can generate ILVs without ESCRT involvement:
- Tetraspanin-mediated pathways: Tetraspanins (CD63, CD9, CD37, CD82, CD81) participate in extracellular vesicle biogenesis and are essential for secretion and uptake [1]
- Lipid-dependent mechanisms: Neutral sphingomyelinase 2 (nSMase2) contributes to ILV formation through lipid metabolism [3]
- Additional effectors: Flotillins and cholesterol also participate in the budding process [3]
MVB Fate Determination and Exosome Release
MVBs face one of two potential fates: degradation through fusion with lysosomes or autophagosomes, or release of exosomes through fusion with the plasma membrane [1] [3] [5]. The molecules responsible for MVB docking and fusion with the plasma membrane include:
- Rab GTPase proteins: Particularly Rab27, which regulates vesicle trafficking and membrane fusion [5]
- SNARE proteins: Facilitate membrane fusion events [2]
- Calcium channels and cellular pH: Modulate the exocytosis process [2]
This final fusion event releases the ILVs as exosomes into the extracellular space, where they can interact with recipient cells [1].
Table 1: Key Molecular Regulators of Exosome Biogenesis
| Biogenesis Stage | Regulatory Molecules | Primary Function |
|---|---|---|
| Early Endosome Formation | Clathrin, Caveolin-1, Rab5a | Membrane invagination and vesicle formation |
| MVB Formation | ESCRT complexes (0, I, II, III), VPS4, Alix | Cargo sorting and ILV formation |
| ESCRT-Independent Formation | Tetraspanins (CD63, CD9, CD81), nSMase2 | Alternative biogenesis pathways |
| MVB Fate Determination | Rab GTPases (Rab27), SNARE proteins | Vesicle trafficking and membrane fusion |
Figure 1: Exosome Biogenesis Pathway. This diagram illustrates the sequential stages of exosome formation, from early endosome generation to eventual exosome release or degradation, highlighting key regulatory molecules at each step.
Exosome Cargo Composition and Sorting Mechanisms
Exosomes carry a diverse molecular payload that reflects their cellular origin and physiological state. This cargo is strategically sorted through specific mechanisms that ensure appropriate composition and function.
Molecular Constituents of Exosomes
Exosomes contain complex biomolecular arrays that can be categorized into several classes:
Protein Content:
- Transmembrane proteins: Tetraspanins (CD9, CD63, CD81), MHC classes I and II, adhesion molecules [1] [2] [4]
- Cytosolic proteins: Heat shock proteins (HSP70, HSP90), transcription factors, enzymes (ATPase, phosphoglycerate kinase) [1] [2]
- Biogenesis-related proteins: ALIX, TSG101, ESCRT machinery components [2] [5]
- Cytokines and chemokines: Various signaling molecules [4]
Nucleic Acid Composition:
- RNA species: microRNA (miRNA), messenger RNA (mRNA), non-coding RNAs including circular and long noncoding RNAs [1] [2] [4]
- DNA components: Mitochondrial DNA (mtDNA), single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), genomic DNA fragments [6] [5]
Lipid Profile: Exosomal membranes are enriched in specific lipids including sphingomyelin (SM), desaturated phosphatidylethanolamine, phosphatidylserine (PS), desaturated phosphatidylcholine (PC), cholesterol (CHOL), GM3, and ganglioside [1]. This unique lipid composition contributes to membrane rigidity and stability while facilitating cellular uptake.
Cargo Sorting Mechanisms
The selective packaging of molecules into exosomes occurs through sophisticated sorting mechanisms:
ESCRT-Mediated Sorting: The ESCRT machinery not only facilitates ILV formation but also participates in cargo selection, particularly for ubiquitinated proteins [1] [3].
Tetraspanin-Organized Microdomains: Tetraspanins create specialized membrane platforms that recruit specific client proteins for incorporation into exosomes [1].
Lipid-Dependent Sorting: Ceramide and other lipids contribute to the formation of lipid rafts that facilitate the sorting of specific proteins into exosomes [1].
RNA-Binding Protein Coordination: RNA motifs and RNA-binding proteins (such as hnRNPs) mediate the selective packaging of RNA species into exosomes [4].
Table 2: Major Exosome Cargo Components and Their Functions
| Cargo Category | Specific Examples | Biological Functions |
|---|---|---|
| Transmembrane Proteins | CD9, CD63, CD81, MHC-I/II | Vesicle identification, antigen presentation |
| Intracellular Proteins | HSP70, HSP90, ALIX, TSG101 | Stress response, biogenesis regulation |
| Nucleic Acids | miRNA, mRNA, circRNA, mtDNA | Genetic regulation, horizontal gene transfer |
| Lipids | Cholesterol, sphingomyelin, phosphatidylserine | Membrane stability, signaling, cellular uptake |
| Signaling Molecules | Cytokines, chemokines, growth factors | Intercellular communication, immune modulation |
Experimental Methodologies for Exosome Research
Isolation and Purification Techniques
Multiple approaches have been developed for exosome isolation, each with distinct advantages and limitations:
Ultracentrifugation: Considered the gold standard technique, differential ultracentrifugation involves sequential separation based on size and density [7] [2] [4]. While it provides relatively high purity and requires minimal reagents, it is time-consuming, requires expensive instrumentation, and may cause damage to exosomes or co-isolate lipoproteins [2] [4].
Size-Based Techniques:
- Size-exclusion chromatography (SEC): Separates exosomes based on size differences, preserving integrity and bioactivity but potentially co-isolating similar-size vesicles [7]
- Tangential flow filtration (TFF): Uses parallel flow dynamics to reduce clogging potential, suitable for large-scale applications [2]
Polymer Precipitation: Utilizes hydrophilic polymers to force exosomes out of solution, offering ease of use but potential contamination with non-vesicular components [7].
Immunoaffinity Capture: Employs antibodies against exosome surface markers (CD9, CD63, CD81) for high-purity isolation, though it depends on surface antigen expression and may miss subpopulations [7] [2] [4].
Microfluidics-Derived Techniques: Emerging approaches that offer rapid processing with small sample volumes, though not yet widely established [7].
Combining multiple complementary methods often performs better in reducing contamination, improving separation purity, and maintaining natural exosome characteristics [7].
Characterization and Validation Methods
Comprehensive exosome characterization requires multi-parametric analysis:
Physical Characterization:
- Nanoparticle tracking analysis (NTA): Determines particle size distribution and concentration
- Dynamic light scattering (DLS): Measures size distribution of particles in suspension
- Transmission electron microscopy (TEM): Visualizes exosome morphology and structure
Biochemical Characterization:
- Western blotting: Detects specific protein markers (CD9, CD63, CD81, TSG101, ALIX)
- Flow cytometry: Analyzes surface markers, though size limitations require special instrumentation
- Proteomic/lipidomic/genomic analysis: Comprehensive profiling of molecular cargo
Researchers should adhere to MISEV (Minimal Information for Studies of Extracellular Vesicles) guidelines to ensure reproducibility and quality in exosomal research [7].
MSC Exosome Uptake by Keratinocytes and Endothelial Cells
The therapeutic potential of MSC-derived exosomes is particularly relevant for skin regeneration and vascular repair, processes dependent on efficient uptake by keratinocytes and endothelial cells.
Uptake Mechanisms
Exosomes utilize multiple pathways to enter recipient cells, with preference depending on exosome characteristics and the target cell type:
Fusion: Direct merging with the plasma membrane, resulting in release of contents intracellularly, mediated by SNARE and Rab proteins [2].
Endocytosis:
- Clathrin-mediated endocytosis: A stepwise process involving clathrin-coated vesicle formation [3] [2]
- Caveolin-mediated endocytosis: Dependent on caveolin proteins, particularly caveolin-1 in epithelial cells [2]
- Lipid raft-mediated endocytosis: Utilizes sphingolipid- and cholesterol-enriched membrane microdomains [2]
- Macropinocytosis: Actin-driven process creating membrane extensions that internalize extracellular materials [2]
Phagocytosis: Primarily observed in professional phagocytes like macrophages, involving membrane deformation and phagosome formation [2].
Receptor-Mediated Interactions: Specific ligand-receptor interactions facilitate targeted binding and subsequent internalization [4].
Functional Consequences of Uptake
Upon internalization by keratinocytes and endothelial cells, MSC exosomes exert pleiotropic effects:
Keratinocyte Responses:
- Enhanced proliferation and migration: Critical for re-epithelialization during wound healing [6] [8]
- Modulation of differentiation: Influences keratinocyte maturation and barrier function
- Cytoprotection: Increased resistance to oxidative stress and apoptosis
Endothelial Cell Responses:
- Angiogenic stimulation: Promotes tube formation and vascular network development [6] [9]
- Barrier function enhancement: Improves endothelial integrity
- Proliferation and migration: Facilitates vascular repair and regeneration
The molecular mechanisms underlying these effects involve the delivery of regulatory miRNAs, proteins, and lipids that modulate key signaling pathways including PI3K/AKT, Wnt/β-catenin, and TGF-β/Smad [6].
Figure 2: MSC Exosome Uptake Mechanisms and Functional Outcomes in Target Cells. This diagram illustrates the various pathways through which MSC-derived exosomes enter keratinocytes and endothelial cells, and the subsequent biological effects that promote tissue repair and regeneration.
Research Reagent Solutions for Exosome Studies
Table 3: Essential Research Tools for Exosome Isolation, Characterization, and Functional Analysis
| Research Tool Category | Specific Examples | Primary Applications | Technical Considerations |
|---|---|---|---|
| Isolation Kits | Polymer-based precipitation kits, Immunoaffinity columns (CD9/CD63/CD81) | Rapid exosome isolation from biological fluids | Potential co-precipitation of contaminants; antibody specificity critical |
| Characterization Antibodies | Anti-CD9, CD63, CD81, TSG101, ALIX, HSP70 | Exosome validation by western blot, flow cytometry, immunofluorescence | Confirm specificity for species of interest; optimize concentration |
| Tracking Dyes | PKH67, PKH26, DiI, DiD, CFSE, GFP-labeled markers | Exosome labeling for uptake and trafficking studies | Potential dye aggregation; validate non-toxic concentrations |
| Cell Culture Supplements | Exosome-depleted FBS, defined growth factors | Production of exosomes in controlled conditions | Verify exosome depletion efficiency; maintain cell viability |
| Knockdown/CRISPR Tools | siRNA against Rab27a, nSMase2, ESCRT components; CRISPR for tetraspanins | Functional studies of biogenesis mechanisms | Confirm knockdown efficiency; monitor compensatory mechanisms |
| Analysis Kits | BCA protein assay, RNA extraction kits optimized for exosomes | Cargo quantification and analysis | Account for low RNA yields; use sensitive detection methods |
Experimental Protocols for Key Investigations
Protocol: MSC Exosome Uptake by Keratinocytes
Objective: Quantify and visualize internalization of MSC-derived exosomes by human keratinocytes.
Materials:
- MSC-derived exosomes isolated via ultracentrifugation or SEC
- Primary human keratinocytes
- Fluorescent membrane dyes (e.g., PKH67)
- Confocal microscopy equipment
- Flow cytometer with appropriate detectors
Procedure:
- Exosome Labeling:
- Resuspend exosome pellet (100 μg protein) in 1 mL Diluent C
- Add PKH67 ethanol dye solution (2 μM final concentration)
- Incubate 5 minutes at room temperature, protected from light
- Add equal volume of 1% BSA to stop staining reaction
- Isolate labeled exosomes by ultracentrifugation (100,000 × g, 70 minutes)
- Wash with PBS and repeat centrifugation to remove unincorporated dye
Uptake Assay:
- Plate keratinocytes in 24-well plates (5 × 10^4 cells/well) and culture until 70% confluent
- Add labeled exosomes (20 μg/mL) to cells and incubate for 0, 15, 30, 60, 120 minutes
- For inhibition studies, pre-treat cells with endocytosis inhibitors:
- Chlorpromazine (10 μM, clathrin-mediated inhibition)
- Filipin (5 μg/mL, caveolae-mediated inhibition)
- Cytochalasin D (10 μM, macropinocytosis inhibition)
Analysis:
- Flow cytometry: Trypsinize cells, wash with PBS, and analyze fluorescence intensity (Ex/Em: 490/502 nm)
- Confocal microscopy: Fix cells with 4% PFA, stain actin with phalloidin-TRITC, mount with DAPI-containing medium, and image using appropriate filter sets
Protocol: Functional Angiogenesis Assay with Endothelial Cells
Objective: Assess pro-angiogenic effects of MSC exosomes on endothelial tube formation.
Materials:
- Human umbilical vein endothelial cells (HUVECs)
- Growth factor-reduced Matrigel
- MSC exosomes (0.35-1.75 μg/mL concentration range) [7]
- Tubule formation analysis software
Procedure:
- Matrigel Preparation:
- Thaw Matrigel overnight at 4°C
- Coat 96-well plates (50 μL/well) and polymerize at 37°C for 30 minutes
Tube Formation Assay:
- Serum-starve HUVECs for 4 hours
- Trypsinize, count, and resuspend cells in exosome-containing medium (2 × 10^4 cells/100 μL)
- Plate cells on Matrigel-coated plates
- Incubate at 37°C, 5% CO₂ for 4-8 hours
Quantification:
- Capture images using phase-contrast microscopy (4× objective)
- Analyze tube parameters: total tube length, number of branch points, number of meshes
- Compare experimental groups to positive (complete medium) and negative (serum-free) controls
Exosome biogenesis and cargo composition represent fundamental biological processes with significant implications for therapeutic development. The intricate molecular machinery governing exosome formation, cargo sorting, and cellular uptake provides multiple points for scientific investigation and potential intervention. MSC-derived exosomes offer particular promise as cell-free therapeutic agents, leveraging their natural trafficking capabilities to deliver complex molecular payloads to target cells like keratinocytes and endothelial cells. As research methodologies continue to advance, particularly in single-vesicle analysis and engineered exosome technologies, our understanding of these sophisticated nanoscale communicators will undoubtedly expand, opening new avenues for regenerative medicine and targeted therapeutic applications.
Exosomes, nanoscale extracellular vesicles (30-150 nm), are fundamental mediators of intercellular communication, transferring functional proteins, lipids, and nucleic acids between cells [10]. Their uptake by recipient cells is a critical step for eliciting biological effects and is of paramount importance for developing exosome-based therapeutic applications [11] [12]. In the context of regenerative medicine, understanding how mesenchymal stem cell (MSC)-derived exosomes are internalized by specific target cells like keratinocytes and endothelial cells is a central research focus [13] [14]. These uptake processes are not random but are highly regulated by the exosome's cellular origin, surface composition, and the recipient cell's type and state [10] [12]. This whitepaper provides an in-depth technical overview of the universal mechanisms—membrane fusion, endocytosis, and receptor-mediated internalization—that govern exosome uptake, with a specific frame of reference for research involving MSC exosome interactions with keratinocytes and endothelial cells.
Major Exosome Uptake Pathways
Exosomes utilize a complex array of pathways to deliver their cargo into recipient cells. The primary mechanisms are membrane fusion, various forms of endocytosis, and phagocytosis, each leading to distinct intracellular fates for the vesicle and its cargo [10] [13] [12].
Table 1: Major Pathways of Exosome Uptake by Recipient Cells
| Uptake Mechanism | Key Molecular Regulators | Intracellular Fate | Implications for Cargo Delivery |
|---|---|---|---|
| Membrane Fusion | SNARE proteins [11] | Direct release of cargo into cytoplasm | Avoids endolysosomal degradation; direct access to cytosolic targets |
| Clathrin-Mediated Endocytosis | Clathrin, Dynamin, Chlorpromazine-sensitive pathways [15] [16] | Trafficking to early endosomes, then to lysosomes | Cargo can be degraded; requires escape from endosomes for bioactive delivery |
| Caveolae-Mediated Endocytosis | Caveolin-1, Dynamin, Nystatin-sensitive pathways [16] [13] | Trafficking to caveosomes | Bypasses classical endolysosomal pathway; alternative delivery route |
| Macropinocytosis | Actin, Na+/H+ exchangers, Amiloride-sensitive pathways [13] | Trafficking to macropinosomes, then to lysosomes | Non-specific uptake of extracellular fluid and vesicles; cargo subject to degradation |
| Phagocytosis | Actin cytoskeleton (primarily in phagocytes) [13] | Trafficking to phagolysosomes | Primarily in specialized cells; strong degradation environment |
| Clathrin-Independent Endocytosis | Galectin-3, Lysosome-associated membrane protein-2B (LAMP2B), Dynamin [16] | Recycling pathways [16] | Facilitated by paracrine adhesion signaling; may avoid degradation |
The following diagram illustrates the logical progression of these key uptake mechanisms and their intracellular trajectories.
Receptor-Mediated Internalization and Adhesion
The initial attachment of exosomes to the recipient cell plasma membrane is a critical, receptor-mediated step that often dictates the subsequent internalization pathway [11] [16]. This adhesion is far from a simple docking event; it can trigger active signaling within the recipient cell that facilitates the ultimate uptake of the vesicle.
Table 2: Key Molecules in Exosome Adhesion and Receptor-Mediated Internalization
| Adhesion Molecule Category | Specific Molecules on Exosome / Recipient Cell | Function in Uptake | Cell Type / Context |
|---|---|---|---|
| Integrins | αvβ3/β5, β1 (CD29), α3 (CD49c), αL (CD11a) [11] | Mediate firm adhesion; trigger intracellular signaling; determine organotropism [11] | Endothelial cells, Keratinocytes, Cancer cells |
| Tetraspanins | CD9, CD63, CD81, CD82 [11] | Form platforms that spatially organize receptors; influence signal induction [11] | Ubiquitous exosome markers; various cell types |
| Immunoglobulin Superfamily | ICAM-1 (on exosome) / LFA-1 (on cell) [11] | Critical for initial binding/docking, especially in immune contexts [11] | Dendritic cells, T cells |
| Lectin Families | Galectin-3 [16] | Binds glycoproteins; mediates clathrin-independent endocytosis [16] | Tumor cells, Endothelial cells |
| Other Adhesion Proteins | CD169 (sialoadhesin) on macrophages [11]; Heparin sulfate proteoglycans [11] | Capture and internalize exosomes [11] | Macrophages, Glioblastoma cells, Kidney cells |
| MHC & Antigen Presentation | MHC Class I & II with antigen [11] | Directs exosomes to antigen-specific T cells [11] | Antigen-presenting cells, T cells |
A pivotal finding is that the adhesion of paracrine exosomes (derived from different cells) to recipient cells can trigger intracellular Ca2+ mobilization via activation of Src family kinases and phospholipase Cγ (PLCγ). This Ca2+ signal subsequently activates the calcineurin–dynamin machinery, which directly promotes exosome internalization, often routing them into recycling pathways [16]. This indicates that the recipient cell is an active participant in the uptake process, not a passive vessel.
Experimental Protocols for Studying Uptake
Elucidating the specific pathway used in a given biological context requires carefully designed experiments involving chemical inhibition, genetic manipulation, and advanced imaging.
Pharmacological Inhibition of Specific Pathways
A standard methodology to dissect the contribution of different endocytic routes is the use of specific pharmacological inhibitors [15].
Protocol: Inhibitor-Based Pathway Analysis
- Cell Seeding: Plate recipient cells (e.g., keratinocytes or endothelial cells) in multi-well plates or on glass coverslips and allow them to adhere and grow to ~70% confluency.
- Inhibitor Pre-treatment: Incubate cells with optimized concentrations of pathway-specific inhibitors in serum-free media for 30 minutes to 1 hour prior to exosome addition. Common inhibitors include:
- Exosome Incubation: Add fluorescently labeled exosomes (e.g., DiO, DiD, PKH67) to the pre-treated cells. Incubate for a defined period (e.g., 1-4 hours) at 37°C.
- Wash and Analysis: Thoroughly wash cells with PBS to remove non-internalized exosomes. Analyze internalization using:
- Imaging Flow Cytometry: Provides quantitative, high-throughput data on the percentage of cells with internalized exosomes and the fluorescence intensity per cell [15].
- Confocal Microscopy: Allows visual confirmation of intracellular localization of exosomes, typically appearing as punctate dots within the cell cytoplasm, and can be used to perform Z-stack analysis to confirm internalization versus surface binding [15].
Single-Particle Tracking and Super-Resolution Imaging
To overcome the limitations of bulk population assays and directly observe the behavior of individual exosomes, state-of-the-art imaging techniques are employed.
Protocol: Single-SEV Particle Tracking [16]
- Exosome Labeling: Label exosomes with lipophilic dyes (e.g., DiO) or genetically engineer donor cells to express tetraspanins (CD63, CD9, CD81) fused with fluorescent proteins (e.g., mGFP, Halo7-tag).
- Image Acquisition: Use high-speed, high-sensitivity microscopy such as Total Internal Reflection Fluorescence Microscopy (TIRFM) or single-particle tracking setups to visualize individual exosome particles in living cells.
- Simultaneous Imaging: Combine single-particle tracking of exosomes with Photoactivated Localization Microscopy (PALM) to simultaneously observe membrane invaginations and other structural changes in the recipient cell's plasma membrane with super-resolution.
- Data Analysis: Analyze trajectories to determine the kinetics of binding, mode of entry (e.g., confined diffusion before internalization), and co-localization with markers of specific pathways (e.g., clathrin, caveolin).
Signaling Pathways in Exosome Uptake
The internalization of exosomes is not merely a mechanical process but can be regulated by, and in turn regulate, specific intracellular signaling cascades. Research in the context of MSC exosomes and their target cells has highlighted several key pathways.
Table 3: Key Signaling Pathways in Exosome Uptake and Function
| Signaling Pathway | Role in Uptake / Subsequent Function | Cell Type / Context |
|---|---|---|
| Src / PLCγ / Ca²⁺ Signaling | Paracrine exosome binding triggers Ca²⁺ mobilization, activating calcineurin and dynamin to drive internalization [16]. | Recipient cells (e.g., endothelial cells) |
| Wnt/β-catenin | MSC exosomes can deliver Wnt4, activating this pathway in recipient cells to promote proliferation and migration [17]. | Keratinocytes, Endothelial cells |
| MAPK Signaling | Uptake of MSC exosomes can modulate p38 MAPK and ERK pathways, influencing inflammatory responses and cell survival [17]. | Chondrocytes, Macrophages |
| Oxidative Stress (NRF2) | MSC exosomes reduce oxidative stress in recipient cells, a key mechanism in their regenerative effects [17]. | Keratinocytes, Neuronal cells |
The following diagram summarizes the key signaling pathway triggered by paracrine exosome adhesion.
The Scientist's Toolkit: Key Research Reagents
Successful investigation of exosome uptake mechanisms relies on a suite of essential reagents and tools.
Table 4: Essential Reagents for Exosome Uptake Research
| Reagent / Tool Category | Specific Examples | Function / Application |
|---|---|---|
| Fluorescent Labels | DiO, DiD, PKH67, PKH26, CellTracker CM-Dil [15] | Label exosome membranes for visualization and tracking by flow cytometry and microscopy. |
| Pharmacological Inhibitors | Chlorpromazine, Nystatin, Bafilomycin A1, Amiloride, Dynasore [15] | Selectively block specific endocytic pathways to determine their contribution to uptake. |
| Genetic Tools | siRNAs/shRNAs (e.g., against clathrin, caveolin, dynamin); Plasmid vectors for fluorescent protein-tagged tetraspanins (CD63-GFP) [16] | Knock down or visualize components of the uptake machinery to study their role. |
| Antibodies for Staining | Anti-CD63, Anti-CD9, Anti-CD81, Anti-TSG101, Anti-Alix [14] | Characterize exosomes and confirm their identity via Western Blot, ELISA, or immunostaining. |
| Advanced Microscopy Systems | Imaging Flow Cytometry [15], Confocal Microscopy, TIRF Microscopy, dSTORM/PALM Super-Resolution Microscopy [16] | Visualize, quantify, and track exosome uptake at both population and single-particle levels. |
The universal mechanisms of exosome uptake—membrane fusion, endocytosis, and receptor-mediated internalization—are complex, dynamic, and context-dependent. For researchers focusing on MSC exosome interactions with keratinocytes and endothelial cells, the key takeaways are that uptake is an active process heavily influenced by surface adhesion molecules like integrins and tetraspanins, and that it can trigger specific signaling cascades within the recipient cell. The choice of experimental methodology, from pharmacological inhibition to sophisticated single-particle tracking, is critical for accurately delineating these pathways. A deep understanding of these mechanisms is the foundation for rationally designing engineered exosomes with enhanced targeting and delivery efficiency for therapeutic applications in wound healing, vascular regeneration, and beyond.
Keratinocyte-Specific Uptake Pathways and Post-Uptake Signaling Activation (e.g., PI3K/Akt, ERK)
Keratinocytes, the predominant cell type in the epidermis, rely on sophisticated intracellular signaling networks to regulate their core functions: proliferation, differentiation, migration, and programmed cell death. The phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) and mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathways represent two crucial signaling axes that determine keratinocyte fate in both physiological and pathological contexts. These pathways integrate signals from the extracellular environment—including those from mesenchymal stem cell-derived exosomes (MSC-Exos)—to coordinate appropriate cellular responses during processes such as wound healing and epidermal homeostasis [18] [19] [20].
The PI3K/Akt pathway functions as a critical regulator balancing keratinocyte differentiation against apoptotic death, while the MAPK/ERK pathway primarily directs proliferative responses and capillary morphogenesis. Understanding the precise mechanisms through which these pathways are activated following the uptake of external cues, such as extracellular vesicles, provides fundamental insights for developing novel therapeutic strategies in regenerative medicine and dermatological disorders [21] [19] [20]. This technical guide examines the molecular machinery governing keratinocyte-specific uptake pathways and the subsequent signaling activation, with particular emphasis on their relevance to MSC exosome research.
PI3K/Akt Signaling in Keratinocytes
Pathway Mechanism and Regulation
The PI3K/Akt pathway serves as a central signaling node that determines the fate choice between keratinocyte differentiation and death. This pathway is activated during early stages of keratinocyte differentiation both in vitro and in intact epidermis in vivo [19]. Pathway activation initiates when extracellular signals stimulate receptor tyrosine kinases such as the epidermal growth factor receptor (EGFR) and Src families, leading to PI3K recruitment and conversion of phosphatidylinositol (4,5)-bisphosphate (PIP2) to phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the plasma membrane [18] [19].
Akt (protein kinase B) is then recruited to the membrane through its pleckstrin homology domain, where it undergoes phosphorylation and activation. Importantly, research demonstrates that PI3K/Akt activation in keratinocyte differentiation depends on E-cadherin-mediated adhesion, with PI3K increasingly associating with cadherin-catenin protein complexes bearing tyrosine-phosphorylated YXXM motifs during this process [19]. This membrane-proximal signaling complex integrates adhesion signals with growth factor signaling to fine-tune keratinocyte responses.
Functional Consequences of Pathway Activation
PI3K/Akt signaling promotes keratinocyte growth arrest and differentiation while protecting against premature apoptosis during this transition. Experimental evidence confirms that expression of active Akt in keratinocytes directly promotes growth arrest and differentiation, whereas pharmacological blockade of PI3K inhibits expression of late differentiation markers and leads to death of cells that would otherwise differentiate [19]. This pathway therefore represents a critical survival signal during keratinocyte differentiation, ensuring that cells complete their differentiation program rather than undergoing apoptotic death.
The functional outcomes of PI3K/Akt activation extend to wound healing contexts, where keratinocyte migration and re-epithelialization are essential. Keratinocyte-derived extracellular vesicles contain proteins that influence these processes, including integrins, growth factors, and matrix metalloproteinases that interact with PI3K/Akt signaling outputs [20].
Experimental Modulation and Assessment
Investigating PI3K/Akt signaling in keratinocytes requires specific methodological approaches, as outlined in Table 1. These include pharmacological inhibitors, genetic manipulation techniques, and assessment methodologies for evaluating pathway activity and functional outcomes.
Table 1: Experimental Approaches for Studying PI3K/Akt Signaling in Keratinocytes
| Method Category | Specific Approach | Key Reagents/Tools | Output Measurements |
|---|---|---|---|
| Pharmacological Inhibition | PI3K pathway blockade | LY294002, Wortmannin | Differentiation marker expression, apoptosis assays |
| Genetic Manipulation | Constitutive activation | Active Akt constructs | Growth arrest, differentiation markers |
| Pathway Assessment | Phosphorylation status | Phospho-specific Akt antibodies (Ser473, Thr308) | Western blot, immunofluorescence |
| Functional Assays | Differentiation capacity | Calcium-induced differentiation model | Late differentiation markers (involucrin, loricrin) |
| Adhesion Studies | E-cadherin engagement | Calcium switch assays | Co-immunoprecipitation of cadherin-catenin complexes |
MAPK/ERK Signaling in Keratinocytes
Pathway Mechanism and Regulation
The MAPK/ERK pathway represents another crucial signaling cascade in keratinocytes, particularly in contexts of angiogenesis and wound healing. This pathway is activated in response to various stimuli, including growth factors, cytokines, and physical cues such as electric fields (EF) [21]. The canonical RAF-MEK-ERK phosphorylation cascade begins with RAS activation, progressing through sequential phosphorylation of RAF, MEK, and ultimately ERK, which then translocates to the nucleus to regulate gene expression.
In microvascular endothelial cells, which share signaling similarities with keratinocytes during wound responses, EF exposure has been shown to enhance capillary morphogenesis and promote MEK-cRaf complex formation along with subsequent MEK and ERK phosphorylation [21]. This activation occurs in a frequency-dependent manner, with high-frequency EF (7.5 GHz) proving more effective than low-frequency (60 Hz) stimulation. Importantly, EF-induced MEK phosphorylation can be reversed by MEK and Ca²⁺ inhibitors, reduced by endothelial nitric oxide synthase (eNOS) inhibition, and operates independently of PI3K pathway activation [21].
Functional Consequences of Pathway Activation
MAPK/ERK signaling drives keratinocyte functions essential for wound healing, including proliferation, migration, and the secretion of factors that support angiogenesis. Activation of this pathway enhances vascular endothelial growth factor (VEGF) release, a key angiogenic factor that promotes neovascularization in healing tissues [21]. The ERK pathway also influences keratinocyte interactions with other cell types, including fibroblasts and immune cells, through regulation of cytokine and chemokine production.
Notably, the endothelial response to EF that activates MAPK/ERK does not require VEGF binding to its receptor VEGFR2, indicating that this pathway can be initiated through alternative mechanisms relevant to tissue regeneration strategies [21]. This finding has significant implications for understanding how physical stimulation approaches might enhance healing in compromised wound environments.
Experimental Modulation and Assessment
Research into MAPK/ERK signaling employs distinct methodological approaches, particularly when investigating responses to physical stimuli like electric fields. Table 2 outlines key experimental parameters and assessment methods for studying this pathway.
Table 2: Experimental Approaches for MAPK/ERK Signaling Investigation
| Parameter | High-Frequency EF | Low-Frequency EF | Assessment Methods |
|---|---|---|---|
| Frequency | 7.5 GHz | 60 Hz | Phospho-ERK/MEK immunoblotting |
| Field Intensity | 156 mV/mm | 209 mV/mm | Capillary morphogenesis assays |
| Setup | Cavity resonator | Parallel-plate capacitor | VEGF measurement (ELISA) |
| Key Inhibitors | MEK inhibitors (U0126), Ca²⁺ inhibitors | MEK inhibitors, eNOS inhibitors | Raf-MEK co-immunoprecipitation |
| Biological Effects | Enhanced capillary formation, VEGF release | Reduced response | Cell proliferation/migration assays |
Extracellular Vesicle Uptake and Signaling Activation
Keratinocyte-Derived Extracellular Vesicles
Keratinocytes actively secrete extracellular vesicles (EVs), including exosomes and microvesicles, that carry diverse molecular cargo capable of influencing recipient cell behavior. Keratinocyte-derived EVs contain characteristic membrane proteins (ITGA6, CD9, CD63) and cytoplasmic proteins (HSPA5, eEF1A1, SDCBP) that facilitate skin development and repair [20]. These EVs also carry specialized proteins including transforming growth factor beta (TGF-β), epidermal growth factor (EGF), involucrin, kallikrein 7 (KLK7), jagged 1 (JAG1), plasminogen activator inhibitor 1 (PAI-1), and multiple matrix metalloproteinases (MMP-1, -3, -8, -9) that collectively influence wound re-epithelialization, extracellular matrix remodeling, and cellular adhesion/migration [20].
The biological state of keratinocytes determines EV composition, with differentiated versus undifferentiated keratinocytes releasing distinct exosomal populations containing different isoforms of 14-3-3 proteins [20]. Similarly, activated migrating keratinocytes secrete EVs containing cathepsin B, which participates in intracellular proteolysis during wound healing. This state-dependent variation in EV content represents a mechanism for fine-tuning cellular responses during tissue repair.
MSC Exosomes as Signaling Modulators
Mesenchymal stem cell-derived exosomes (MSC-Exos) have emerged as powerful acellular therapeutic tools that can modify regenerative programs in recipient cells, including keratinocytes, by delivering functional RNAs, proteins, and other signaling elements [22] [6]. These nanoscale vesicles precisely regulate inflammatory responses, angiogenesis, and tissue repair processes by targeting central signaling pathways in keratinocytes, including PI3K/Akt, JAK/STAT, TGF-β/Smad, and Wnt/β-catenin cascades [6].
MSC-Exos offer significant advantages over whole-cell therapies, including low immunogenicity, efficient biological barrier penetration, stable storage characteristics, and reduced risks of tumorigenicity [22]. Their capacity to regulate macrophage activation, stimulate angiogenesis, and promote keratinocyte and dermal fibroblast proliferation and migration makes them particularly valuable for dermatological applications and wound healing [6]. Worldwide, 64 registered clinical trials have preliminarily validated the safety and applicability of MSC-EVs across various diseases, showing significant progress in treating complex wound healing, among other conditions [22].
Integration of Signaling Pathways in Keratinocyte Functions
Cross-Talk Between PI3K/Akt and MAPK/ERK Pathways
While often studied separately, the PI3K/Akt and MAPK/ERK pathways exhibit significant cross-talk in keratinocytes, creating a signaling network that integrates multiple inputs to determine cellular responses. Both pathways can be simultaneously activated by common upstream signals, including receptor tyrosine kinase engagement and integrin-mediated adhesion events. The balanced activation of these pathways likely determines whether keratinocytes primarily undergo differentiation (favored by PI3K/Akt) versus proliferation (favored by MAPK/ERK).
This cross-talk becomes particularly relevant in the context of MSC exosome therapy, as these vesicles deliver complex cargo that may simultaneously modulate multiple signaling pathways. Understanding the integrated response of these pathways to exosomal components is essential for predicting and optimizing therapeutic outcomes in regenerative applications.
Therapeutic Implications for Skin Disorders and Wound Healing
The strategic manipulation of keratinocyte signaling pathways holds significant promise for treating various dermatological conditions and enhancing wound healing. Dysregulated PI3K/Akt signaling contributes to pathological conditions such as psoriasis, while proper activation of this pathway supports keratinocyte differentiation and barrier formation [23]. Similarly, controlled MAPK/ERK activation promotes the re-epithelialization crucial for healing chronic wounds, including those in diabetic patients [21] [24].
MSC exosomes represent a promising vehicle for delivering targeted modulation of these pathways, as they can be engineered to enrich specific miRNA or protein cargo that preferentially activates desired signaling outcomes. Current research focuses on enhancing exosome targeting, optimizing production processes, and understanding long-term biodistribution to facilitate clinical translation of these approaches [22].
Visualization of Signaling Pathways
Keratinocyte Signaling Pathway Diagram
Diagram Title: Keratinocyte Signaling Pathways Integration
This diagram illustrates the integrated signaling network in keratinocytes, highlighting how external stimuli including MSC exosomes, electric fields, growth factors, and cell adhesion events converge on the PI3K/Akt and MAPK/ERK pathways. The visualization emphasizes key activation steps and demonstrates potential cross-talk between these crucial signaling axes that determine keratinocyte fate decisions.
Experimental Workflow for Signaling Studies
Diagram Title: Experimental Workflow for Signaling Studies
This workflow outlines a systematic approach for investigating keratinocyte signaling pathways, from initial cell culture through treatment application, molecular analysis, functional assessment, and final data integration. The methodology supports comprehensive evaluation of how MSC exosomes and other stimuli modulate PI3K/Akt and MAPK/ERK signaling networks.
Research Reagent Solutions
Table 3: Essential Research Reagents for Keratinocyte Signaling Studies
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| PI3K/Akt Inhibitors | LY294002, Wortmannin | Selective PI3K inhibition | Confirm specificity via downstream phosphorylation |
| Akt Activators | SC79 | Allosteric Akt activation | Validate with phosphorylation-specific antibodies |
| MAPK Pathway Inhibitors | U0126 (MEK inhibitor) | Blocks ERK phosphorylation | Use appropriate concentrations to avoid off-target effects |
| Calcium Modulators | BAPTA-AM, Thapsigargin | Modulate intracellular Ca²⁺ | Essential for EF studies and adhesion signaling |
| eNOS Inhibitors | L-NAME | Reduces nitric oxide production | Important for mechanotransduction studies |
| Keratinocyte Culture Models | Primary keratinocytes, HaCaT cells | Physiological relevance vs. immortalized line | Primary cells better reflect in vivo differentiation |
| EV Isolation Tools | Ultracentrifugation, Size-exclusion chromatography, Immunoprecipitation | Isolation of keratinocyte or MSC-derived EVs | Method affects yield, purity, and biological activity |
| Differentiation Inducers | High-calcium medium | Induces keratinocyte differentiation | Essential for studying differentiation-linked signaling |
| Phospho-Specific Antibodies | Anti-pAkt (Ser473, Thr308), Anti-pERK, Anti-pMEK | Detection of pathway activation | Validate with appropriate controls and inhibition |
| Adhesion Molecules | Recombinant E-cadherin, Anti-E-cadherin antibodies | Study adhesion-mediated signaling | Critical for investigating mechanotransduction pathways |
Keratinocyte PI3K/Akt and MAPK/ERK signaling pathways represent sophisticated regulatory networks that integrate diverse external cues to determine cellular fate decisions. The PI3K/Akt pathway critically balances differentiation versus apoptotic death, while MAPK/ERK signaling directs proliferative and migratory responses essential for tissue repair. These pathways can be activated through multiple mechanisms, including MSC exosome uptake, electric field exposure, growth factor receptor engagement, and adhesion-mediated signaling.
Understanding the precise molecular mechanisms governing these pathways provides a foundation for developing targeted therapeutic strategies in regenerative medicine and dermatology. MSC exosomes represent particularly promising delivery vehicles for modulating these pathways, offering natural targeting capabilities, low immunogenicity, and complex cargo that can simultaneously engage multiple signaling nodes. Future research focusing on exosome engineering, pathway cross-talk, and in vivo validation will further enhance our ability to harness these signaling networks for therapeutic benefit.
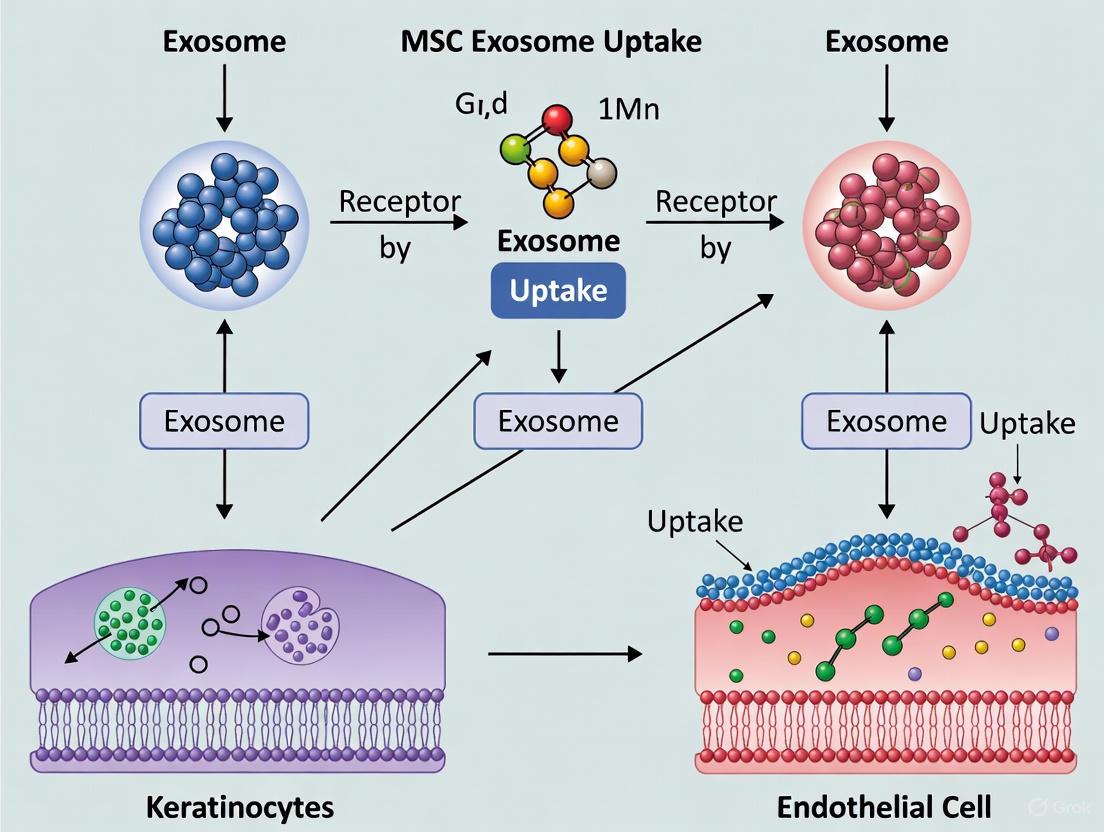
Endothelial Cell Uptake Dynamics and Induction of Pro-angiogenic Responses
The uptake of mesenchymal stromal cell (MSC)-derived exosomes by endothelial cells represents a crucial mechanistic pathway in therapeutic angiogenesis. As natural nanoscale vesicles, exosomes facilitate intercellular communication by transferring bioactive cargo—including proteins, lipids, and nucleic acids—from donor MSCs to recipient endothelial cells, initiating a cascade of pro-angiogenic responses [25] [4]. This process is particularly relevant in the context of wound healing and tissue regeneration, where the formation of new blood vessels is essential for restoring oxygen and nutrient supply to damaged tissues [25] [8]. Understanding the precise dynamics of exosome uptake and the subsequent intracellular signaling events in endothelial cells provides a foundation for developing novel therapeutic strategies for conditions characterized by impaired angiogenesis, such as diabetic wounds, ischemic diseases, and other vascular insufficiencies [26] [27].
Mechanisms of Exosome Uptake by Endothelial Cells
The process of exosome internalization by endothelial cells is a coordinated sequence of events that leads to the delivery of exosomal cargo and the initiation of downstream signaling pathways. The journey begins with the initial contact and docking, where exosomes present surface molecules that interact with recipient endothelial cells [4]. Key surface proteins involved in this recognition include tetraspanins (CD9, CD63, CD81), integrins, and other adhesion molecules that facilitate binding to the endothelial cell membrane [20] [4].
Following initial contact, exosomes enter endothelial cells through multiple endocytic pathways. The predominant mechanism involves endocytosis, where the exosomes are engulfed through membrane invagination to form early sorting endosomes [4]. These early endosomes then mature into late sorting endosomes and subsequently form multivesicular bodies (MVBs) after a second indentation [4]. The final critical step involves the release of exosomal contents into the endothelial cell cytoplasm through the fusion of MVBs with the cell membrane or through direct membrane fusion, allowing the bioactive cargo to access intracellular compartments and modulate cellular functions [4].
The entire lifecycle from exosome biogenesis to uptake and intracellular signaling can be tracked using fluorescent, luminescent, and radioactive techniques, providing researchers with tools to visualize and quantify these dynamic processes [4].
Diagram 1: Endothelial cell exosome uptake and signaling pathway. This diagram illustrates the sequential process from MSC exosome release through cellular uptake mechanisms to final pro-angiogenic activation in endothelial cells.
Pro-angiogenic Responses in Endothelial Cells
Upon successful internalization and cargo release, MSC-derived exosomes initiate comprehensive pro-angiogenic programming in endothelial cells. This multifaceted response encompasses several critical processes that collectively contribute to new blood vessel formation.
Enhancement of Endothelial Cell Migration and Proliferation
The fundamental processes of endothelial cell migration and proliferation are significantly enhanced by exosomal exposure. Research has demonstrated that exosomes stimulate endothelial cell migration, inducing coverage of scratched surface areas up to 110 ± 31% compared to 47 ± 13% in negative controls based on scratch test assays [25]. This enhanced migratory capacity is essential for the initial stages of angiogenesis, allowing endothelial cells to navigate toward angiogenic stimuli. Simultaneously, exosomes promote endothelial cell proliferation through the delivery of growth factors and regulatory miRNAs that stimulate cell cycle progression and mitogenic signaling pathways [25] [27].
Induction of Tube Formation and Vascular Network Assembly
The most functionally significant outcome of exosome-mediated pro-angiogenic activation is the induction of capillary-like tube formation. In vitro tube formation assays using "ECM Gel Matrix" have quantified this effect, demonstrating that exosomes generate tube-like structures with complexity similar to VEGF-positive controls [25]. The pro-angiogenic effect is quantified through multiple parameters, including the number of junctions and meshes, as well as total tube length, all of which show significant enhancement following exosome treatment [25]. This structured assembly into tubular networks represents the culmination of the angiogenic process, resulting in the creation of new vascular structures capable of supporting blood flow.
Molecular Signaling Pathways Activated by Exosomal Cargo
The pro-angiogenic effects of MSC-derived exosomes are mediated through the activation of key molecular signaling pathways within endothelial cells. The VEGF-VEGFR signaling axis serves as a central regulator of this process [28]. When vascular endothelial growth factor receptors (VEGFRs) are activated, they recruit PI3K, initiating the PI3K/Akt pathway which directs cell growth, survival, and migration [28]. Simultaneously, VEGFR stimulation activates the MAPK cascade, including ERK, which is essential for endothelial cell proliferation and movement [28]. VEGFR signaling also upregulates endothelial nitric oxide synthase (eNOS) and matrix metalloproteinases (MMPs), both of which support vascular growth, cell motility, and new vessel formation [28].
Table 1: Quantitative Pro-angiogenic Effects of MSC-Derived Exosomes on Endothelial Cells
| Angiogenic Parameter | Experimental Results | Experimental Method | Significance |
|---|---|---|---|
| Cell Migration | 110 ± 31% surface coverage | Scratch wound assay | Enhanced capacity for endothelial cell movement toward angiogenic stimuli |
| Tube Formation | Increased junctions, meshes, and total tube length | ECM Gel Matrix tube formation assay | Promotion of capillary-like structure assembly |
| Comparative Angiogenic Potential | Similar to VEGF-positive control | Comparative assay with VEGF control | Demonstration of potent pro-angiogenic activity |
Experimental Protocols for Studying Uptake and Angiogenic Responses
Exosome Isolation and Characterization
The investigation of endothelial cell uptake dynamics and pro-angiogenic responses requires standardized methodologies for exosome isolation and characterization. The current gold standard for exosome extraction is ultracentrifugation, which involves sequential centrifugation steps to separate exosomes from other extracellular vesicles and contaminants [4]. For enhanced purity, immunoaffinity chromatography utilizing antibodies against exosomal surface markers (CD9, CD63, CD81) provides high specificity, though it requires known surface antigen expression [4]. Additional techniques include size-exclusion chromatography and precipitation-based methods, each with distinct advantages and limitations in terms of yield, purity, and scalability [4].
Comprehensive characterization of isolated exosomes should include:
- Nanoparticle Tracking Analysis: To determine size distribution and concentration, typically showing exosomes in the 30-150 nm range [25] [4]
- Transmission Electron Microscopy: To confirm cup-shaped morphology characteristic of exosomes [25]
- Western Blot Analysis: To detect exosomal markers (CD9, CD63, CD81, TSG101, Alix) and exclude contaminants such as calnexin from endoplasmic reticulum membranes [25] [29]
- Flow Cytometry: To verify surface markers and quantify specific antigen presence [25]
Tracking Exosome Uptake Dynamics
Visualizing and quantifying exosome internalization by endothelial cells is essential for establishing uptake dynamics. Fluorescent labeling using lipophilic dyes such as PKH67 provides a robust method for tracking exosomes over time [25]. Following incubation with labeled exosomes, endothelial cells are fixed at predetermined time points and analyzed using confocal microscopy to determine the efficiency and kinetics of uptake. Additionally, techniques such as fluorescence anisotropy and fluorescence correlation spectroscopy can provide quantitative data on exosome-binding interactions [29].
Functional Angiogenesis Assays
The functional consequences of exosome uptake are evaluated through a series of standardized angiogenesis assays:
- Tube Formation Assay: Endothelial cells are seeded on "ECM Gel Matrix" and monitored for their ability to form capillary-like structures. Key parameters include the number of junctions, number of meshes, and total tube length, which are quantified using image analysis software [25]
- Scratch Wound Migration Assay: A standardized scratch is created in a confluent endothelial cell monolayer, and cell migration to close the wound is measured over 12-24 hours with and without exosome treatment [25]
- Proliferation Assays: Endothelial cell proliferation in response to exosome treatment is quantified using metabolic activity assays (e.g., MTT, CCK-8) or by directly counting cell numbers over time [25]
Diagram 2: Experimental workflow for exosome uptake and angiogenesis studies. This diagram outlines the sequential methodology from initial exosome isolation through characterization and functional assessment of pro-angiogenic effects.
Research Reagent Solutions for Angiogenesis Studies
Table 2: Essential Research Reagents for Studying Exosome-Mediated Angiogenesis
| Research Reagent/Category | Specific Examples | Research Application | Function in Experimental Design |
|---|---|---|---|
| Exosome Isolation Tools | Ultracentrifugation, Immunoaffinity (CD9/CD63/CD81), Size-exclusion chromatography | Exosome purification | Separation of exosomes from conditioned media or biological fluids based on physical properties or surface markers |
| Characterization Antibodies | Anti-CD9, Anti-CD63, Anti-CD81, Anti-TSG101, Anti-Alix, Anti-calnexin (negative) | Exosome validation | Confirmation of exosomal identity and purity through Western blot, flow cytometry, or immunofluorescence |
| Tracking Dyes | PKH67, PKH26, other lipophilic dyes | Uptake visualization | Fluorescent labeling of exosome membranes to track internalization by endothelial cells over time |
| ECM Matrices | ECM Gel Matrix, Matrigel | Tube formation assay | Providing a basement membrane substitute that supports endothelial cell organization into capillary-like structures |
| Angiogenesis Assay Kits | Tube formation assay kits, Migration assay kits | Functional assessment | Standardized systems for quantifying pro-angiogenic responses of endothelial cells following exosome treatment |
| Endothelial Cell Markers | CD31, VE-cadherin, vWF | Cell type validation | Confirmation of endothelial cell identity and assessment of phenotypic changes during angiogenesis |
The uptake dynamics of MSC-derived exosomes by endothelial cells and the subsequent induction of pro-angiogenic responses represent a sophisticated biological process with significant therapeutic implications. The molecular mechanisms involve precise exosome-receptor interactions, efficient internalization, and activation of key signaling pathways including VEGF-VEGFR, PI3K/Akt, and MAPK cascades [28]. The functional outcomes—enhanced migration, proliferation, and tube formation—collectively contribute to the formation of new vascular networks essential for tissue repair and regeneration [25]. As research in this field advances, the potential for harnessing these mechanisms for therapeutic angiogenesis in conditions characterized by vascular insufficiency continues to grow, offering promising avenues for the development of novel treatments for wound healing, ischemic diseases, and other vascular disorders.
Key Exosomal Cargo (miRNAs, Proteins) Implicated in Functional Modulation of Recipient Cells
Exosomes, nanoscale extracellular vesicles secreted by nearly all cell types, have emerged as pivotal mediators of intercellular communication, fundamentally advancing our understanding of cellular crosstalk in tissue homeostasis and repair. These lipid-bilayer enclosed vesicles transport a sophisticated cargo of proteins, lipids, and nucleic acids from donor to recipient cells, thereby modulating recipient cell function and phenotype [30] [4]. Within the context of skin biology and the specific research focus on MSC exosome uptake mechanisms by keratinocytes and endothelial cells, exosomal cargo orchestrates critical wound healing processes including keratinocyte migration, angiogenesis, and inflammatory modulation [9] [31]. This technical guide provides an in-depth analysis of the key functional cargos, with a particular emphasis on microRNAs (miRNAs), their quantitative profiles, mechanisms of action, and the experimental methodologies essential for elucidating their role in recipient cell modulation.
Exosomal Cargo: Composition and Functional Significance
Exosomes encapsulate a diverse array of biomolecules that reflect the physiological state of their parental cells and confer functional specificity. The cargo includes conserved tetraspanin proteins (CD9, CD63, CD81), heat shock proteins (HSP70, HSP90), biogenesis-related proteins (ALIX, TSG101), and major histocompatibility complexes, which serve as canonical exosomal markers [32] [4]. Beyond these structural components, the functionally active cargo includes:
- Nucleic Acids: DNA, mRNAs, microRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) that can regulate gene expression in target cells [30] [32].
- Proteins: Growth factors (VEGF, TGF-β, EGF), cytokines, and enzymes that directly activate signaling pathways [30].
- Lipids: Ceramide, sphingomyelin, phosphatidylserine, and cholesterol that contribute to membrane stability and biological activity [30].
The molecular composition of exosomes varies significantly based on cellular origin and extracellular environment. For instance, exosomes from three-dimensional dermal papilla spheroids contain elevated pro-hair growth miRNAs compared to those from 2D monolayers, while serum exosomes from psoriasis patients show elevated miR-199a-3p correlated with disease severity [30]. This compositional plasticity enables exosomes to perform context-specific functions, making them particularly valuable as therapeutic agents and diagnostic biomarkers.
Key Exosomal miRNAs and Their Functional Roles
MicroRNAs represent one of the most extensively studied and functionally significant exosomal cargo components. These small non-coding RNAs typically regulate gene expression by binding to target mRNAs, leading to translational repression or mRNA degradation. The table below summarizes key exosomal miRNAs implicated in skin repair and their specific effects on keratinocytes and endothelial cells.
Table 1: Key Exosomal miRNAs in Skin Repair and Recipient Cell Modulation
| miRNA | Exosome Source | Target Cells | Molecular Targets/Pathways | Functional Outcomes | Reference |
|---|---|---|---|---|---|
| miR-27b | Human Umbilical Cord Mesenchymal Stem Cells (HUMSCs) | Keratinocytes, Fibroblasts | ITCH/JUNB/IRE1α signaling | Activates keratinocytes and fibroblasts in vitro, accelerates wound healing in vivo | [30] |
| miR-181c | HUMSCs | Immune cells, Endothelial cells | TLR4/NF-κB pathway | Reduces inflammatory cytokine production | [30] |
| miR-21-3p | Mesenchymal Stem Cells (MSCs) | Endothelial cells, Fibroblasts | PI3K/Akt and ERK1/2 signaling | Promotes angiogenesis and enhances fibroblast function | [30] [33] |
| miR-223 | Bone Marrow MSCs (BMSCs) | Macrophages | Undefined | Promotes M2 polarization of macrophages | [30] |
| miR-146a | Adipose-derived MSCs (ADMSCs) | Endothelial cells | Src kinase | Mitigates endothelial cell senescence, promotes angiogenesis in diabetic models | [30] |
| miR-135a | Human Amnion MSCs | Keratinocytes, Fibroblasts | LATS2 (Hippo pathway kinase) | Inhibits LATS2, activates YAP/TAZ signaling, enhances keratinocyte migration and proliferation | [31] |
| miR-126 | Mesenchymal Stem Cells | Keratinocytes, Endothelial cells | PI3K/Akt and MAPK pathways | Promotes epithelial cell survival and proliferation, enhances angiogenesis | [31] |
| miR-4505 | Keratinocytes (VDR-deficient) | Macrophages | Undefined | Promotes macrophage proliferation and M1 polarization (psoriasis pathogenesis) | [32] |
| miR-291a-3p | Embryonic Stem Cells (ESCs) | Dermal Fibroblasts | TGF-β receptor 2 | Reduces cellular senescence markers, suppresses TGF-β signaling | [31] |
| miR-199a-3p | Serum (Psoriasis patients) | Skin cells | Undefined | Elevated in psoriasis, correlates with disease severity | [30] |
The mechanistic actions of these miRNAs illustrate sophisticated regulatory networks. For instance, miR-135a-mediated inhibition of LATS2 kinase leads to subsequent activation of YAP/TAZ signaling, a crucial pathway for cell proliferation and migration [31]. Similarly, miR-146a targeting of Src kinase mitigates endothelial senescence, particularly relevant in diabetic wound healing where cellular senescence is prevalent [30]. The functional specificity of these exosomal miRNAs enables precise modulation of recipient cell behavior, making them potent therapeutic candidates.
Key Exosomal Proteins and Their Functional Roles
While miRNAs provide sophisticated gene regulation, exosomal proteins often deliver immediate functional signals to recipient cells. The protein cargo includes surface receptors, enzymes, and growth factors that directly activate signaling pathways in target cells.
Table 2: Key Exosomal Proteins in Skin Repair and Recipient Cell Modulation
| Protein Cargo | Exosome Source | Target Cells | Molecular Targets/Pathways | Functional Outcomes | Reference |
|---|---|---|---|---|---|
| VEGF | Multiple cell sources | Endothelial cells | VEGF Receptor | Promotes angiogenesis, endothelial cell proliferation and migration | [33] [9] |
| TGF-β | Mesenchymal Stem Cells | Fibroblasts, Keratinocytes | SMAD pathway | Modulates cell proliferation, differentiation, and immune regulation | [30] |
| EGF | Multiple cell sources | Keratinocytes, Fibroblasts | EGFR pathway | Promotes epithelial cell proliferation and migration | [30] |
| Wnt4 | Mesenchymal Stem Cells | Endothelial cells | β-catenin pathway | Promotes angiogenesis | [30] |
| Cytoplasmic PLA2 | Mast Cells (IFN-α treated) | T cells | CD1a lipid presentation | Generates neo-lipid antigens, induces IL-22 and IL-17A production (psoriasis) | [32] |
| Olfactomedin 4 | Neutrophils (Generalized Pustular Psoriasis) | Keratinocytes | MAPK and NF-κB pathways | Induces inflammatory gene expression (IL-36G, TNF-α, IL-1β) | [32] |
| Tetraspanins (CD9, CD63, CD81) | Virtually all exosomes | Recipient cells | Cell adhesion, fusion, and uptake | Facilitates exosome uptake by recipient cells, mediates recipient cell targeting | [30] [32] |
The synergistic action of protein and miRNA cargo within individual exosomes creates powerful combinatorial effects. For instance, exosomes may simultaneously deliver miR-21-3p to activate PI3K/Akt signaling and VEGF protein to directly stimulate VEGF receptors, producing a potent angiogenic response [30] [33]. This multi-component signaling approach enhances the efficacy and specificity of exosomal communication compared to single-factor therapies.
Experimental Protocols for Cargo Analysis and Functional Validation
Exosome Isolation and Characterization
Standardized methodologies are crucial for reproducible exosome research. The following protocols represent current best practices:
Isolation Methods:
- Differential Ultracentrifugation: Considered the gold standard, this method involves sequential centrifugation steps (300 × g for 10 min to remove cells; 2,000 × g for 20 min to remove debris; 10,000 × g for 30 min to remove larger vesicles; and 100,000 × g for 70 min to pellet exosomes) [4] [34]. A washing step with phosphate-buffered saline followed by a final 100,000 × g centrifugation for 70 minutes improves purity by removing co-isolated proteins.
- Size-Exclusion Chromatography (SEC): Separates exosomes from contaminating proteins based on size using porous polymer beads. Quick and suitable for large-scale applications, though potential pore clogging and exosome loss require consideration [4].
- Immunoaffinity Capture: Uses antibodies against exosomal surface markers (CD9, CD63, CD81) for high-purity isolation. Ideal for specific exosome subpopulations but may not capture all exosome varieties [4].
Characterization Techniques:
- Nanoparticle Tracking Analysis (NTA): Determines exosome size distribution and concentration [4].
- Transmission Electron Microscopy (TEM): Visualizes exosome morphology and ultrastructure [4] [34].
- Western Blotting: Confirms presence of exosomal markers (CD9, CD63, CD81, TSG101, ALIX) and absence of negative markers (calnexin, GM130) [4] [34].
- Flow Cytometry: With appropriate instrumentation, can detect and quantify exosomes based on surface markers [4].
The International Society for Extracellular Vesicles (ISEV) MISEV (Minimal Information for Studies of Extracellular Vesicles) guidelines provide essential standardization for these procedures [34].
Cargo Profiling and Functional Validation
miRNA Profiling:
- RNA Extraction: Use commercial kits specifically validated for small RNA isolation from exosomes.
- Next-Generation Sequencing: Provides comprehensive, unbiased miRNA profiling. Bioinformatics analysis identifies differentially expressed miRNAs.
- qRT-PCR Validation: Confirm sequencing results using TaqMan assays specifically designed for miRNA quantification.
Protein Analysis:
- Mass Spectrometry: Liquid chromatography with tandem mass spectrometry (LC-MS/MS) enables proteomic profiling of exosomal cargo.
- Western Blotting: Validates presence of specific proteins of interest.
- Antibody Arrays: High-throughput screening for specific protein classes (cytokines, growth factors).
Functional Validation:
- Gain/Loss-of-Function Studies: Transfer candidate miRNAs into exosomes via engineered parent cells or direct loading. Inhibit specific miRNAs using antagomiRs.
- Luciferase Reporter Assays: Validate direct miRNA-mRNA interactions by cloning putative target sequences downstream of a luciferase gene.
- In Vitro Functional Assays:
- Keratinocyte Migration Scratch Assay: Measure wound closure rates with/without exosome treatment.
- Endothelial Tube Formation Assay: Assess angiogenic potential on Matrigel.
- Macrophage Polarization: Flow cytometry analysis of M1/M2 markers (CD86, CD206) following exosome treatment.
Diagram 1: Experimental workflow for exosomal cargo analysis and functional validation, covering isolation to in vivo studies.
Signaling Pathways Regulated by Exosomal Cargo
Exosomal miRNAs and proteins converge on several key signaling pathways that regulate fundamental processes in skin biology. The following diagram illustrates the major pathways implicated in keratinocyte and endothelial cell modulation:
Diagram 2: Key signaling pathways in keratinocytes and endothelial cells modulated by MSC exosomal cargo.
The pathway diagram illustrates how exosomal cargo coordinates multiple processes simultaneously. For instance, while miR-135a promotes keratinocyte proliferation through Hippo pathway inhibition, miR-146a concurrently reduces endothelial senescence through Src targeting, and miR-181c dampens inflammation through NF-κB inhibition [30] [32] [31]. This multi-target approach explains the superior efficacy of exosome therapies compared to single-factor treatments.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Reagents for Exosome Cargo Research
| Reagent/Category | Specific Examples | Primary Function/Application | Key Considerations |
|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Kits (various vendors) | Rapid precipitation-based isolation from cell media or biofluids | Can co-precipitate contaminants; suitable for screening but may require validation with standard methods |
| Characterization Antibodies | Anti-CD63, Anti-CD9, Anti-CD81, Anti-TSG101, Anti-ALIX, Negative markers: Anti-Calnexin | Western blot confirmation of exosomal identity and purity | Essential for MISEV compliance; validate species reactivity |
| miRNA Analysis Kits | Small RNA Isolation Kits, miRNA Sequencing Kits, TaqMan MicroRNA Assays | Extraction, profiling, and validation of exosomal miRNAs | Select kits optimized for low-concentration small RNA; include spike-in controls for normalization |
| Proteomic Tools | Mass Spectrometry kits, Antibody Arrays for growth factors/cytokines | Comprehensive protein cargo profiling | Require specialized equipment; consider core facility collaboration |
| Functional Assay Kits | Endothelial Tube Formation Assay (Matrigel), Cell Migration Assay (Boyden chamber), Cell Proliferation Assays (CCK-8, EdU) | In vitro validation of exosome functional effects | Standardize exosome quantification (particle number vs. protein amount) |
| Engineering Tools | Transfection reagents (for parent cell modification), Electroporation systems (for direct loading), Click-chemistry kits for tracking | Modify exosomal cargo for gain/loss-of-function studies | Optimization critical for efficiency and maintaining exosome integrity |
| Tracking Reagents | Lipophilic dyes (DiI, DiD), Membrane dyes (PKH67, PKH26), Quantum dots | Label exosomes for uptake and biodistribution studies | Potential dye aggregation; include proper controls to distinguish membrane labeling from uptake |
This toolkit provides the foundational resources for conducting rigorous exosome cargo research. Selection should be guided by specific research questions, with particular attention to standardization across experiments to ensure reproducibility.
The sophisticated cargo of exosomes, particularly miRNAs and proteins, represents a powerful mechanism for functional modulation of recipient cells that is highly relevant to MSC exosome uptake by keratinocytes and endothelial cells. The precise targeting of key signaling pathways including PI3K/Akt, Hippo/YAP, Wnt/β-catenin, and NF-κB enables exosomes to coordinate complex processes such as re-epithelialization, angiogenesis, and inflammation resolution. As research advances, engineered exosomes with enhanced or specific cargo loading represent the next frontier in therapeutic development [33] [4]. The standardization of isolation protocols, functional assays, and analytical techniques remains crucial for translating these findings into clinical applications for wound healing, skin regeneration, and the treatment of inflammatory skin diseases.
From Observation to Application: Methodologies for Tracking and Leveraging Uptake
In the field of regenerative medicine, understanding the mechanisms by which recipient cells internalize mesenchymal stem cell-derived exosomes (MSC-Exos) is paramount for advancing therapeutic applications. Research focusing on keratinocytes and endothelial cells—key players in skin regeneration and vascular repair—has highlighted the need for sophisticated methodological approaches to visualize and quantify exosome uptake. This technical guide details three cornerstone techniques: electron microscopy for ultrastructural analysis, PKH26/lipophilic dye labeling for membrane integration studies, and immunofluorescence for specific antigen detection. Each method offers unique insights into the dynamics of MSC exosome uptake, a process critical for mediating therapeutic effects in conditions ranging from radiation-induced skin injury to diabetic wound healing [31] [8].
Electron Microscopy for Ultrastructural Visualization
Transmission Electron Microscopy (TEM) provides nanometer-scale resolution, enabling researchers to visualize the precise subcellular compartments involved in exosome internalization. For studying MSC exosome uptake by keratinocytes, a powerful specific technique involves combining the lipophilic dye PKH26 with diaminobenzidine (DAB) photo-oxidation [35].
Detailed Protocol: PKH26 Staining and DAB Photo-Oxidation for TEM
The following procedure allows for the correlation of fluorescence microscopy observations with high-resolution electron microscopy images.
- Cell Culture and Staining: Plate human keratinocyte cells (e.g., HaCaT cell line) or endothelial cells on glass coverslips. Upon reaching 70-80% confluence, stain the cells using the PKH26 Red Fluorescent Cell Linker Kit according to the manufacturer's instructions. After staining, replace the dye-containing medium with fresh culture medium and incubate for various time points (e.g., 0 min, 30 min, 1 h, 3 h) to capture different stages of internalization [35].
- Fixation: Wash the cells with phosphate-buffered saline (PBS) and fix with a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 1 hour at 4°C [35].
- DAB Photo-Oxidation: Following fixation, incubate the cells with a DAB solution (2 mg/mL in 0.05 M Tris-HCl, pH 7.6) while simultaneously irradiating with two 8W Osram Blacklite 350 lamps for 2 hours at room temperature. The emission peaks of these lamps (550 and 580 nm) align with the excitation maximum of PKH26 (551 nm). This process generates free oxygen radicals that oxidize DAB, resulting in finely granular, electron-dense precipitates at the site of the dye [35].
- Post-processing and Embedding: Post-fix the samples with 1% osmium tetroxide for 1 hour at room temperature. Subsequently, dehydrate the cells through a graded acetone series and embed in Epon resin [35].
- Sectioning and Imaging: Prepare thin sections (60-80 nm) using an ultramicrotome. To enhance the contrast of the osmicated samples while preserving the visibility of the DAB precipitates, stain the sections weakly with a 2.5% aqueous uranyl acetate solution for 1-2 minutes or with 2.5-5% gadolinium triacetate for 10 minutes. Observe the sections using a TEM operating at 80 kV [35].
Expected Results and Interpretation
This technique allows for the precise localization of PKH26-labeled membranes within the cell. At early time points, the electron-dense DAB reaction product is typically visible along the plasma membrane, including invaginations and small vesicles just beneath the cell surface, illustrating the initial stages of uptake. At later time points (e.g., 1-3 hours), the label is predominantly found within multivesicular bodies (MVBs) and multilamellar bodies, indicating endosomal trafficking and downstream processing [35]. The central role of MVBs in the endocytotic pathway makes them a key organelle for confirming successful exosome internalization.
Lipophilic Dye Labeling (PKH26)
Lipophilic dyes like PKH26 are widely used for tracking exosomes in live cells due to their strong fluorescence and stable integration into lipid bilayers.
Protocol: Labeling and Tracking of MSC Exosomes
- Exosome Isolation and Labeling: Isolate MSC exosomes from conditioned culture media via sequential ultracentrifugation or density gradient purification. Resuspend the purified exosome pellet in Diluent C (provided with the PKH26 kit). Add an equal volume of PKH26 dye (pre-diluted in Diluent C) to the exosome suspension and incubate for 5-20 minutes. The staining process must be optimized to minimize dye aggregation [36] [37].
- Removing Free Dye and Contaminants: To separate labeled exosomes from unincorporated dye and potential PKH26 nanoparticle contaminants, ultracentrifugation alone is insufficient. The most effective method is purification via a sucrose density gradient. This process separates PKH26-labeled exosomes from less dense PKH26 nanoparticles, albeit sometimes at the expense of final exosome recovery [36].
- Cell Uptake Assay: Seed keratinocytes or endothelial cells in culture plates. Incubate the cells with the purified, PKH26-labeled MSC exosomes for desired time periods. Following incubation, wash the cells thoroughly with PBS to remove any non-internalized exosomes [37].
- Visualization and Analysis: Fix the cells with 4% paraformaldehyde and mount for microscopy. The internalized exosomes will appear as red fluorescent puncta within the cytoplasm when visualized using a confocal microscope with a He/Ne laser (543 nm excitation). Image analysis can quantify the number or fluorescence intensity of these puncta per cell [35] [37].
Critical Considerations and Validation
A major technical challenge is the propensity of PKH26 dyes to form micelles or nanoparticles that are similar in size to exosomes and can be internalized by cells, leading to false-positive signals [36]. Studies using Nanoparticle Tracking Analysis (NTA) have consistently shown that PKH labeling can artificially increase the apparent size of exosomes, which may alter their uptake kinetics and biodistribution [38]. Therefore, rigorous controls are essential, including:
- PKH Dye-only controls: Process a sample without exosomes through the entire labeling and purification procedure.
- Sucrose gradient purification: This is the recommended method to obtain a PKH26-exosome preparation devoid of contaminating nanoparticles [36].
- Validation with alternative methods: Confirm key findings using a different labeling technique, such as immunofluorescence.
The table below summarizes the quantitative findings from systematic evaluations of PKH labeling effects on extracellular vesicles.
Table 1: Impact of PKH26 Labeling on Extracellular Vesicle Size and Integrity
| Analysis Method | Key Finding | Experimental Condition | Implication for Uptake Studies |
|---|---|---|---|
| Nanoparticle Tracking Analysis (NTA) | Significant increase in EV size mode observed [38]. | All tested PKH:EV ratios, even below fluorescent detection limits [38]. | Altered size may affect uptake efficiency and tropism, as cellular uptake is size-dependent. |
| Asymmetrical-Flow Field-Flow Fractionation | PKH26 nanoparticles are almost indistinguishable from PKH26-labeled exosomes in size and fluorescence [36]. | Ultracentrifugation-based staining protocols [36]. | High potential for false-positive signals in uptake experiments. |
| Fluorescence Microscopy & Internalization | PKH26 nanoparticles are internalized by primary astrocytes into similar compartments as genuine exosomes [36]. | Presence of contaminating nanoparticles from labeling reaction [36]. | Compromised interpretation of EV internalization without proper purification. |
| NTA Comparison with CFSE dye | CFSE luminal labeling showed no significant shift in EV size distribution [38]. | EVs labeled with luminal binding dye CFSE [38]. | Suggests CFSE as a potential alternative for tracking studies where size preservation is critical. |
Immunofluorescence (IF) for Specific Detection
Immunofluorescence allows for the highly specific detection of exosomes based on their surface or intravesicular protein markers, overcoming the specificity limitations of lipophilic dyes.
Direct Staining of Exosomes and Recipient Cells
This protocol is ideal for visualizing isolated exosomes and tracking their uptake in recipient cells without the use of non-specific dyes.
- Isolation and Permeabilization of Exosomes: Isolate MSC exosomes using standard methods (e.g., PEG precipitation or ultracentrifugation). To stain intravesicular antigens (e.g., TSG101, HSP70) or transmembrane proteins (e.g., CD63, CD81), permeabilize the exosomes by incubating with 0.001% Triton X-100 for 5 minutes. This low concentration is critical for allowing antibody access while preserving vesicle integrity [39].
- Immune-Labeling: Incubate the permeabilized exosomes with primary antibodies against specific exosomal markers (e.g., anti-CD63, anti-TSG101) for 1 hour at room temperature or overnight at 4°C. Wash the exosomes via ultracentrifugation to remove unbound antibodies. Subsequently, incubate with fluorophore-conjugated secondary antibodies for 1 hour at room temperature, protected from light. Remove excess secondary antibodies through another washing step [39].
- Uptake Assay and Imaging: Add the stained exosomes to cultures of keratinocytes or endothelial cells. After an appropriate incubation period, wash the cells, fix with 4% PFA, and process for IF staining of cellular markers. For instance, to visualize keratinocytes, use an antibody against cytokeratin, and for endothelial cells, use an antibody against CD31 or VEGFR2. Finally, mount the samples and image using confocal or super-resolution microscopy. The internalized exosomes will be visible as distinct fluorescent puncta within the cytoplasm of the recipient cells [39].
Controls and Methodological Advantages
This method offers high specificity by targeting authentic exosomal proteins. Key controls include:
- Isotype controls: To assess non-specific antibody binding.
- "No primary antibody" controls: To confirm the specificity of the secondary antibody.
- Unstained exosomes: To account for cellular autofluorescence.
The primary advantage of this technique is the avoidance of lipophilic dye-related artifacts, providing a more reliable assessment of exosome uptake. Furthermore, it enables multiplexing, allowing researchers to simultaneously track exosomes and analyze subsequent changes in the recipient cell, such as activation of signaling pathways or expression of specific proteins [40] [39].
The Scientist's Toolkit: Essential Research Reagents
Successful visualization of exosome uptake relies on a suite of specific reagents and tools. The following table catalogs the essential solutions for the techniques described in this guide.
Table 2: Key Research Reagent Solutions for Visualizing Exosome Uptake
| Reagent / Tool | Function / Application | Technical Notes |
|---|---|---|
| PKH26 Dye Kit | Lipophilic fluorescent dye for long-term labeling of exosome and cell membranes [35]. | Requires careful purification (sucrose gradient) to remove contaminating dye nanoparticles that cause false positives [36]. |
| Diaminobenzidine (DAB) | Enzyme substrate used in photo-oxidation to convert PKH26 fluorescence into an electron-dense precipitate for TEM [35]. | The reaction product is osmiophilic, providing high contrast for EM. Must be handled with care as it is a suspected carcinogen. |
| Sucrose Density Gradient | Gold-standard method for purifying PKH26-labeled exosomes away from free dye and PKH26 nanoparticles [36]. | Essential for ensuring that observed fluorescence signals originate from exosomes and not dye artifacts. |
| Anti-Tetraspanin Antibodies (CD63, CD81, CD9) | Primary antibodies for the specific immunofluorescence detection of exosomes via common surface markers [39]. | Validation for specific cell types (MSCs, keratinocytes) is recommended, as exosome surface cargo can be heterogeneous. |
| Anti-Internal Antigen Antibodies (TSG101, HSP70) | Primary antibodies for immunofluorescence detection of intravesicular exosomal proteins [39]. | Requires a brief permeabilization step (e.g., 0.001% Triton X-100) for antibody access before the uptake assay. |
| Nanoparticle Tracking Analysis (NTA) | Instrumentation for determining the size distribution and concentration of isolated exosomes pre- and post-labeling [38]. | Critical for quantifying the effect of PKH26 labeling on exosome size and for characterizing preparations. |
Integrated Workflow and Pathway Diagrams
To achieve robust and interpretable results, researchers should employ an integrated workflow that combines the strengths of multiple techniques while accounting for the limitations of each. A recommended strategy is to use immunofluorescence as the primary method for specific uptake quantification, supported by PKH26 tracking for dynamic live-cell imaging, and validated with EM for ultrastructural confirmation. Crucially, any experiment using PKH26 must include stringent purification and control procedures to mitigate artifacts.
The following diagram illustrates the critical decision points and procedures in the integrated experimental workflow for visualizing MSC exosome uptake, with a special emphasis on the PKH26 labeling and validation pathway.
Diagram 1: Integrated Workflow for Visualizing MSC Exosome Uptake. This flowchart outlines the critical steps from exosome isolation to data interpretation, highlighting key methodological considerations for PKH26 labeling (yellow), immunofluorescence (green), and artifact avoidance (red).
The internalization of MSC exosomes by recipient cells triggers functional changes that underpin their therapeutic mechanism. The following diagram summarizes the key signaling pathways modulated by exosomal cargo in keratinocytes and endothelial cells, processes that can be investigated once uptake is confirmed.
Diagram 2: Functional Consequences of MSC Exosome Uptake. This diagram maps the key signaling pathways and functional outcomes in keratinocytes and endothelial cells following the internalization of MSC exosomes, linking uptake to downstream therapeutic effects like proliferation, angiogenesis, and senescence inhibition.
Within the field of regenerative medicine and drug development, a central challenge is establishing a direct, causative link between the cellular uptake of a therapeutic agent and its subsequent biological effect. This is particularly critical for complex biological nanoparticles like Mesenchymal Stem Cell (MSC) exosomes, which are emerging as a potent cell-free therapeutic strategy. The therapeutic potential of MSC exosomes, especially those derived from adipose tissue (ADSC-EVs), is well-documented in promoting wound healing by modulating keratinocyte and endothelial cell behavior [41] [31]. However, the full translation of these findings into reliable clinical applications requires robust experimental frameworks that explicitly correlate uptake with functional outcomes.
The core premise is that for an exosome to exert its influence on a recipient cell—be it a keratinocyte, fibroblast, or endothelial cell—it must first be successfully internalized. This guide provides an in-depth technical framework for researchers aiming to quantitatively link the uptake of MSC exosomes to three fundamental biological endpoints: proliferation, migration, and tube formation. By integrating precise uptake quantification with standardized functional assays, scientists can move beyond observational correlations to establish mechanistic causal relationships, thereby strengthening the validation of exosome-based therapies and their uptake mechanisms.
Core Functional Assays for Biological Outcomes
To systematically evaluate the therapeutic effects of MSC exosomes on target cells, standardized assays for proliferation, migration, and tube formation are essential. The quantitative data from these assays provide the critical link to uptake metrics. The table below summarizes the key findings from foundational studies.
Table 1: Summary of MSC Exosome Effects on Functional Outcomes in Vitro
| Functional Assay | Target Cell Type | Exosome Source | Key Quantitative Findings | Signaling Pathways Modulated |
|---|---|---|---|---|
| Proliferation | Dermal fibroblasts (from normal donors and chronic wound patients) [42] | Bone Marrow MSC | Dose-dependent enhancement of proliferation [42] | Akt, ERK, STAT3 activation [42] |
| Migration | Dermal fibroblasts (from normal donors and chronic wound patients) [42] | Bone Marrow MSC | Dose-dependent enhancement of migration [42] | Akt, ERK, STAT3 activation [42] |
| Migration | Senescent HUVECs [43] | MSC (sEV) | Remarkable increase in migration in transwell and scratch assays [43] | miR-146a/Src pathway [43] |
| Tube Formation | Human Umbilical Vein Endothelial Cells (HUVECs) [42] | Bone Marrow MSC | Dose-dependent increases in tube formation [42] | Akt, ERK, STAT3 activation [42] |
| Tube Formation | Senescent HUVECs [43] | MSC (sEV) | Rescued tube formation ability in vitro and blood vessel formation in vivo [43] | miR-146a/Src pathway [43] |
Proliferation Assay
Overview and Rationale: The proliferation assay measures the ability of MSC exosomes to stimulate cell division, a critical process in wound healing and tissue regeneration. This is typically assessed using resazurin assays, which measure metabolic activity as a surrogate for cell number and viability [44]. The underlying principle is that the internalization of pro-regenerative exosomal cargo (e.g., microRNAs like miR-135a and miR-126) can activate key proliferative signaling pathways such as PI3K/Akt and MAPK, and inhibit pathways like Hippo, leading to increased cell numbers [31].
Detailed Experimental Protocol:
- Cell Seeding: Plate keratinocytes or endothelial cells (e.g., HUVECs) in a 96-well plate at a low density (e.g., 5,000 cells/well) in complete growth medium and allow them to adhere overnight.
- Exosome Treatment: The following day, replace the medium with serum-free or low-serum medium containing varying concentrations of MSC exosomes (e.g., 0, 25, 50, 100, 200 µg/mL). Include a negative control (vehicle only) and a positive control (e.g., a known mitogen).
- Incubation: Incubate the cells for a desired period (e.g., 24, 48, or 72 hours) under standard culture conditions (37°C, 5% CO₂).
- Viability/Profileration Measurement:
- Prepare a working solution of resazurin by diluting it in culture medium (e.g., 1:10 ratio).
- Carefully remove the exosome-containing medium from the wells and add the resazurin working solution.
- Incubate the plate for 1-4 hours at 37°C, protected from light.
- Measure the fluorescence of the reduced product, resorufin, using a microplate reader (excitation 530–570 nm, emission 580–620 nm).
- Data Analysis: The fluorescence intensity is directly proportional to the number of metabolically active cells. Normalize the data to the negative control to calculate the percentage increase in proliferation.
Migration Assay
Overview and Rationale: Cell migration is essential for re-epithelialization and angiogenesis during wound healing. The scratch wound assay is a common, straightforward method to evaluate this parameter in vitro [43]. When MSC exosomes are internalized by cells at the wound edge, they deliver cargo that enhances migratory capacity. For instance, exosomal miR-146a has been shown to promote the migration of senescent endothelial cells by modulating the Src signaling pathway [43].
Detailed Experimental Protocol:
- Cell Seeding: Plate a high-density monolayer of keratinocytes or endothelial cells in a 12- or 24-well plate and incubate until 90-100% confluent.
- Scratch Creation: Using a sterile pipette tip (200 µL), gently and slowly create a straight "scratch" through the center of each well. Use a ruler underneath the plate as a guide for consistency.
- Wash and Treatment: Gently wash the wells with PBS 2-3 times to remove detached cells. Add serum-free medium containing MSC exosomes at the desired concentration. Include a control well with exosome-free medium.
- Image Acquisition: Immediately after adding treatment (t=0h), take a micrograph of the scratch in each well using a microscope with a 4x or 10x objective. Mark the bottom of the plate to ensure imaging the same locations at each time point.
- Incubation and Final Imaging: Incubate the plate for 12-24 hours, then acquire images from the same pre-marked locations.
- Data Analysis: Use image analysis software (e.g., ImageJ) to measure the area of the scratch at 0h and at the final time point. Calculate the percentage of wound closure:
% Closure = [(Area_t=0 - Area_t=final) / Area_t=0] * 100.
Tube Formation Assay
Overview and Rationale: The tube formation assay is a fundamental in vitro model for assessing angiogenesis—the formation of new blood vessels. In this assay, endothelial cells (like HUVECs) are plated on a basement membrane matrix (e.g., Matrigel) and their inherent capacity to form capillary-like tubular structures is evaluated [42] [43]. The internalization of pro-angiogenic MSC exosomes, which are rich in factors like miR-146a or those that activate Akt and ERK signaling, can significantly enhance this tubulogenesis process [42] [43].
Detailed Experimental Protocol:
- Matrix Preparation: Thickly coat the wells of a pre-chilled 96-well plate with Matrigel (approximately 50-60 µL per well). Avoid creating air bubbles. Incubate the plate at 37°C for 30-60 minutes to allow the Matrigel to polymerize.
- Cell Preparation and Treatment: Trypsinize, count, and resuspend HUVECs in serum-free medium. Pre-treat the cells with MSC exosomes (e.g., at 200 ng/µL) for a few hours or directly resuspend them in exosome-containing medium.
- Cell Seeding: Carefully seed the HUVEC suspension (e.g., 1x10^4 to 5x10^4 cells) on top of the polymerized Matrigel.
- Incubation: Incubate the plate at 37°C, 5% CO₂ for 4-8 hours.
- Image Acquisition and Analysis: After incubation, capture images of the tubular networks using an inverted microscope at 4x magnification. Analyze multiple fields per well. Key parameters to quantify include:
- Total Tube Length: The combined length of all tubular structures.
- Number of Meshes: The count of enclosed polygons in the network.
- Number of Branch Points: The points where three or more tubes intersect.
Correlating Uptake with Functional Outcomes
Establishing a quantitative relationship between exosome uptake and the resulting biological effect is the cornerstone of validating the mechanism of action. This requires precise measurement of internalization.
Quantifying Exosome Uptake
A standard method for quantifying uptake involves fluorescently labeling exosomes and tracking their internalization over time.
- Exosome Labeling: Isolated MSC exosomes can be labeled with lipophilic membrane dyes such as PKH26 (red fluorescence) or DiO (green fluorescence) [42] [43]. A common protocol involves incubating 7.5 µg of exosomes in 1 mL of Diluent C with a 2×10^−6 M concentration of PKH26 for 5 minutes. The excess dye is then removed using 100k Amicon Ultra centrifugal filters or via ultracentrifugation [43].
- Visualization and Quantification: Labeled exosomes are added to recipient cells (keratinocytes or endothelial cells) at a final concentration of 200 ng/µL. Internalization is observed using fluorescence microscopy or, more quantitatively, via flow cytometry.
- For microscopy, cells are imaged at various time points (e.g., 2, 4, 8, 24 hours) to visualize the accumulation of fluorescent exosomes within the cytoplasm [43].
- For flow cytometry, after incubating with labeled exosomes, cells are washed with PBS, trypsinized, and resuspended in flow cytometry staining buffer. The median fluorescence intensity (MFI) of the cell population is measured, which is directly proportional to the degree of exosome uptake.
Integrated Experimental Workflow
To reliably correlate uptake with function, a sequential, integrated workflow is recommended. The following diagram illustrates the key stages of this process, from exosome preparation to final correlation analysis.
Data Correlation and Analysis
With quantitative data from both uptake and functional assays, statistical correlation can be performed.
- Data Normalization: Normalize both uptake (Median Fluorescence Intensity, MFI) and functional outcome data (e.g., % proliferation increase, % wound closure, total tube length) to their respective control values.
- Statistical Analysis: Perform a linear regression analysis or calculate the Pearson correlation coefficient (r) to determine the strength and direction of the relationship between the normalized MFI (independent variable) and the normalized functional outcome (dependent variable). A strong positive correlation (r close to 1) provides compelling evidence that the biological effect is a direct consequence of exosome internalization.
Molecular Mechanisms and Signaling Pathways
The functional outcomes observed upon MSC exosome uptake are mediated by the activation of specific intracellular signaling pathways, driven by the exosomal cargo. The diagram below summarizes the key pathways involved in modulating proliferation, migration, and tube formation in recipient cells.
Key mechanistic insights include:
- Proliferation and Migration: MSC exosomes activate pro-proliferative and pro-migratory signaling pathways, including Akt, ERK, and STAT3 [42]. Furthermore, exosomal microRNAs like miR-135a can inhibit the Hippo pathway kinase LATS2, leading to activation of YAP/TAZ signaling, which enhances keratinocyte and fibroblast migration and proliferation [31].
- Angiogenesis and Senescence Reversal: A key mechanism for improving endothelial cell function involves the transfer of miR-146a via MSC-sEV. This miRNA suppresses Src phosphorylation, which in turn modulates downstream targets like VE-cadherin and Caveolin-1. This pathway mitigates oxidative stress-induced senescence, thereby rescuing the capacity of endothelial cells to migrate and form tubes [43].
The Scientist's Toolkit: Essential Reagents and Materials
Successful execution of these functional assays requires a suite of reliable reagents and specialized materials. The following table catalogues the key solutions and tools required for the experiments described in this guide.
Table 2: Research Reagent Solutions for Uptake and Functional Assays
| Reagent/Material | Function/Application | Example Catalog Number/Reference |
|---|---|---|
| PKH26 Fluorescent Dye | Lipophilic dye for labeling and tracking exosome membranes for uptake studies. | Protocol described in [43] |
| Propidium Iodide (PI) | Membrane-impermeant viability dye to exclude dead cells from flow cytometry analysis. | R&D Systems #00-6990 [45] [46] |
| Fixable Viability Dyes (FVD) | Covalently labels dead cells for exclusion in intracellular staining or fixed-cell experiments. | Invitrogen eFluor 780 (#65-0865) [45] |
| Calcein AM | Cell-permeant dye used to stain live cells for viability and proliferation assays. | Thermo Fisher (#65-0853) [45] |
| Resazurin Sodium Salt | Cell-permeable compound reduced to fluorescent resorufin in viable cells, used in proliferation assays. | Used in [44] |
| Basement Membrane Matrix | Extracellular matrix extract (e.g., Matrigel) used as a substrate for the tube formation assay. | Used in [42] [43] |
| Flow Cytometry Staining Buffer | Buffer containing BSA and often sodium azide for washing and resuspending cells during flow cytometry. | Thermo Fisher (#00-4222) [45] [46] |
| Ultracentrifugation Equipment | Standard method for isolating and purifying exosomes from conditioned cell media. | Beckman Optima L-XP [42] [43] |
Concluding Remarks
The experimental framework outlined in this guide provides a robust pathway for establishing a quantitative correlation between MSC exosome uptake and critical biological outcomes. By meticulously combining flow cytometry-based uptake quantification with standardized assays for proliferation, migration, and tube formation, researchers can generate compelling data that bridges the gap between cellular internalization and therapeutic function.
This approach is particularly powerful when integrated with molecular analyses of the signaling pathways activated by exosomal cargo. As the field progresses, the application of these correlated functional assays will be instrumental in optimizing exosome dosing, engineering exosomes for enhanced targeting and potency, and ultimately accelerating the translation of MSC exosome-based therapies from the laboratory bench to the clinical bedside.
Engineering MSC Exosomes for Enhanced Targeting and Delivery to Skin and Vasculature
Mesenchymal stem cell (MSC)-derived exosomes represent a groundbreaking cell-free therapeutic platform with immense potential for targeting skin and vascular tissues. These nano-sized extracellular vesicles (30-150 nm in diameter) naturally facilitate intercellular communication by shuttling proteins, lipids, and nucleic acids between cells [47] [48]. Their intrinsic low immunogenicity, high biocompatibility, and innate ability to cross biological barriers make them superior to synthetic nanoparticles for drug delivery applications [47] [49]. For researchers focusing on the uptake mechanisms by keratinocytes and endothelial cells, engineered exosomes offer a versatile tool to precisely deliver therapeutic cargo to these specific cell types, thereby modulating pathways critical for dermatological and cardiovascular applications. The transition from MSC-based therapies to MSC-derived exosomes addresses significant clinical challenges, including the risk of immune rejection, tumorigenicity, and the low survival rate of transplanted cells [50] [51]. This technical guide explores the methodologies for engineering these natural nanocarriers to enhance their targeting specificity and therapeutic efficacy for skin and vascular systems.
Fundamental Biology of MSC Exosomes
Biogenesis and Composition
The biogenesis of MSC exosomes begins with the inward budding of the plasma membrane to form early endosomes, which mature into late endosomes. Subsequent invagination of the endosomal membrane generates intraluminal vesicles (ILVs) within multivesicular bodies (MVBs). Upon fusion of MVBs with the plasma membrane, ILVs are released into the extracellular space as exosomes [47] [52] [48]. This process incorporates specific biological cargo—including proteins, lipids, DNA, and various RNA species—from the parent MSC, defining the exosome's initial biological activity [47]. The resulting exosomes exhibit a characteristic lipid bilayer membrane enriched with tetraspanins (CD9, CD63, CD81) and other marker proteins such as TSG101 and Alix, which serve as identification standards [47] [52].
Innate Therapeutic Mechanisms
MSC exosomes exert their therapeutic effects through multiple mechanisms. They can directly fuse with the target cell membrane, be internalized via endocytosis, or engage in ligand-receptor binding to initiate downstream signaling cascades [47]. Their cargo, particularly microRNAs (miRNAs) and proteins, modulates key cellular processes. For instance, in skin regeneration, miR-21-3p in exosomes derived from human umbilical cord MSCs (hucMSC-exos) stimulates fibroblast proliferation and migration by inhibiting PTEN and SPRY1, while the 14-3-3ζ protein activates SIRT1-dependent antioxidant pathways to mitigate oxidative stress [53]. In the vasculature, MSC exosomes promote angiogenesis by delivering pro-angiogenic factors like VEGF and miR-210 to endothelial cells [47].
Engineering Strategies for Enhanced Targeting
Surface Modification for Active Targeting
Engineering the exosomal surface to display targeting ligands is a principal strategy for achieving cell-specific delivery. This is primarily accomplished through two methods:
- Parent Cell Engineering (Pre-loading): Transducing parent MSCs with viral vectors encoding targeting peptides or proteins fused to exosomal surface markers (e.g., Lamp2b). This approach allows the engineered proteins to be naturally incorporated into the exosome membrane during biogenesis [51]. For example, genetic modification of MSCs to express the RGD peptide, which binds integrins highly expressed on endothelial cells, can enhance exosome homing to the vasculature.
- Direct Surface Functionalization (Post-loading): Chemically conjugating targeting moieties—such as antibodies, aptamers, or peptides—to the purified exosome surface via click chemistry or streptavidin-biotin interactions. While this method offers direct control, it risks damaging the exosome's structural integrity [51] [48].
The table below summarizes promising targeting ligands for skin and vascular delivery:
Table 1: Targeting Ligands for Engineering MSC Exosomes
| Target Cell | Targeting Ligand | Target Receptor | Potential Application |
|---|---|---|---|
| Keratinocytes | Laminin-332 peptide | α6β4 Integrin | Skin regeneration, wound healing |
| Endothelial Cells | RGD peptide | αvβ3 Integrin | Angiogenesis, ischemic disease |
| Endothelial Cells | VCAM-1 targeting peptide | VCAM-1 | Atherosclerosis, inflammation |
Cargo Loading Techniques
Precise loading of therapeutic molecules is crucial for efficacy. Cargo loading strategies can be categorized as follows:
- Endogenous Loading: Co-incubating parent MSCs with desired therapeutic agents (e.g., small molecule drugs, nucleic acids) or genetically engineering MSCs to overexpress specific miRNAs or mRNAs. This leverages the cell's natural machinery to package cargo into forming exosomes [48].
- Exogenous Loading: Isolating pure exosomes first, then loading cargo using techniques such as:
- Electroporation: Applying an electric field to create transient pores in the exosomal membrane for nucleic acid entry.
- Sonication: Using ultrasound waves to disrupt the membrane and allow drug diffusion.
- Simple Incubation: Suitable for small hydrophobic molecules that can passively diffuse across the lipid bilayer [51] [48].
Experimental Protocols for Uptake and Mechanism Studies
Isolating and Characterizing MSC Exosomes
Protocol: Isolation via Ultracentrifugation Ultracentrifugation remains the "gold standard" for laboratory-scale exosome isolation [47] [52].
- Cell Culture: Culture MSCs in exosome-depleted serum until 70-80% confluent.
- Collection: Collect conditioned medium and perform sequential centrifugation: 300 × g for 10 min (remove cells), 2,000 × g for 20 min (remove dead cells), 10,000 × g for 30 min (remove cell debris and larger vesicles).
- Ultracentrifugation: Centrifuge the supernatant at 100,000 × g for 70 minutes at 4°C to pellet exosomes.
- Washing: Resuspend the pellet in phosphate-buffered saline (PBS) and centrifuge again at 100,000 × g for 70 minutes.
- Resuspension: Finally, resuspend the purified exosome pellet in a small volume of PBS and store at -80°C [47] [52].
Characterization:
- Nanoparticle Tracking Analysis (NTA): Determines particle size distribution and concentration [47].
- Transmission Electron Microscopy (TEM): Visualizes exosome morphology and confirms cup-shaped structure [47] [48].
- Western Blot: Detects positive protein markers (CD9, CD63, CD81, TSG101) and absence of negative markers (e.g., Calnexin) [47].
Tracking Exosome Uptake by Keratinocytes and Endothelial Cells
Protocol: Fluorescent Labeling and Confocal Microscopy This protocol is essential for visualizing the internalization of exosomes by target cells.
- Labeling: Incubate isolated MSC exosomes with a lipophilic fluorescent dye, such as PKH67 or DiD, for 20-30 minutes. Remove excess dye using a size-exclusion chromatography column or ultracentrifugation [51].
- Cell Seeding: Culture target cells (e.g., HaCaT keratinocytes or Human Umbilical Vein Endothelial Cells - HUVECs) on glass-bottom confocal dishes.
- Treatment: Apply the labeled exosomes to the cells and incubate for a predetermined time (e.g., 1-24 hours).
- Staining: Fix the cells, stain actin filaments with phalloidin, and counterstain nuclei with DAPI.
- Imaging: Analyze using confocal laser scanning microscopy. The internalization of green fluorescent exosomes can be confirmed via Z-stack imaging to verify intracellular localization [53].
Functional Assays
- In Vitro Angiogenesis Assay: To assess pro-angiogenic effects, plate HUVECs on Matrigel and treat with engineered exosomes. Quantify tube formation by measuring total tube length and number of branching points [47].
- Keratinocyte Migration (Scratch Assay): Create a scratch in a confluent monolayer of keratinocytes. Treat with exosomes and monitor wound closure over time using live-cell imaging to evaluate enhanced migration [53] [50].
- Gene Expression Analysis: Post-treatment, extract RNA from target cells and perform qRT-PCR to quantify changes in the expression of genes related to angiogenesis (VEGF, Ang-1), extracellular matrix synthesis (COL1A1), or inflammation (IL-6, TNF-α) [53] [49].
Diagram 1: Experimental workflow for developing targeted MSC exosome therapies, from isolation to functional validation.
Signaling Pathways in Target Tissues
Signaling in Keratinocytes and Skin Regeneration
Engineered MSC exosomes target specific molecular pathways in keratinocytes and fibroblasts to promote skin repair and combat ageing. Key mechanisms include:
- Inhibition of MAPK/AP-1 Pathway: UV radiation-induced ROS activate MAPK pathways (ERK, p38, JNK), leading to AP-1 formation and subsequent upregulation of Matrix Metalloproteinases (MMPs). HucMSC-exos have been shown to inhibit this pathway, reducing MMP expression and preventing collagen degradation [53].
- Activation of PI3K/Akt Pathway: Stimulation of this pathway by exosomal miRNAs (e.g., miR-21-3p) fosters keratinocyte and fibroblast proliferation and migration, thereby enhancing re-epithelialization during wound healing [53] [50].
- Antioxidant via SIRT1: The transfer of the 14-3-3ζ protein from exosomes activates SIRT1, mitigating oxidative stress and promoting autophagy in skin cells exposed to UV or H₂O₂ [53].
Signaling in Endothelial Cells and Angiogenesis
For vascular targeting, MSC exosomes promote angiogenesis primarily through:
- HIF-1α/VEGF Signaling: Exosomes from MSCs grown in hypoxic conditions are enriched with HIF-1α and VEGF, which directly stimulate endothelial cell proliferation and new vessel formation [47] [50].
- miRNA-Mediated Regulation: Exosomal miRNAs like miR-126 and miR-210 enhance angiogenesis by activating pro-angiogenic signals and stabilizing HIF-1α, respectively. They also reduce apoptosis in endothelial cells, ensuring vessel stability [47] [48].
Diagram 2: Core signaling pathways in skin and vasculature targeted by engineered MSC exosomes.
The Scientist's Toolkit: Key Research Reagents
Successful research into engineered MSC exosomes requires a suite of specific reagents and tools. The following table catalogues essential materials for key experimental procedures.
Table 2: Essential Research Reagents for Targeted Exosome Studies
| Reagent / Tool | Function / Application | Key Considerations |
|---|---|---|
| PKH67 / DiD Fluorescent Dyes | Lipophilic dyes for labeling and tracking exosome uptake in vitro and in vivo. | PKH67 is green (λex/~490 nm), DiD is far-red (λex/~640 nm); ideal for co-localization studies. |
| Anti-CD63 / CD81 / CD9 Antibodies | Immunoaffinity capture for isolation; characterization via Western Blot/Flow Cytometry. | Confirm species reactivity; used for validating exosomal identity and purity. |
| Laminin-332 / RGD Peptides | Targeting ligands for engineering exosomes to bind keratinocytes or endothelial cells. | Can be fused to exosomal membrane proteins (e.g., Lamp2b) via genetic engineering. |
| Matrigel Matrix | In vitro assessment of exosome-induced angiogenesis via HUVEC tube formation assay. | Keep on ice; polymerization is temperature-sensitive. |
| qRT-PCR Assays | Quantifying changes in gene expression (e.g., VEGF, COL1A1, IL-6) in target cells post-treatment. | Use TaqMan assays for specific miRNA quantification from exosomal cargo. |
Quantitative Data and Efficacy Metrics
The therapeutic potential of engineered exosomes is quantified through specific, measurable outcomes in both in vitro and in vivo models. The table below summarizes key efficacy metrics from the literature.
Table 3: Quantitative Efficacy Metrics of MSC Exosomes in Preclinical Models
| Application / Model | Key Efficacy Metrics | Reported Outcomes |
|---|---|---|
| Skin Wound Healing (Diabetic Mouse Model) | Wound closure rate, Re-epithelialization, Granulation tissue thickness. | Up to 90% wound closure acceleration; significant increase in collagen deposition and epithelial thickness [50] [51]. |
| UV-Induced Skin Photoaging (In Vitro) | Fibroblast proliferation, Collagen I synthesis, MMP-1 reduction. | hucMSC-exos increased cell proliferation by ~40% and reduced MMP-1 expression by >50% [53]. |
| Ischemic Heart Disease (Mouse Model) | Infarct size reduction, Capillary density, Cardiac function (Ejection Fraction). | MSC-exos reduced infarct size by ~30% and increased capillary density by >40% [47] [48]. |
| In Vitro Angiogenesis (HUVEC) | Tube length, Branching points. | 2- to 3-fold increase in total tube length and number of nodes compared to controls [47]. |
| Targeting Efficiency (Cellular Uptake) | Fluorescence intensity in target cells vs. non-target cells. | Engineered exosomes showed 5- to 8-fold higher uptake in target cells compared to naive exosomes [51]. |
The strategic engineering of MSC exosomes represents the forefront of precision nanomedicine for dermatological and vascular applications. By combining targeted surface modifications with controlled cargo loading, researchers can transform these natural vesicles into powerful, specific therapeutic tools for delivering drugs, genes, and proteins to keratinocytes and endothelial cells. While challenges in scalable manufacturing, standardized characterization, and comprehensive safety profiling remain, the continued refinement of engineering protocols and a deeper understanding of exosome-cell interactions are paving the way for clinical translation. The future of this field lies in developing even more sophisticated multi-functional exosomes capable of responding to specific disease microenvironment stimuli, ultimately offering unprecedented efficacy in regenerative medicine and targeted drug delivery.
Biomaterial-Assisted Delivery Systems (e.g., Hydrogels) for Sustained Release and Improved Local Uptake
The therapeutic application of mesenchymal stem cell-derived exosomes (MSC-Exos) represents a paradigm shift in regenerative medicine and drug delivery. These natural nanoparticles inherit the regenerative and immunomodulatory potential of their parental cells, demonstrating significant capabilities in promoting angiogenesis, modulating inflammatory responses, and facilitating tissue repair [4]. However, the clinical translation of MSC-Exos faces substantial challenges related to their short half-life, rapid clearance from administration sites, and lack of targeted delivery [54]. Biomaterial-assisted delivery systems have emerged as a powerful strategy to overcome these limitations by providing controlled release kinetics, protecting exosomal integrity, and enhancing localization at target sites [55].
Within the specific context of keratinocyte and endothelial cell research, precise spatiotemporal delivery of exosomes is critical for investigating uptake mechanisms and therapeutic efficacy. Keratinocytes, as the major constituent of the epidermis, and endothelial cells, which form the vascular network, are key players in cutaneous wound healing and regeneration [56] [57]. Biomaterial scaffolds, particularly hydrogels, create a three-dimensional microenvironment that can mimic native tissue architecture while serving as reservoirs for sustained exosome release [57]. This technical guide comprehensively examines the current state of biomaterial-assisted exosome delivery systems, with specific emphasis on methodologies relevant to studying uptake mechanisms by keratinocytes and endothelial cells.
Biomaterial Platforms for Exosome Delivery
Hydrogel-Based Delivery Systems
Hydrogels represent the most extensively investigated biomaterial platform for exosome delivery due to their high water content, tunable physical properties, and excellent biocompatibility. These cross-linked polymer networks can be engineered to control the diffusion and release kinetics of encapsulated exosomes through modulation of mesh size, degradation rate, and polymer-exosome interactions [55].
Gelatin Methacryloyl (GelMA) hydrogels have demonstrated particular promise for exosome delivery in cutaneous applications. In a recent investigation focused on diabetic wound healing, GelMA hydrogels successfully served as a sustained-release vehicle for keratinocyte-derived extracellular vesicles [56]. The porous structure of GelMA effectively preserved the bioactivity of encapsulated vesicles while prolonging their release profile, resulting in enhanced microvascular regeneration and accelerated wound closure in diabetic murine models through activation of the PDGF/PI3K/AKT signaling pathway [56]. The versatility of GelMA allows for precise control over mechanical properties and degradation kinetics through adjustment of the degree of methacrylation and polymer concentration.
Hyaluronic acid (HA) based hydrogels offer inherent bioactive properties that support wound healing and tissue regeneration. The integration of MSC-derived exosomes within HA hydrogels has shown enhanced retention and stability of the vesicles while providing a bioactive matrix that facilitates cellular infiltration and tissue integration [54]. The viscoelastic properties of HA hydrogels can be tailored to match those of native skin, creating a more physiologically relevant microenvironment for studying keratinocyte and endothelial cell interactions with delivered exosomes.
Other natural polymer hydrogels, including chitosan, alginate, and collagen-based systems, have also been investigated for exosome delivery. Each material offers distinct advantages: chitosan possesses inherent antimicrobial properties, alginate provides highly tunable gelation kinetics, and collagen offers natural cell-adhesion motifs [58] [57]. The selection of hydrogel matrix should be guided by the specific research objectives and the particular cellular uptake mechanisms under investigation.
Table 1: Hydrogel Biomaterials for Exosome Delivery in Cutaneous Research
| Biomaterial | Key Properties | Release Kinetics | Advantages for Uptake Studies |
|---|---|---|---|
| GelMA | Tunable mechanical properties, RGD adhesion motifs | 5-21 days (concentration-dependent) | Supports both keratinocyte and endothelial cell adhesion |
| Hyaluronic Acid | inherent bioactivity, viscoelasticity | 7-28 days (cross-linking dependent) | Mimics native extracellular matrix composition |
| Chitosan | Antimicrobial, hemostatic | 3-14 days (degree of deacetylation dependent) | Reduces infection risk in in vivo models |
| Alginate | Mild gelation conditions, high porosity | 7-21 days (calcium concentration dependent) | Gentle encapsulation preserves exosome integrity |
| Collagen | Natural skin component, excellent biocompatibility | 5-10 days (rapid degradation) | Physiologically relevant for skin models |
Alternative Biomaterial Scaffolds
Beyond hydrogels, several other scaffold architectures have been utilized for exosome delivery in tissue engineering applications. Nanofibrous scaffolds created through electrospinning provide high surface area-to-volume ratios that can enhance exosome loading and create topographic cues that guide cellular behavior [57]. 3D-printed scaffolds offer precise control over spatial distribution of exosomes within complex architectures, enabling the creation of gradient release systems [58]. Microparticle and nanoparticle systems serve as injectable carriers that can be localized to specific tissue compartments through minimally invasive administration [55].
Characterization Techniques for Exosome-Loaded Biomaterials
Physicochemical Characterization
Comprehensive characterization of exosome-loaded biomaterials is essential for understanding structure-function relationships and interpreting uptake study results. Scanning Electron Microscopy (SEM) provides high-resolution visualization of scaffold morphology and porosity, while Transmission Electron Microscopy (TEM) confirms exosome integrity following encapsulation [56]. Nanoparticle Tracking Analysis (NTA) enables quantitative assessment of exosome size distribution and concentration within both the initial preparation and release media [56].
The mechanical properties of biomaterial scaffolds significantly influence cellular behavior and exosome release kinetics. Rheological analysis quantifies storage and loss moduli of hydrogel systems, while compressive testing determines mechanical strength under physiological loads. These properties should be tailored to match the target tissue environment—skin typically exhibits elastic moduli in the 0.5-20 kPa range, depending on anatomical location and hydration state.
Release Kinetics and Bioactivity Assessment
Quantifying exosome release profiles from biomaterial systems provides critical data for experimental design and interpretation. Fluorescent labeling of exosomes with lipophilic dyes (e.g., DiI, DiR) enables real-time tracking of release kinetics through fluorescence measurement of collection media [56]. The bicinchoninic acid (BCA) assay provides an alternative method for quantifying protein content associated with released vesicles [56].
Confirming retention of biological activity following encapsulation and release is paramount. In vitro functional assays including endothelial cell tube formation, keratinocyte proliferation, and macrophage polarization provide robust assessment of exosome bioactivity [56]. Specific marker expression analysis through Western blot (e.g., CD9, CD63, CD81, ALIX) confirms the preservation of exosomal integrity throughout the loading and release process [56].
Table 2: Standard Characterization Methods for Exosome-Loaded Biomaterials
| Parameter | Characterization Method | Key Outputs | Quality Standards |
|---|---|---|---|
| Exosome Identity | Western Blot | CD9, CD63, CD81, ALIX expression | Positive for tetraspanins, negative for contaminants |
| Exosome Size Distribution | Nanoparticle Tracking Analysis | Size profile, concentration | 30-150 nm diameter, monomodal distribution |
| Scaffold Morphology | Scanning Electron Microscopy | Pore size, structure, homogeneity | Interconnected porous structure |
| Release Kinetics | Fluorescence measurement/BCA assay | Cumulative release profile, release rate | Sustained release over 1-3 weeks |
| Bioactivity | Endothelial tube formation assay | Tube length, branching points | Significant enhancement vs. negative control |
| Cellular Uptake | Confocal microscopy with labeled exosomes | Internalization efficiency, localization | Time-dependent increase in fluorescence |
Experimental Protocols for Uptake Mechanism Studies
Isolation and Characterization of MSC-Derived Exosomes
Protocol: Exosome Isolation via Ultracentrifugation
- Cell Culture and Conditioning: Culture MSCs in appropriate growth medium until 70-80% confluence. Replace with exosome-depleted serum medium for 48-hour conditioning [56].
- Conditioned Media Collection: Collect conditioned media and perform sequential centrifugation:
- 300 × g for 10 minutes to remove cells
- 2,000 × g for 20 minutes to remove apoptotic bodies
- 10,000 × g for 30 minutes to remove cell debris [56]
- Ultracentrifugation: Transfer supernatant to ultracentrifugation tubes and centrifuge at 100,000 × g for 70 minutes at 4°C [56].
- Washing and Resuspension: Wash pellet with phosphate-buffered saline (PBS) and repeat ultracentrifugation. Resuspend final exosome pellet in PBS for immediate use or storage at -80°C [56].
- Characterization: Validate exosomes through NTA, TEM, and Western blot analysis for positive markers (CD9, CD63, ALIX) and negative markers (calnexin) [56].
Biomaterial Loading and Release Kinetics Assessment
Protocol: GelMA Hydrogel Loading with Exosomes
- GelMA Solution Preparation: Disspose GelMA polymer in PBS at desired concentration (typically 5-10% w/v) with 0.5% photoinitiator (Irgacure 2959) [56].
- Exosome Incorporation: Mix concentrated exosome preparation with GelMA solution to achieve target loading concentration (typically 50-200 μg/mL).
- Hydrogel Crosslinking: Transfer exosome-GelMA mixture to molds and expose to UV light (365 nm, 5-10 mW/cm²) for 30-60 seconds to form crosslinked hydrogels [56].
- Release Kinetics Study: Immerse loaded hydrogels in PBS or cell culture medium at 37°C. Collect release medium at predetermined time points and replace with fresh medium. Quantify released exosomes through BCA assay or fluorescence measurement [56].
In Vitro Uptake Studies with Keratinocytes and Endothelial Cells
Protocol: Tracking Cellular Uptake via Confocal Microscopy
- Exosome Labeling: Label isolated exosomes with lipophilic fluorescent dye (e.g., PKH26, DiI) according to manufacturer's protocol, followed by purification to remove unincorporated dye [56].
- Cell Culture Setup: Seed keratinocytes (e.g., HaCaT cells) or endothelial cells (e.g., HUVECs) on chambered coverslips at appropriate density.
- Treatment with Exosome-Loaded Biomaterials: Apply labeled exosome-loaded hydrogels to transwell inserts or directly immerse in culture medium.
- Fixation and Staining: At predetermined time points, fix cells with 4% paraformaldehyde, permeabilize with 0.1% Triton X-100, and stain actin cytoskeleton with phalloidin and nuclei with DAPI.
- Imaging and Analysis: Visualize using confocal microscopy with appropriate filter sets. Quantify uptake through fluorescence intensity measurements or by counting vesicles within cell boundaries [56].
Diagram 1: Experimental Workflow for Cellular Uptake Studies. This diagram illustrates the comprehensive protocol for preparing exosome-loaded biomaterials and assessing cellular uptake by keratinocytes and endothelial cells.
Signaling Pathways in Exosome-Mediated Effects
PDGF/PI3K/AKT Pathway in Angiogenesis
Recent research has elucidated the molecular mechanisms through which biomaterial-delivered exosomes influence endothelial cell behavior. The platelet-derived growth factor (PDGF)/phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway has been identified as a critical signaling axis in exosome-mediated angiogenesis [56]. In a diabetic wound healing model, keratinocyte-derived extracellular vesicles delivered via GelMA hydrogels upregulated PDGF expression in wound tissues, subsequently activating PI3K/AKT signaling and promoting microvascular network reconstruction [56].
The PI3K/AKT pathway influences endothelial cell function through multiple downstream effectors. Activation of AKT phosphorylates endothelial nitric oxide synthase (eNOS), increasing nitric oxide production and promoting vasodilation. Additionally, AKT signaling inhibits pro-apoptotic proteins while activating mTOR, coordinating cell survival with proliferative responses [56]. This pathway represents a promising target for enhancing the therapeutic efficacy of exosome-based therapies for vascularization.
Immunomodulatory Pathways
MSC-derived exosomes modulate inflammatory responses through complex signaling networks that influence both innate and adaptive immunity. A key mechanism involves shifting macrophage polarization from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype [57]. This transition is mediated through exosomal transfer of regulatory miRNAs and proteins that modulate signaling pathways including NF-κB, STAT, and PPARγ [57].
In keratinocytes, exosome-mediated immunomodulation can reduce the production of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-1β while enhancing expression of anti-inflammatory factors [57]. This creates a more favorable microenvironment for tissue repair and regeneration, particularly in chronic inflammatory conditions such as diabetic ulcers or psoriasis.
Diagram 2: PDGF/PI3K/AKT Signaling Pathway in Angiogenesis. This diagram illustrates the key molecular events through which exosomes activate angiogenic signaling in endothelial cells following uptake.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Biomaterial-Exosome Studies
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Biomaterial Polymers | GelMA, Hyaluronic Acid, Chitosan, Alginate | Scaffold formation for exosome encapsulation | Degree of functionalization affects mechanical properties |
| Crosslinking Agents | Irgacure 2959, Calcium Chloride, Genipin | Hydrogel solidification | Crosslinking method impacts exosome bioactivity |
| Exosome Isolation Kits | Total Exosome Isolation, ExoQuick | Alternative to ultracentrifugation | Yield and purity variations between methods |
| Fluorescent Labels | PKH26, DiI, DiD, CFSE | Exosome tracking and visualization | Potential dye aggregation; purification required |
| Cell Culture Models | HaCaT keratinocytes, HUVECs, MSC lines | Target cells for uptake studies | Primary cells more physiological but higher variability |
| Antibodies for Characterization | Anti-CD9, CD63, CD81, ALIX, TSG101 | Exosome identification and validation | Species specificity and validation required |
| Signal Pathway Inhibitors | LY294002 (PI3K inhibitor), AKT inhibitor VIII | Mechanistic studies | Concentration optimization critical for specificity |
| Analysis Kits | BCA protein assay, CCK-8 proliferation kit | Quantitative assessment | Standard curve establishment essential for accuracy |
Biomaterial-assisted delivery systems represent a transformative approach for investigating MSC exosome uptake mechanisms by keratinocytes and endothelial cells. The integration of exosomes with advanced biomaterials addresses critical challenges in bioavailability and localization while providing controlled release kinetics that mimic physiological processes. The methodologies and experimental frameworks outlined in this technical guide provide researchers with comprehensive tools for designing rigorous studies in this rapidly evolving field.
Future directions in biomaterial-assisted exosome delivery will likely focus on the development of "smart" responsive systems that release their cargo in response to specific physiological triggers such as pH changes, enzyme activity, or reactive oxygen species. Additionally, the integration of CRISPR-based technologies with exosome delivery platforms holds promise for genetic manipulation of target cells [59]. As these technologies mature, standardized protocols for exosome loading, release kinetics, and functional assessment will be essential for comparing results across studies and advancing toward clinical translation.
The continued refinement of biomaterial systems for exosome delivery will undoubtedly expand our understanding of cellular uptake mechanisms and enhance the therapeutic potential of exosome-based therapies for cutaneous regeneration and beyond.
Dosage and Administration Route Optimization for Preclinical Models
Mesenchymal stem cell-derived exosomes (MSC-Exos) have emerged as promising cell-free therapeutics in regenerative medicine, recapitulating the therapeutic benefits of their parent cells through sophisticated paracrine signaling [60]. These nano-sized extracellular vesicles (30-150 nm) function as biological messengers, transferring proteins, lipids, and nucleic acids to recipient cells [61]. Within the context of skin biology and vascular repair, optimizing the delivery of MSC-Exos to keratinocytes and endothelial cells represents a critical research frontier. The therapeutic efficacy of MSC-Exos hinges on overcoming biological barriers and achieving sufficient uptake by target cells, necessitating systematic optimization of dosage parameters and administration routes in preclinical models [62] [31].
Current evidence indicates that MSC-Exos exert their effects through multiple mechanisms including reducing cellular senescence, promoting angiogenesis, modulating inflammation, and enhancing tissue regeneration [31]. For keratinocytes, exosomal cargo can accelerate re-epithelialization by promoting proliferation and migration, while for endothelial cells, exosomes stimulate angiogenic responses crucial for wound healing and tissue repair [31] [41]. Understanding the precise uptake mechanisms and optimizing delivery strategies for these target cells forms the foundation for effective therapeutic translation.
Uptake Mechanisms of MSC Exosomes by Target Cells
Molecular Pathways in Keratinocyte and Endothelial Cell Uptake
The internalization of MSC-Exos by keratinocytes and endothelial cells occurs through multiple endocytic pathways, significantly influencing their functional outcomes in skin repair and angiogenesis.
Keratinocyte Uptake Mechanisms:
- miRNA-Mediated Proliferation: MSC exosomes deliver specific microRNAs that promote keratinocyte migration and proliferation. Notably, miR-135a inhibits the Hippo pathway kinase LATS2, leading to activation of pro-proliferative YAP/TAZ signaling [31]. Similarly, exosomal miR-126 activates PI3K/Akt and MAPK pathways, essential for keratinocyte survival and proliferation [31].
- Re-epithelialization Support: MSC-Exos accelerate re-epithelialization – the process where keratinocytes migrate and proliferate to restore the epidermal barrier. This process is crucial for wound closure and is significantly enhanced by exosomal cargo [31].
Endothelial Cell Uptake Mechanisms:
- Angiogenic Programming: MSC-Exos promote angiogenesis through multiple pathways. They have demonstrated the ability to enhance DNA repair in hypoxic environments through modulation of hypoxia-inducible factor-1 signaling, which is crucial for endothelial cell survival under stressful conditions [31].
- Microvascular Protection: Exosomes provide protection to the dermal microvasculature, countering radiation-induced endothelial apoptosis and senescence-driven TGF-β1 signaling that compromises vascular integrity [31].
The following diagram illustrates the key molecular mechanisms involved in MSC exosome uptake by keratinocytes and endothelial cells:
Diagram 1: Molecular mechanisms of MSC exosome uptake by keratinocytes and endothelial cells
Quantitative Dosing Parameters for Preclinical Models
Evidence-Based Dosing Ranges Across Disease Models
Extensive preclinical studies have established preliminary dosing parameters for MSC-Exos across various disease models. The table below summarizes effective dosing ranges identified in preclinical studies for conditions relevant to keratinocyte and endothelial cell targeting:
Table 1: MSC-Exosome Dosing Parameters in Preclinical Models
| Disease Model | Effective Dose Range | Administration Route | Dosing Frequency | Primary Outcomes | Evidence Source |
|---|---|---|---|---|---|
| Radiation-Induced Skin Injury | 10^8 - 10^11 particles | Topical, intradermal | Single to multiple doses (varies by severity) | Improved healing, reduced senescence, enhanced angiogenesis | [31] |
| Diabetic Wounds | 10^9 - 10^11 particles | Topical with scaffolds | Every 2-3 days until closure | Accelerated re-epithelialization, angiogenesis modulation | [41] |
| Skin Rejuvenation | 10^8 - 10^10 particles | Topical, microneedling | Single application | Improved texture, elasticity, hydration | [61] [22] |
| General Wound Healing | 100-400 μg protein content | Local injection, topical | Weekly for 2-4 weeks | Enhanced collagen deposition, reduced inflammation | [63] [41] |
The dosing parameters demonstrate significant variation depending on the specific model, administration route, and desired therapeutic outcome. For skin-specific applications targeting keratinocytes, evidence suggests that topical administration requires higher particle counts (10^8-10^11) compared to systemic routes, though direct comparative studies are limited [61] [31].
Route-Dependent Dosage Optimization
Analysis of preclinical and clinical data reveals that administration route significantly influences the effective dosage of MSC-Exos. The following table compares route-specific dosing parameters:
Table 2: Route-Dependent Dosage Optimization for MSC-Exosomes
| Administration Route | Optimal Dose Range | Target Cells/Tissues | Advantages | Limitations |
|---|---|---|---|---|
| Topical Application | 10^9-10^11 particles | Keratinocytes, superficial dermal cells | Direct delivery, minimal systemic exposure | Limited penetration, requires formulations |
| Intradermal Injection | 10^8-10^10 particles | Dermal fibroblasts, endothelial cells | Localized delivery, bypasses epidermal barrier | Invasive, potential for local reactions |
| Intravenous Injection | 10^10-10^12 particles | Systemic delivery, endothelial cells | Whole-body distribution, reaches multiple organs | Rapid clearance, potential off-target effects |
| Aerosolized Inhalation | ~10^8 particles | Pulmonary endothelium, epithelial cells | Lower effective dose, targets respiratory system | Technical complexity, variable deposition |
Recent evidence indicates that nebulization therapy achieved therapeutic effects at doses around 10^8 particles, significantly lower than those required for intravenous routes, suggesting a narrow and route-dependent effective dose window [64]. This has important implications for optimizing delivery to skin cells, where localized administration may achieve higher target tissue concentrations with lower total doses.
Administration Route Selection and Optimization
Comparative Efficacy of Delivery Routes
The selection of administration route profoundly impacts the bioavailability of MSC-Exos to target keratinocytes and endothelial cells. Current evidence from preclinical models supports several optimized delivery approaches:
Topical Delivery for Keratinocytes:
- Standard Application: Direct application of exosome formulations to skin surfaces shows variable efficacy dependent on formulation. Absorption is limited to superficial layers without penetration enhancers [61].
- Microneedling Enhancement: Combination with microneedling creates microchannels that significantly improve delivery to viable epidermis and dermis, enhancing keratinocyte and endothelial cell exposure [61]. Studies report improved skin texture, elasticity, and hydration with MSC-Exos following microneedling delivery [61].
Local Injection for Dermal Targets:
- Intradermal Injection: Direct intradermal administration ensures precise delivery to both epidermal and dermal compartments, benefiting keratinocytes and dermal endothelial cells simultaneously [41].
- Perilesional Injection: Circumferential injection around wound perimeters creates a gradient favorable for both re-epithelialization and angiogenesis [31] [41].
Systemic Administration Limitations:
- Intravenous Challenges: While enabling whole-body distribution, intravenous administration requires significantly higher doses (10^10-10^12 particles) to achieve therapeutic effects in skin, with rapid clearance limiting bioavailability to target cells [64].
The following workflow outlines a systematic approach for selecting and optimizing administration routes in preclinical studies:
Diagram 2: Systematic workflow for administration route selection and optimization
Experimental Protocols for Uptake and Efficacy Studies
Standardized Isolation and Characterization Protocols
Exosome Isolation Protocol:
- Source Material Preparation: Culture MSCs from bone marrow, adipose tissue, or umbilical cord in exosome-depleted media for 48-72 hours [62]. Consistent passage numbers (P3-P5) are recommended to maintain exosome characteristics [60] [65].
- Differential Ultracentrifugation: Centrifuge conditioned media at 300 × g for 10 minutes (remove cells), 2,000 × g for 20 minutes (remove dead cells), 10,000 × g for 30 minutes (remove cell debris), and 100,000 × g for 70 minutes (pellet exosomes) [62].
- Alternative Isolation Methods: Density gradient centrifugation, size exclusion chromatography, or ultrafiltration may be employed based on downstream applications [62].
- Quality Control: Resuspend exosome pellets in PBS and characterize using nanoparticle tracking analysis (size distribution and concentration), transmission electron microscopy (morphology), and Western blot (CD63, CD81, CD9, TSG101 markers) [62] [64].
In Vitro Uptake Studies:
- Fluorescent Labeling: Label isolated exosomes with lipophilic dyes (e.g., PKH67, DiI) or membrane-permeant tracers according to manufacturer protocols [31].
- Co-culture with Target Cells: Seed keratinocytes (HaCaT cell line or primary) and endothelial cells (HUVECs) in appropriate media. Add labeled exosomes at optimized particle-to-cell ratios (typically 10^3-10^4 particles per cell) [31] [41].
- Uptake Quantification: After 4-24 hours incubation, fix cells and analyze using fluorescence microscopy, flow cytometry, or confocal imaging. Inhibitors (chlorpromazine for clathrin-mediated endocytosis, filipin for caveolae-mediated uptake) can determine specific uptake mechanisms [31].
In Vivo Dosing and Efficacy Assessment
Animal Model Dosing Protocol:
- Dose Calculation: Calculate exosome doses based on particle number (determined by NTA) or protein content (BCA assay). For mouse models, common doses range from 10^8-10^11 particles per administration [63] [31].
- Formulation for Administration: For topical delivery, formulate exosomes in hydrogels or creams for sustained release. For injection, use sterile PBS with protein stabilizers (e.g., 0.1% HSA) [31] [41].
- Dosing Schedule: Optimize frequency based on disease kinetics. Acute wounds may benefit from single or bi-weekly administration, while chronic conditions may require weekly dosing for 4-8 weeks [41].
Efficacy Endpoint Assessment:
- Histological Analysis: Process tissue samples for H&E staining (general morphology), Masson's trichrome (collagen), and immunohistochemistry for Ki-67 (proliferation), CD31 (angiogenesis), and cytokeratin (epithelialization) [31] [41].
- Molecular Analysis: Isulate RNA or protein from treated tissues for qRT-PCR analysis of genes associated with proliferation, angiogenesis, and inflammation [31].
- Functional Assessments: For wound healing models, measure wound closure rates daily. For skin rejuvenation, assess biomechanical properties and hydration [61] [41].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for MSC Exosome Studies
| Reagent Category | Specific Products/Assays | Research Application | Technical Notes |
|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Kit, ExoQuick-TC | Rapid isolation from conditioned media | Quality varies; validate against ultracentrifugation |
| Characterization Tools | Nanoparticle Tracking Analysis (NTA), TEM, Western Blot | Size distribution, morphology, marker confirmation | Follow MISEV2018 guidelines for standardization |
| Uptake Tracking | PKH67, DiI, CFSE, ExoGlow | Fluorescent labeling for uptake studies | Optimize staining to avoid aggregation |
| Cell Culture Models | HaCaT keratinocytes, HUVECs, primary epidermal keratinocytes | Target cell uptake and functional assays | Use low passage numbers for primary cells |
| Animal Models | Diabetic (db/db) mice, radiation-induced skin injury models | In vivo efficacy and biodistribution | Select models that recapitulate human disease pathophysiology |
| Analysis Kits | BCA protein assay, RNA isolation kits, ELISA arrays | Quantification and cargo analysis | Standardize normalization methods across experiments |
Optimizing dosage and administration routes for MSC exosomes in preclinical models requires a systematic approach that considers target cell biology, disease pathophysiology, and pharmacokinetic principles. The accumulating evidence demonstrates that route selection profoundly influences effective dosing, with localized administration generally requiring lower doses for skin and endothelial targets. Future research should focus on standardized reporting of exosome characterization, dose metrics, and administration parameters to enable meaningful cross-study comparisons.
Emerging strategies including exosome engineering for enhanced target cell specificity, development of advanced delivery systems for sustained release, and combination therapies with biomaterial scaffolds hold promise for improving therapeutic efficacy while reducing dose requirements [62] [41]. As the field advances, rigorous dose-response studies and mechanistic investigations of uptake pathways will be essential for translating MSC exosome therapies from preclinical models to clinical applications for skin repair and vascular regeneration.
Navigating Challenges: Troubleshooting and Optimizing Uptake Efficiency
Abstract The therapeutic efficacy of mesenchymal stem cell-derived exosomes (MSC-Exos) is contingent upon their efficient uptake by target cells, such as keratinocytes and endothelial cells. However, significant heterogeneity in MSC-Exos, driven by donor age, health status, and tissue source, introduces substantial variability in their uptake and function. This whitepaper synthesizes current research to delineate the impact of these donor characteristics on exosomal properties. It provides a framework of standardized experimental protocols and analytical tools to quantify and mitigate this variability, aiming to enhance the reproducibility and efficacy of exosome-based therapies in regenerative medicine and drug development.
Mesenchymal stem cell-derived exosomes have emerged as promising acellular therapeutic agents, offering advantages over whole-cell therapies, including low immunogenicity, high stability, and a reduced risk of tumorigenesis [22] [1] [66]. Their efficacy in promoting tissue repair, modulating immune responses, and facilitating regenerative processes is largely dependent on their uptake by recipient cells, such as keratinocytes in wound healing and endothelial cells in angiogenesis [8] [66].
A critical, yet often overlooked, challenge in the clinical translation of MSC-Exos is their inherent heterogeneity. MSC populations are not uniform; they exhibit significant donor-to-donor variation influenced by age, health, and anatomical source [67] [68]. This variability is directly reflected in the molecular cargo and biological activity of the exosomes they produce [69]. Consequently, exosomes from different donors can exhibit disparate uptake efficiencies by the same target cell type, leading to inconsistent therapeutic outcomes. This whitepaper examines the sources of this variability and provides a technical guide for researchers to address it, ensuring that exosome uptake studies and therapeutic development are built upon a foundation of rigorous and reproducible science.
Impact of Donor Characteristics on MSC and Exosome Properties
The biological "fitness" of the parent MSCs fundamentally shapes the characteristics of their secreted exosomes. Key donor factors systematically influence MSC proliferation, differentiation potential, and paracrine secretion, thereby modulating the exosome cargo that dictates uptake and function.
Donor Age
Donor age is a primary factor contributing to MSC senescence and functional decline. Evidence indicates that MSCs from older donors exhibit:
- Reduced Proliferative Capacity: A study on bovine MSCs showed that fetal and calf-derived cells from Holstein Friesian breeds demonstrated a high proliferation capacity, with most donors surpassing 30 population doublings, a trait generally diminished in adult-derived cells [67].
- Altered Differentiation Potential: The adipogenic potential of bovine MSCs was higher in fetal and adult donors compared to calves, while no clear linear relationship with age was observed for osteogenic potential, which was more affected by breed [67]. This suggests age-related shifts in lineage commitment.
- Accumulation of Senescence Markers: Aging MSCs show enlargement, telomere shortening, accumulation of DNA damage, and elevated levels of reactive oxygen species (ROS) [68]. The senescent secretome can subsequently alter exosome content, potentially impairing their communication with keratinocytes and endothelial cells.
Tissue Source
The anatomical origin of MSCs is a major determinant of their exosomal molecular signature. A thorough analysis of publicly available omic datasets has revealed that MSC-Exos from adipose tissue, bone marrow, and umbilical cord possess unique protein and miRNA profiles, leading to distinct functional pathways [69]. For instance:
- Adipose-Derived MSCs (AD-MSCs) are widely accessible and have demonstrated efficacy in wound healing and plastic surgery [68].
- Bone Marrow-Derived MSCs (BM-MSCs) have been extensively studied, but their quantity and osteogenic potential decrease with donor age [68].
- Umbilical Cord-Derived MSCs (UC-MSCs) are considered more primitive and may exhibit enhanced proliferative and immunomodulatory capacities without donor age-related effects [66].
The choice of tissue source must therefore be aligned with the intended therapeutic application, as the exosomal cargo will dictate the specific signals delivered to recipient keratinocytes or endothelial cells.
Donor Health and Breed/Genetics
Underlying health conditions and genetic background introduce another layer of complexity.
- Health Status: Systemic diseases like diabetes can impede the regenerative functions of MSCs [8]. MSCs isolated from individuals with metabolic disorders or chronic inflammation will likely produce exosomes with altered cargo, affecting their ability to promote keratinocyte migration or endothelial tube formation.
- Genetic Background (Breed): In animal models, breed-specific differences significantly impact MSC characteristics. Belgian Blue bovine MSCs demonstrated a superior osteogenic differentiation potential compared to Holstein Friesian breeds, independent of age [67]. Furthermore, immunophenotype differences, such as the percentage of CD34+ cells, were also breed-dependent [67]. This genetic influence underscores the potential for similar inter-individual variability in human populations.
Table 1: Impact of Donor Characteristics on MSC and Exosome Properties
| Donor Characteristic | Impact on MSCs | Potential Impact on Exosome Cargo & Uptake |
|---|---|---|
| Age (Fetal/Neonatal) | High proliferation, enhanced differentiation capacity, low senescence [67] [68] | May carry pro-regenerative miRNAs/proteins, potentially enhancing uptake in target cells. |
| Age (Adult/Aged) | Reduced proliferation, skewed differentiation, increased senescence markers [67] [68] | May contain inflammatory or senescence-associated cargo, potentially impairing uptake or delivering conflicting signals. |
| Tissue Source (e.g., UC, AD, BM) | Unique transcriptional and functional profiles based on tissue of origin [69] | Distinct miRNA, protein, and lipid compositions that determine tropism and signaling to recipient cells [69]. |
| Health Status (e.g., Diabetic) | Functional impairment, reduced reparative potential [8] | Altered cargo may fail to activate necessary pathways in keratinocytes/endothelial cells, reducing therapeutic efficacy. |
| Genetic Background | Affects immunophenotype (e.g., CD34 expression) and differentiation potential [67] | Could influence surface integrins and tetraspanins, directly modulating binding and uptake by recipient cells. |
Experimental Protocols for Characterizing Donor-Dependent Variability
To systematically evaluate the impact of donor variability on exosome uptake, a standardized set of characterization protocols is essential. The following methodologies are critical for pre-qualifying MSC sources and their derived exosomes.
MSC Isolation and Donor Stratification
- Cell Isolation: MSCs should be isolated from target tissues (e.g., adipose tissue, bone marrow) using a standardized enzymatic digestion method. For subcutaneous adipose tissue, this involves dissecting the tissue, digesting it with a solution like Liberase, filtering the cell suspension, and culturing the adherent fraction [67].
- Donor Stratification: Design studies to include MSCs from stratified donors: different age groups (fetal, young, old), healthy vs. disease-state donors (e.g., diabetic), and distinct tissue sources (UC, AD, BM). A minimum of n=7 donors per group is recommended for robust statistical analysis [67].
- Core MSC Characterization: All MSC batches must be characterized according to International Society for Cell & Gene Therapy (ISCT) criteria: plastic adherence, expression of surface markers (CD105, CD73, CD90 ≥95%; CD45, CD34, CD14, CD11b, CD79a, CD19, HLA-DR ≤2%), and tri-lineage differentiation potential (osteogenic, adipogenic, chondrogenic) [68] [67].
Functional Assays to Gauge MSC "Fitness"
- Proliferation and Senescence Assays:
- Population Doublings: Track cumulative population doublings over serial passages to assess long-term proliferative capacity [67].
- Senescence-Associated β-Galactosidase (SA-β-Gal) Staining: A standard histochemical method to detect β-galactosidase activity at pH 6.0, a hallmark of senescent cells. An increased percentage of SA-β-Gal positive cells indicates reduced cellular fitness [67] [68].
- Tri-lineage Differentiation Assays:
- Osteogenic Differentiation: Culture MSCs in osteo-inductive media containing dexamethasone, ascorbate-2-phosphate, and β-glycerophosphate for 2-3 weeks. Differentiate using Alizarin Red S staining to detect calcium deposits [67] [68].
- Adipogenic Differentiation: Culture MSCs in adipogenic media containing dexamethasone, indomethacin, and insulin for 1-2 weeks. Differentiate using Oil Red O staining to visualize lipid vacuoles [67].
- Chondrogenic Differentiation: Pellet culture in chondrogenic media containing TGF-β for 3-4 weeks. Differentiate using Alcian Blue or Safranin O staining to detect sulfated glycosaminoglycans.
Exosome Isolation and Characterization
- Isolation via Ultracentrifugation: The most common method involves serial centrifugation steps to remove cells, debris, and larger vesicles, followed by ultracentrifugation at ≥100,000 × g to pellet exosomes [70] [1].
- Characterization: Isolated exosomes must be characterized for size, concentration, and identity.
- Nanoparticle Tracking Analysis (NTA): Determines the particle size distribution and concentration [70].
- Transmission Electron Microscopy (TEM): Provides visual confirmation of exosome morphology (cup-shaped vesicles) [70].
- Flow Cytometry/Western Blot: Detects the presence of exosomal surface markers (e.g., CD63, CD81, CD9, TSG101) and the absence of negative markers (e.g., calnexin) [70] [1].
Quantifying Exosome Uptake by Target Cells
- Fluorescent Labeling and Tracking: Label purified exosomes with a lipophilic dye (e.g., PKH67, DiD) or a cell-permeant dye (e.g., CFSE). Incubate labeled exosomes with target keratinocytes or endothelial cells for a defined period.
- Flow Cytometry and Confocal Microscopy:
- Flow Cytometry: Quantifies the percentage of cells that have taken up exosomes and the mean fluorescence intensity, providing a quantitative measure of uptake efficiency.
- Confocal Microscopy: Provides visual, high-resolution confirmation of exosome internalization within the cytoplasm of target cells, allowing for co-localization studies.
Table 2: The Scientist's Toolkit: Essential Reagents for Variability Studies
| Research Reagent / Tool | Function in Experimental Protocol |
|---|---|
| Liberase | Enzymatic blend for high-yield isolation of viable MSCs from tissue [67]. |
| Flow Cytometry Antibodies (CD105, CD73, CD90, CD34, CD45) | Immunophenotyping of MSCs to confirm identity and purity per ISCT criteria [67] [68]. |
| Osteo-/Adipo-/Chondro-Induction Media | Directed differentiation kits to validate MSC multipotency, a key quality indicator [67] [68]. |
| Senescence-Associated β-Galactosidase Staining Kit | Histochemical detection of cellular senescence in MSC populations [67] [68]. |
| PKH67 / DiD Fluorescent Cell Linker Kits | Lipophilic dyes for stable and quantitative labeling of exosome membranes for uptake tracking. |
| Nanoparticle Tracking Analyzer (e.g., NanoSight) | Measures the size and concentration of isolated exosome preparations [70]. |
| Antibodies for CD63, CD81, TSG101 | Western Blot or Flow Cytometry markers for confirming the exosomal identity of isolates [70] [1]. |
Visualization of Experimental Workflow and Key Signaling
The following diagrams outline the core experimental workflow for assessing donor-impacted variability and a generalized signaling mechanism of exosome uptake and action.
Diagram 1: Experimental Workflow for Assessing Donor Impact. This workflow systematically links donor characteristics to MSC fitness and subsequent exosome function.
Diagram 2: Exosome Signaling and Functional Impact. Donor-modulated exosome cargo is internalized by target cells, triggering signaling pathways that drive functional outcomes relevant to therapy.
The systematic addressing of donor-related variability is not merely an academic exercise but a fundamental prerequisite for the successful clinical translation of MSC exosome therapies. The age, tissue source, and health status of the donor are intrinsic variables that define the identity and functional capacity of both MSCs and their exosomes, directly influencing critical processes like uptake by keratinocytes and endothelial cells.
Future efforts must focus on several key areas. First, the development of more sophisticated potency assays that can predict exosome efficacy based on donor MSC profiles is crucial. Second, bioengineering strategies, such as preconditioning MSCs or directly modifying exosomes, offer promising avenues to standardize and enhance exosome function irrespective of donor variance [22] [49]. Finally, establishing large-scale, well-characterized donor MSC banks with comprehensive metadata will provide the consistent starting material necessary for reproducible therapy development. By integrating the detailed characterization and standardized protocols outlined in this guide, researchers and drug developers can transform the challenge of heterogeneity into an opportunity for creating more precise, reliable, and effective exosome-based therapeutics.
Optimizing Production and Isolation Protocols (e.g., TFF vs. UC) for High-Quality, Potent Exosomes
The transition from mesenchymal stem cell (MSC) therapies to cell-free approaches utilizing MSC-derived exosomes, or small extracellular vesicles (sEVs), represents a paradigm shift in regenerative medicine and drug delivery [22] [71]. These nanoscale vesicles (30-150 nm) mediate the therapeutic effects of their parent cells by transferring functional proteins, lipids, and nucleic acids to recipient cells, thereby precisely modulating processes like inflammation, angiogenesis, and tissue repair [22]. For research focusing on MSC exosome uptake by keratinocytes and endothelial cells—key players in skin regeneration and vascular function—the biological potency of the exosome preparation is paramount [8]. However, this potency is critically dependent on the methods used for exosome production and isolation. Inconsistent or suboptimal protocols can compromise exosome yield, purity, and integrity, leading to irreproducible results and hindering both mechanistic studies and clinical translation [72] [73]. This guide provides an in-depth analysis of current optimization strategies to empower researchers in generating high-quality, potent exosomes for reliable scientific and therapeutic applications.
Exosome Biogenesis and Cargo Loading
A foundational understanding of exosome biogenesis is essential for optimizing their production. Exosomes originate as intraluminal vesicles (ILVs) within the endosomal system, formed through the inward budding of the limiting membrane of multivesicular bodies (MVBs) [73]. These MVBs subsequently fuse with the plasma membrane, releasing the ILVs into the extracellular space as exosomes [73]. This intricate process ensures that exosomes are loaded with a specific molecular cargo—including proteins (e.g., CD63, CD81, TSG101), lipids, DNA, and various RNA species (mRNA, miRNA)—from their parent cell [22]. The cargo reflects the physiological state of the source cell and dictates the exosome's function upon delivery to a recipient cell [73]. Therefore, the primary goal of optimization is not merely to maximize particle count, but to preserve this native cargo and the vesicle's structural integrity throughout the production and isolation pipeline.
Optimizing Cell Culture for High Exosome Yield
The first variable in securing a potent exosome product is the condition of the source cells. Evidence indicates that cell culture parameters significantly impact the quantity and quality of secreted sEVs [72].
Culture Media Selection
- Media Formulation: A comparative study demonstrated that Bone Marrow-MSCs (BM-MSCs) cultured in Alpha Minimum Essential Medium (α-MEM) showed a higher expansion ratio and yielded more sEVs per cell (4,318.72 ± 2,110.22 particles/cell) compared to those cultured in Dulbecco's Modified Eagle Medium (DMEM) (3,751.09 ± 2,058.51 particles/cell) [72]. While not statistically significant in this study, the trend underscores the media's influence.
- Serum Supplements: The use of human platelet lysate (hPL) is recommended for GMP-compliant, xeno-free culture systems, supporting robust cell growth and sEV production [72]. Serum-free media or media supplemented with exosome-depleted fetal bovine serum (FBS) are crucial to avoid contamination with bovine EVs.
Culture System Advancedments
- Two-Dimensional (2D) vs. Three-Dimensional (3D) Culture: Traditional 2D culture is practical but may not replicate the native cellular microenvironment. Emerging 3D dynamic culture systems and bioreactors can enhance MSC proliferation and paracrine secretion, potentially leading to higher exosome yields and altered, potentially more therapeutically relevant, cargo profiles [22].
Table 1: Key Research Reagents for MSC Culture and Exosome Production
| Reagent/Solution | Function in Protocol | Key Considerations for Optimization |
|---|---|---|
| α-MEM Culture Medium | Supports MSC expansion and sEV secretion. | Formulation can influence both cell proliferation and the particle yield of derived sEVs [72]. |
| Human Platelet Lysate (hPL) | Xeno-free supplement for cell culture media. | Supports GMP-compliant manufacturing; preferable to FBS for clinical translation [72]. |
| Serum-Free / EV-Depleted Media | Used during the conditioning phase to collect sEVs. | Prevents contamination of the conditioned medium with exogenous serum-derived EVs [72]. |
Comparative Analysis of Exosome Isolation Techniques
Selecting an isolation method is a critical decision that directly impacts the yield, purity, and functional integrity of exosomes. The optimal choice depends on the specific application, sample volume, and required purity [74].
Ultracentrifugation (UC)
- Principle: This traditional "gold standard" method uses sequential centrifugation steps at increasing forces (up to >100,000 × g) to pellet exosomes based on their size and density [74] [73].
- Performance: UC is known for high purity, but the process is time-consuming and can lead to low recovery rates (as low as 30%) due to vesicle aggregation and damage during the high-force spin [73]. The yield is typically medium [74].
Tangential Flow Filtration (TFF)
- Principle: TFF circulates the sample tangentially across a membrane, separating particles based on size and concentrating the sample simultaneously. This method is highly scalable and gentler than UC [74] [72].
- Performance: A direct comparative study found that TFF provided a statistically higher particle yield than UC when isolating BM-MSC-sEVs [72]. It offers high scalability and medium purity, making it suitable for processing large volumes for therapeutic applications [74].
Size-Exclusion Chromatography (SEC)
- Principle: SEC separates exosomes from smaller contaminants like proteins by passing the sample through a column of porous beads. Smaller molecules enter the pores and are delayed, while larger exosomes elute faster [74].
- Performance: SEC is renowned for maintaining exosome structural integrity and providing high purity. It is highly reproducible and ideal for downstream analytical workflows, though its scalability can be limited [74].
Immunoaffinity Capture
- Principle: This technique uses antibodies against specific exosomal surface markers (e.g., CD9, CD63, CD81) immobilized on a solid support to capture exosome subpopulations [74].
- Performance: It offers very high purity and selectivity for specific exosome subtypes. However, it has low yield and throughput, is costly, and the elution step may damage exosomes [74].
Polymer-Based Precipitation
- Principle: Chemicals like polyethylene glycol (PEG) are used to force exosomes out of solution by altering their solubility [74].
- Performance: This method is simple and offers high yield, but the resulting purity is low due to co-precipitation of non-exosomal contaminants like lipoproteins and protein aggregates [74].
Table 2: Quantitative Comparison of Major Exosome Isolation Techniques
| Method | Purity | Yield | Scalability | Processing Time | Key Advantages | Key Limitations |
|---|---|---|---|---|---|---|
| Ultracentrifugation (UC) | High [74] | Medium [74] | Medium [74] | Long (6-18 hours) [73] | Considered the gold standard; no additional reagents needed [73]. | Time-consuming; potential for vesicle damage; requires expensive equipment [72] [73]. |
| Tangential Flow Filtration (TFF) | Medium [74] | High [74] [72] | High [74] | Moderate to Fast | Gentle on vesicles; excellent for large volumes; scalable [74] [72]. | Lower purity than UC or SEC; membrane fouling can occur [74]. |
| Size-Exclusion Chromatography (SEC) | Medium–High [74] | Medium [74] | High (for analytical scale) [74] | Moderate | Preserves vesicle integrity and function; high reproducibility [74]. | Sample dilution can occur; limited capacity for very large volumes [74]. |
| Immunoaffinity Capture | Very High [74] | Low [74] | Low [74] | Moderate to Long | High specificity for subpopulations; very high purity [74]. | High cost; low throughput; potential antibody-mediated damage [74]. |
| Polymer-Based Precipitation | Low [74] | High [74] | High [74] | Fast (simple protocol) | Simple protocol; minimal equipment needs; high yield [74]. | Very low purity; difficult to remove polymer contaminants [74]. ``` |
Standardized Characterization and Quality Control
Rigorous characterization is non-negotiable for confirming the identity, purity, and potency of isolated exosomes. The International Society for Extracellular Vesicles (ISEV) advocates for a multifaceted approach using "orthogonal" techniques to comply with Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines [74].
- Nanoparticle Tracking Analysis (NTA): This technique measures the size distribution and concentration of particles in a solution by tracking their Brownian motion. It is essential for quantifying yield and ensuring the preparation is enriched in particles of the expected size (e.g., 30-150 nm) [74] [72].
- Transmission Electron Microscopy (TEM): TEM provides visual confirmation of the classic cup-shaped morphology of exosomes and is a key tool for assessing structural integrity [72].
- Western Blotting: This method detects the presence of exosome-associated protein markers (e.g., tetraspanins CD9, CD63, CD81, and the ESCRT-related protein TSG101). Simultaneously, it should confirm the absence of negative markers from the endoplasmic reticulum (e.g., Calnexin) or other cellular compartments to ensure purity [72].
- Flow Cytometry: Advanced flow cytometry allows for high-throughput phenotyping of exosomes based on their surface markers, which is useful for assessing heterogeneity and confirming the presence of specific subpopulations [74].
- Functional Potency Assays: For research on keratinocytes and endothelial cells, relevant in vitro bioassays are critical. These may include testing the exosome preparation's ability to promote keratinocyte migration in a scratch assay, enhance endothelial tube formation, or modulate the expression of specific inflammatory cytokines [8] [72]. Establishing a correlation between physical characteristics and a relevant biological readout is the key to defining potency.
Optimizing the production and isolation of MSC-derived exosomes is a decisive factor in the success and reproducibility of research into their uptake by keratinocytes and endothelial cells. The evidence strongly suggests that moving beyond traditional ultracentrifugation towards more scalable and gentle methods like Tangential Flow Filtration (TFF), often in combination with high-resolution techniques like Size-Exclusion Chromatography (SEC), can significantly enhance the yield and quality of exosome preparations [74] [72]. Future progress hinges on the widespread adoption of standardized characterization protocols and the development of robust, disease-relevant potency assays. Furthermore, emerging technologies such as genetic engineering of parent cells, dynamic 3D culture systems, and the development of intelligent slow-release biomaterials for delivery promise to further refine exosome therapeutics, potentially transforming them from innate regenerative factors into programmable nanomedicines [22] [8]. For the field to mature, a pragmatic focus on identifying critical quality attributes linked to specific mechanisms of action will be essential for clinical translation [71].
Mesenchymal stem cell-derived exosomes (MSC-Exos) represent a promising cell-free therapeutic paradigm in regenerative medicine, demonstrating significant potential for targeting keratinocytes and endothelial cells to promote skin repair and angiogenesis. These nanoscale vesicles (30-150 nm) mediate therapeutic effects through their cargo of proteins, nucleic acids, and lipids, mirroring the paracrine benefits of their parent cells while potentially minimizing risks associated with whole-cell transplantation [75] [76]. Their inherent properties—including low immunogenicity, ability to cross biological barriers, and high biocompatibility—make them particularly attractive for drug delivery [62] [77]. However, their clinical translation is constrained by significant in vivo pharmacokinetic challenges: rapid clearance by the mononuclear phagocyte system, insufficient stability in circulation, and inadequate biodistribution to target tissues [70] [77]. Overcoming these hurdles is paramount for realizing the therapeutic potential of MSC-Exos in applications requiring efficient uptake by keratinocytes and endothelial cells.
Critical Analysis of In Vivo Hurdles
Rapid Systemic Clearance Mechanisms
Upon administration, MSC-Exos face immediate clearance mechanisms that limit their therapeutic window. The primary elimination pathways include hepatic clearance, renal filtration, and sequestration by resident macrophages in the reticuloendothelial system (RES). The half-life of intravenously administered native exosomes is typically short, often measured in minutes, necessitating strategies to prolong circulation time [77]. Furthermore, enzymatic degradation and opsonization (antibody binding) further accelerate removal from circulation. This rapid clearance directly impacts the bioavailability of exosomes at target sites, such as skin tissues containing keratinocytes or endothelial cells in wound beds.
Stability and Integrity Challenges
Exosome stability is compromised by several factors in the biological environment. Shear forces during circulation, enzymatic degradation (particularly by proteases and nucleases in plasma), and physicochemical instability (aggregation, membrane fusion) can lead to premature cargo release and loss of function [62]. Storage conditions also impact stability; while MSC-Exos can be preserved at -80°C for extended periods, the freeze-thaw process can damage their integrity and reduce biological activity [77]. These stability issues must be addressed to ensure that a sufficient proportion of intact, functional exosomes reach their cellular targets.
Suboptimal Biodistribution and Targeting
The natural biodistribution of MSC-Exos following systemic administration often results in accumulation in clearance organs (liver, spleen, and lungs) rather than at pathological sites requiring intervention [77]. While this natural tropism can be advantageous for treating diseases affecting these organs, it presents a significant challenge for targeting specific cell types like keratinocytes and endothelial cells in cutaneous wounds. The enhanced permeability and retention (EPR) effect, which benefits some nanotherapeutics in tumors, offers limited assistance in well-vascularized tissues. Passive targeting mechanisms are generally inefficient, leading to the need for active targeting strategies to improve specificity and uptake by desired cell populations.
Quantitative Analysis of Administration Routes and Biodistribution
The administration route significantly influences the pharmacokinetic profile and biodistribution of MSC-Exos. Comparative data from clinical trials reveal substantial differences in effective doses and distribution patterns across administration methods.
Table 1: Comparison of MSC Exosome Administration Routes and Biodistribution Patterns
| Administration Route | Typical Effective Dose (Particles) | Primary Distribution Organs/Tissues | Advantages | Limitations |
|---|---|---|---|---|
| Intravenous (IV) | >10^8 particles [70] | Liver, spleen, lungs [77] | Systemic distribution; crosses blood-brain barrier [62] | Rapid clearance; high first-pass metabolism; potential embolism |
| Nebulization/Aerosol | ~10^8 particles [70] | Lungs, respiratory epithelium | Lower effective dose; direct targeting; non-invasive | Primarily limited to respiratory system |
| Local/Topical | Variable (dose-dependent) | Skin, keratinocytes, local endothelium [78] | High local concentration; minimal systemic exposure | Limited diffusion; potential physical clearance |
| Intravitreal | 50 µg/mL (retinal studies) [72] | Retinal pigment epithelium | Direct ocular targeting; bypasses systemic circulation | Invasive procedure; limited to ocular applications |
The data reveal that route-dependent efficacy necessitates careful therapeutic planning. Notably, nebulization achieves therapeutic effects at doses approximately 10-fold lower than intravenous administration for respiratory conditions, highlighting how targeted delivery can optimize dosing and minimize systemic exposure [70].
Table 2: Impact of Isolation Methods on MSC Exosome Characteristics and In Vivo Performance
| Isolation Method | Particle Yield | Purity | Structural Integrity | Scalability | Impact on In Vivo Performance |
|---|---|---|---|---|---|
| Ultracentrifugation (UC) | Lower [72] | Moderate [62] | Potential deformation due to high g-forces [62] | Limited | Variable biodistribution due to aggregate formation |
| Tangential Flow Filtration (TFF) | Higher [72] | High [72] | Better preservation of membrane integrity | Excellent for large-scale GMP production [72] | More consistent pharmacokinetics |
| Size Exclusion Chromatography (SEC) | Moderate | High [62] | Good preservation of native structure | Moderate | Improved stability and reduced protein contamination |
| Density Gradient Centrifugation | Low [62] | Very High [62] | Maintains vesicle integrity | Poor | Clean profiles but limited yield for therapy |
Isolation methodology directly impacts critical quality attributes that influence in vivo behavior. TFF demonstrates advantages in both yield and preservation of exosome integrity, contributing to more predictable pharmacokinetics [72].
Engineering Strategies to Overcome Pharmacokinetic Barriers
Surface Modification for Enhanced Circulation and Targeting
Surface engineering of MSC-Exos represents a powerful approach to modulate their pharmacokinetic properties and cellular targeting specificity.
Diagram 1: Surface Engineering Strategies for Enhanced Circulation and Targeting. This workflow illustrates modification approaches to address rapid clearance and improve cellular specificity.
Polyethylene glycol (PEGylation) creates a hydrophilic shield around exosomes, reducing protein adsorption and recognition by phagocytic cells, thereby extending circulation half-life [77]. For active targeting, incorporation of peptide ligands specific to receptors overexpressed on keratinocytes (e.g., integrins, growth factor receptors) or endothelial cells (e.g., vascular endothelial growth factor receptor, integrins) enhances site-specific accumulation. Antibody fragments or affibodies against cell-surface markers enable precise targeting of specific cell populations within complex tissues [76].
Biomaterial-Assisted Delivery Systems
Incorporating MSC-Exos into advanced biomaterial scaffolds represents another strategic approach to address pharmacokinetic limitations, particularly for local delivery to skin and endothelial tissues.
Diagram 2: Biomaterial-Assisted Delivery Systems for Localized Exosome Delivery. These systems protect exosomes and control their release kinetics for improved therapeutic outcomes.
Hydrogel systems (e.g., hyaluronic acid, chitosan, collagen) provide a hydrated, three-dimensional environment that preserves exosome integrity while controlling their release kinetics [79]. Microneedle patches physically breach stratum corneum barriers to deliver exosomes directly to epidermal and dermal layers containing keratinocytes and microvascular endothelial cells. Nanofiber scaffolds offer high surface area-to-volume ratios for exosome attachment and sustained release, particularly beneficial for wound healing applications [78].
Cellular Uptake Mechanisms and Modulation Strategies
Understanding the fundamental mechanisms by which keratinocytes and endothelial cells internalize MSC-Exos is essential for designing strategies to enhance uptake efficiency.
Diagram 3: Cellular Uptake Mechanisms of MSC Exosomes by Target Cells. Understanding these pathways enables engineering approaches to enhance specific uptake by keratinocytes and endothelial cells.
MSC-Exos utilize multiple entry mechanisms: direct membrane fusion releases content directly into the cytoplasm, while various endocytic pathways (clathrin-mediated, caveolin-mediated, and macropinocytosis) result in endosomal trafficking [77]. Receptor-mediated uptake can be enhanced by engineering exosomes to display ligands for receptors abundant on target cells—for example, integrins on keratinocytes or ICAM-1 on endothelial cells. Modifying the exosomal surface with cell-penetrating peptides (CPPs) can also facilitate more efficient cellular internalization, bypassing conventional endocytic routes that may lead to lysosomal degradation.
Experimental Protocols for Evaluating In Vivo Performance
Biodistribution and Pharmacokinetics Tracking Protocol
Objective: Quantify the temporal and spatial distribution of administered MSC-Exos in vivo.
Materials:
- Purified MSC-Exos (isolated via TFF or UC)
- Lipophilic membrane dyes (DiR, DiD, PKH67)
- Radiolabeling reagents (99mTc, 111In)
- Luciferase reporter constructs
- In vivo imaging system (IVIS)
- Micro-PET/CT scanner
- Animal model (e.g., murine wound healing model)
Procedure:
- Labeling: Label MSC-Exos with near-infrared lipophilic dye (DiR) according to manufacturer's protocol, followed by purification via size exclusion chromatography to remove unincorporated dye.
- Administration: Administer labeled exosomes (1010 particles/animal) via selected route (IV, local, etc.) to animals.
- Imaging: Acquire whole-body fluorescence images at predetermined time points (5 min, 30 min, 2 h, 6 h, 24 h, 48 h) post-administration using IVIS spectrum system.
- Quantification: Quantify fluorescence intensity in regions of interest (skin, liver, spleen, kidneys, lungs) using living image software.
- Validation: Sacrifice animals at endpoint, collect tissues for cryosectioning, and counterstain with cellular markers (cytokeratin for keratinocytes, CD31 for endothelial cells) to confirm cellular localization via confocal microscopy.
- Pharmacokinetic Analysis: Calculate pharmacokinetic parameters (Cmax, Tmax, AUC, t1/2) from time-activity curves derived from imaging data.
Functional Uptake Assay in Keratinocytes and Endothelial Cells
Objective: Evaluate the efficiency and functional consequences of MSC-Exos uptake by target cells.
Materials:
- Human keratinocyte cell line (HaCaT)
- Human umbilical vein endothelial cells (HUVEC)
- MSC-Exos labeled with PKH67 green fluorescent dye
- Cell culture reagents and equipment
- Flow cytometer
- Confocal microscope
- Western blot equipment
- Antibodies for phospho-AKT, total AKT, phospho-ERK, total ERK
Procedure:
- Cell Culture: Culture HaCaT cells and HUVECs in appropriate media until 70% confluence.
- Exosome Treatment: Treat cells with PKH67-labeled MSC-Exos (50 μg/mL) for various durations (15 min, 30 min, 1 h, 2 h, 4 h).
- Flow Cytometry: Harvest cells, analyze fluorescence intensity via flow cytometry to quantify uptake efficiency.
- Confocal Microscopy: Fix cells at each time point, counterstain with Phalloidin (F-actin) and DAPI (nuclei), and visualize using confocal microscopy with z-stack imaging to confirm intracellular localization.
- Inhibition Studies: Pre-treat cells with endocytosis inhibitors (chlorpromazine for clathrin-mediated, filipin for caveolae-mediated, EIPA for macropinocytosis) for 1 h before exosome addition to determine entry mechanisms.
- Signaling Analysis: Perform Western blot analysis of key signaling pathways (AKT, ERK) at 0, 15, 30, and 60 min post-exosome treatment to document functional activation.
- Functional Assays: Assess downstream functional responses—keratinocyte migration (scratch assay) and endothelial tube formation (Matrigel assay)—following exosome treatment.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Research Reagent Solutions for MSC Exosome In Vivo Studies
| Reagent/Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Kit | Rapid precipitation of exosomes from conditioned media | Can co-precipitate contaminants; moderate purity |
| Characterization Instruments | Nanoparticle Tracking Analysis (NTA) | Size distribution and concentration measurement | Requires appropriate dilution and calibration |
| In Vivo Tracking Agents | DiR, DiD, PKH67 lipophilic dyes | Near-infrared labeling for biodistribution studies | Potential dye aggregation; requires purification post-labeling |
| Animal Models | Diabetic mouse wound model | Testing exosome efficacy in impaired healing | Mirrors human pathophysiology with vascular complications |
| Cell Culture Media | α-MEM with human platelet lysate | MSC expansion and exosome production | Higher proliferation and particle yield vs. DMEM [72] |
| Engineering Tools | Click chemistry reagents | Surface modification with targeting ligands | Precise conjugation without damaging membrane integrity |
Overcoming the in vivo hurdles of rapid clearance, stability, and biodistribution is essential for advancing MSC exosome therapies toward clinical application, particularly for targeting keratinocytes and endothelial cells in regenerative contexts. Strategic integration of surface engineering, biomaterial-assisted delivery, and cellular uptake modulation presents a multifaceted approach to enhance pharmacokinetic profiles. As standardization in production and characterization improves, coupled with advanced engineering strategies, MSC exosomes are poised to transition from promising nanotherapeutics to clinically viable "programmable nanomedicines" capable of precise targeting and enhanced therapeutic efficacy for skin and vascular applications. Future research should focus on developing integrated platforms that combine these strategies while maintaining the innate biological activity of MSC exosomes.
Strategies to Mitigate Batch-to-Batch Heterogeneity and Ensure Reproducibility
Batch-to-batch heterogeneity presents a significant challenge in biological manufacturing, particularly in the production of mesenchymal stromal cell-derived exosomes (MSC-Exo) for therapeutic applications. This variability can substantially impact the reproducibility, efficacy, and safety of exosome-based treatments targeting keratinocytes and endothelial cells in wound healing and regenerative medicine. This technical guide comprehensively examines the sources of heterogeneity spanning genetic, non-genetic, and procedural domains, and provides evidence-based mitigation strategies encompassing process control, analytical frameworks, and quality-by-design principles. By implementing robust monitoring systems and standardized protocols, researchers can significantly enhance batch consistency, thereby accelerating the clinical translation of MSC exosome therapies.
In the context of MSC exosome research for keratinocyte and endothelial cell applications, batch-to-batch heterogeneity refers to variations in exosome characteristics across different production runs. These variabilities can manifest in differences in exosome size distribution, cargo composition (proteins, miRNAs, lipids), surface marker expression, and ultimately, biological potency in stimulating target cell responses [25] [4]. Such heterogeneity poses substantial challenges for reproducible research outcomes and reliable therapeutic development, as inconsistent exosome preparations can lead to conflicting experimental results and variable treatment efficacy.
The implications of uncontrolled heterogeneity are particularly profound in preclinical and clinical studies, where batch effects can mask true biological outcomes or lead to irreproducible conclusions [80]. For MSC exosomes intended to enhance keratinocyte migration and proliferation or stimulate endothelial angiogenesis, consistent quality and performance are prerequisites for reliable mechanistic studies and eventual clinical translation [25] [4]. This guide systematically addresses the multifaceted sources of this heterogeneity and provides a comprehensive framework for its mitigation throughout the exosome production pipeline.
Genetic and Non-Genetic Heterogeneity in Source Cells
The foundation of exosome quality begins with the parental MSC population, which can exhibit both genetic and non-genetic heterogeneity. Genetic instability in source cells, including mutations such as single-nucleotide polymorphisms (SNPs) and mobile element transposition, can occur at frequencies ranging from 10⁻⁵ to 10⁻¹⁰ per generation, potentially altering cellular functions over extended culture periods [81]. While these mutations may not directly incorporate into exosomes, they can fundamentally change the MSC secretome and exosome production.
Perhaps more immediately impactful is non-genetic heterogeneity, which occurs at higher frequencies and includes:
- Epigenetic modifications: DNA methylation patterns, particularly adenine methylation of GATC sequences in Escherichia coli, can vary between single cells and influence gene expression if methylation sites overlap with regulatory regions [81].
- Cellular noise: Stochastic fluctuations in transcription, translation, ATP levels, cofactor abundance, and growth rate create phenotypic diversity even in genetically identical populations [81].
- Gene expression multimodality: Driven by positive feedback loops, this phenomenon can create distinct subpopulations with different functional states, potentially affecting exosome production consistency [81].
Process-Induced Heterogeneity
Variations in production processes represent a significant source of batch-to-batch variability, particularly when protocols lack standardization or are sensitive to minor parameter fluctuations.
Table 1: Major Sources of Process-Induced Heterogeneity
| Process Stage | Variability Source | Impact on Exosome Quality |
|---|---|---|
| Cell Culture | Serum lot variations, passage number, confluency at harvest | Alters exosome yield, cargo loading, and surface protein composition |
| Extraction Method | Ultracentrifugation parameters, polymer-based precipitation, tangential flow filtration | Affects exosome size heterogeneity, aggregate formation, and co-isolation of contaminants |
| Purification | Chromatography conditions, density gradient centrifugation, buffer exchange | Influences exosome purity, potency, and stability |
| Storage | Cryoprotectant use, freeze-thaw cycles, temperature fluctuations | Impacts exosome integrity, aggregation, and biological activity |
The challenges of process consistency are exemplified in collagen isolation research, where different processing methods applied to the same source material resulted in pronounced variations in yield and composition between batches [82]. Similarly, in large-scale microbiota studies, reagent lot changes introduced significant batch effects that masked true biological signals [80], highlighting the vulnerability of biological systems to procedural variations.
Analytical Challenges in Characterization
Accurately quantifying heterogeneity requires robust analytical methods, which themselves can introduce variability. Different isolation techniques (ultracentrifugation, immunoaffinity, size-exclusion chromatography) yield exosome populations with varying degrees of purity and recovery [4]. The inherent limitations of each method contribute to challenges in obtaining consistent characterization data across batches and between laboratories. For instance, ultracentrifugation, while considered the gold standard, can cause exosome aggregation and requires significant expertise to achieve reproducible results [4].
Additionally, the dynamic range of analytical techniques may not capture the full spectrum of heterogeneity, particularly for low-abundance components that might nevertheless have significant biological effects on keratinocyte or endothelial cell responses.
Strategic Framework for Heterogeneity Mitigation
Source Cell Management
Establishing a well-characterized and consistent cellular starting material is fundamental to reducing batch variability in MSC exosome production.
Comprehensive Cell Banking: Create extensive master and working cell banks using early passage MSCs characterized for identity (surface marker profile), viability, proliferation capacity, and differentiation potential. Implement rigorous testing for microbial contamination and genetic stability at regular intervals.
Cellular Quality Control: Monitor key cellular attributes throughout culture, including:
- Growth kinetics: Population doubling times and senescence markers
- Morphology: Consistent adherent spindle-shaped appearance
- Marker expression: Confirmation of positive (CD73, CD90, CD105) and negative (CD34, CD45, HLA-DR) markers
- Functional potency: Standardized assays for trilineage differentiation potential
Culture Standardization: Maintain consistent culture conditions including serum lots (or defined serum-free formulations), seeding densities, feeding schedules, and passage protocols. Document any deviation from established protocols, as minor changes can significantly impact exosome characteristics and performance in keratinocyte and endothelial cell assays [81].
Process Control and Optimization
Implementing robust manufacturing processes with defined critical process parameters (CPPs) is essential for minimizing batch-to-batch variation.
Table 2: Process Control Strategies for Consistent Exosome Production
| Process Stage | Control Parameters | Monitoring Approach |
|---|---|---|
| Cell Culture | Dissolved oxygen (40-60%), pH (7.2-7.4), glucose levels, confluence at harvest | Real-time bioreactor monitoring, scheduled metabolic analysis |
| Exosome Collection | Conditioned media collection timepoint, centrifugation speed/duration, filtration pore size | Process validation studies, step yield quantification |
| Isolation | Ultracentrifugation: g-force, duration, rotor type; Tangential flow filtration: flow rates, transmembrane pressure | In-process controls, performance qualification |
| Purification | Density gradient: concentration, centrifugation parameters; Size-exclusion: column calibration, flow rates | Fraction analysis, quality thresholds |
| Formulation | Buffer composition, cryoprotectant concentration, vialing conditions | Stability testing, container compatibility studies |
Implementing a Quality-by-Design (QbD) approach helps identify and control critical process parameters that most significantly impact exosome critical quality attributes (CQAs). This systematic approach to process development establishes a design space with proven acceptable ranges for each parameter, ensuring consistent production of exosomes with desired characteristics for keratinocyte and endothelial cell targeting [83].
Analytical Quality Control Framework
A comprehensive analytical strategy is essential for quantifying and controlling batch-to-batch heterogeneity.
Orthogonal Characterization Methods: Employ multiple complementary techniques to thoroughly assess exosome attributes:
- Size and concentration: Nanoparticle tracking analysis (NTA) with standardized settings and sample preparation
- Surface markers: Flow cytometry with calibrated instruments and controlled antibody lots
- Morphology: Transmission electron microscopy with standardized staining protocols
- Content analysis: Western blot, miRNA profiling, and proteomic analyses with reference standards
Reference Materials and Controls: Include appropriate controls in each batch to ensure analytical consistency:
- Negative controls: Process blanks to identify potential contaminants
- Positive controls: Well-characterized exosome reference materials or spike-in standards
- Biological controls: Consistent responder cell lines (keratinocytes and endothelial cells) for functional assays
Data-Driven Contaminant Identification: For low-biomass samples like exosomes, implement statistical approaches to identify and remove contaminant signals. As demonstrated in microbiome research, a two-tier strategy combining algorithm-based detection (e.g., decontam) with data structure analysis can effectively identify batch-specific contaminants that might otherwise be misinterpreted as biological signals [80].
Experimental Protocols for Consistency Assessment
Protocol: Technical Reproducibility Assessment
Purpose: To evaluate the repeatability and reproducibility of exosome isolation and characterization methods.
Materials:
- MSC-conditioned media from the same production batch
- Standardized exosome isolation reagents
- Phosphate-buffered saline (PBS)
- Nanoparticle tracking instrument
- Western blot equipment
- Keratinocyte (HaCaT) and endothelial cell (HUVEC) cultures
Procedure:
- Split Sample Analysis: Divide a large pool of MSC-conditioned media into multiple aliquots (n≥5) for parallel processing through the entire isolation workflow.
- Inter-operator Validation: Have multiple trained personnel independently process aliquots using the same protocol.
- Inter-instrument Comparison: Analyze identical exosome preparations on different instruments of the same type.
- Functional Consistency: Assess biological activity using standardized keratinocyte migration and endothelial tube formation assays [25].
Data Analysis: Calculate coefficients of variation (CV) for each measured parameter (yield, size, marker expression, potency). Establish acceptable variability thresholds (typically CV<15-20% for technical replicates) for continued process validation.
Protocol: Inter-batch Comparison Study
Purpose: To systematically evaluate consistency across multiple production batches.
Materials:
- MSC exosomes from at least three independent production runs
- Characterized keratinocyte and endothelial cell lines
- Angiogenesis assay kit (e.g., ECM gel matrix)
- Migration assay equipment
- qPCR system for gene expression analysis
Procedure:
- Physical Characterization: Analyze all batches using NTA, TEM, and surface marker profiling (CD63, CD81, CD9).
- Content Consistency: Perform proteomic analysis on all batches to compare cargo profiles.
- Functional Potency Assessment:
- Keratinocyte proliferation: Incubate HaCaT cells with exosomes (donor to acceptor cell ratio 7:1) for 3 days and quantify proliferation using Ki67 staining [25].
- Keratinocyte migration: Perform scratch wound assay with exosome treatment and measure coverage of scratched surface area [25].
- Endothelial tube formation: Seed endothelial cells on ECM gel matrix with exosomes and quantify tube length, junctions, and meshes [25].
- Molecular Response: Analyze gene expression changes in keratinocytes and endothelial cells after exosome exposure.
Data Analysis: Apply multivariate statistical methods (PCA, hierarchical clustering) to identify batch-related clustering. Establish equivalence margins for key parameters based on biological relevance and analytical variability.
Visualization of Quality Control Workflows
Heterogeneity Mitigation Strategy Diagram
Batch Consistency Assessment Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Research Reagent Solutions for Consistent Exosome Research
| Reagent/Material | Function | Consistency Considerations |
|---|---|---|
| Characterized MSC Lines | Source of exosomes | Validated identity, potency, and genetic stability; extensive banking |
| Defined Culture Media | MSC expansion and exosome production | Serum-free formulations preferred; minimal lot-to-lot variation |
| Ultracentrifugation Equipment | Exosome isolation | Calibrated instruments; standardized rotors and protocols |
| Size Exclusion Columns | Exosome purification | Pre-qualified performance; consistent retention characteristics |
| Reference Exosome Standards | Analytical controls | Well-characterized physical and biological properties |
| Validated Antibody Panels | Exosome characterization | Specificity confirmation; controlled lot-to-lot performance |
| Functional Assay Kits | Potency assessment (keratinocyte/endothelial) | Standardized components; predefined performance criteria |
| Stable Cell Lines | Response assessment (HaCaT, HUVEC) | Authenticated origins; consistent passage protocols |
Mitigating batch-to-batch heterogeneity in MSC exosome production requires a systematic, multi-layered approach addressing source cells, process parameters, analytical methods, and data interpretation. By implementing the strategies outlined in this guide—comprehensive cell banking, process control, orthogonal analytics, and robust functional assessment—researchers can significantly enhance the reproducibility of their MSC exosome preparations. This consistency is fundamental for reliable research on exosome uptake mechanisms by keratinocytes and endothelial cells, and ultimately for the successful clinical translation of exosome-based therapies. The framework presented here provides a roadmap for establishing standardized practices that will advance the field toward more reproducible and therapeutically viable exosome applications.
Critical Controls and Standards for Rigorous Uptake Quantification
This technical guide outlines the critical controls and standardization practices essential for rigorous quantification of MSC exosome uptake by keratinocytes and endothelial cells. Adherence to these guidelines is fundamental for generating reproducible, reliable, and interpretable data in drug development and mechanistic research.
Standardization Frameworks and Reporting Guidelines
The field of extracellular vesicle (EV) research has established robust frameworks to address challenges in reproducibility and standardization.
- MISEV Guidelines: The Minimal Information for Studies of Extracellular Vesicles (MISEV), developed by the International Society for Extracellular Vesicles (ISEV), provides a constantly updated framework for EV research. Adherence to MISEV guidelines is critical for rigorous experimental design, reporting, and peer review, strengthening the credibility of data for clinical applications [84].
- EV-TRACK Platform: The EV-TRACK knowledgebase is a centralized repository that allows researchers to document the experimental parameters of their EV experiments in a standardized manner. Utilizing this platform promotes uniformity and enhances the reproducibility of EV research, including uptake studies [84].
Critical Controls for Exosome Isolation and Characterization
Accurate uptake quantification is contingent upon using well-characterized exosome preparations. The following controls are essential to confirm the identity, purity, and functionality of isolated MSC exosomes.
Isolation Method Selection and Validation
The choice of isolation method significantly impacts exosome yield, purity, and biological activity, thereby influencing uptake experiments. Key techniques are compared below.
Table 1: Comparison of Major Exosome Isolation Protocols [62] [74]
| Method | Principle | Purity | Yield | Key Considerations for Uptake Studies |
|---|---|---|---|---|
| Differential Ultracentrifugation | Sequential centrifugation at increasing forces (up to ≥100,000 × g) [62] | High [74] | Medium [74] | Considered a "gold standard"; may cause vesicle damage due to high shear forces [62]. |
| Size-Exclusion Chromatography (SEC) | Separation by hydrodynamic volume [62] | Medium–High [74] | Medium [74] | Preserves vesicle integrity and biological function; excellent reproducibility [62] [74]. |
| Ultrafiltration / Tangential Flow Filtration (TFF) | Size-based separation using membranes [62] | Medium [74] | High [74] | Scalable for production; shear stress may damage vesicles or affect function [62]. |
| Anion Exchange Chromatography (AEC) | Separates vesicles based on surface charge [62] | High (when combined with other methods) [62] | Medium [62] | Useful for isolating specific subpopulations; can be combined with SEC for high purity [62]. |
| Immunoaffinity Capture | Antibody-based binding to surface markers (e.g., CD9, CD63, CD81) [74] | Very High [74] | Low [74] | Isolates specific exosome subtypes; ideal for studying specific uptake mechanisms; limited throughput [74]. |
Pre-uptake Characterization Controls
Before initiating uptake experiments, exosome preparations must be characterized for key physical and biochemical parameters.
- Particle Concentration and Size Distribution: Use Nanoparticle Tracking Analysis (NTA) or similar techniques to determine the particle size distribution (expected 30-200 nm) and concentration (particles/mL) [85] [74]. This quantification is essential for standardizing the dose of exosomes applied to recipient cells.
- Purity Assessment: Measure the total protein content of the preparation. A low particle-to-protein ratio can indicate co-isolation of contaminating proteins or lipoproteins, which could confound uptake results [74].
- Biomarker Validation: Confirm the presence of positive exosomal markers (e.g., tetraspanins CD63, CD81, CD9) and the absence of negative markers (e.g., calnexin, apolipoproteins) via immunoblotting to ensure the vesicular origin of the preparation [85].
- Visualization: Use electron microscopy to visually confirm the morphology and size of the isolated vesicles [74].
- Functionality Check: Validate the biological activity of the exosome preparation in a relevant functional assay prior to uptake studies.
Methodologies for Quantifying Exosome Uptake
Directly quantifying internalized exosomes requires meticulous experimental design to distinguish true uptake from surface adherence.
Experimental Protocol: sEV Uptake Assay and Retrieval
This protocol, adapted from a 2025 study, provides a method to isolate and analyze internalized exosome subpopulations, offering direct evidence for selective uptake [85].
1. Labeling of Exosomes
- Label MSC exosomes using fluorescent lipophilic dyes (e.g., PKH67) or more specific bio-orthogonal click chemistry. For click chemistry, incubate donor MSCs with Ac4ManNAz to introduce azide groups onto cell surfaces, which are incorporated into exosomes. After isolation, label exosomes with AZDye 488-conjugated dibenzyl cyclooctyne (DBCO) [85].
2. Incubation with Recipient Cells
- Culture recipient keratinocytes or endothelial cells (e.g., ( 3 \times 10^5 ) cells) and allow them to adhere for 24 hours.
- Incubate cells with the labeled exosomes (e.g., ( 5 \times 10^9 ) sEVs/mL) for a predetermined time (e.g., 12-72 hours). Determine the optimal incubation time by measuring fluorescence intensity at different time points [85].
3. Washing and Cell Harvesting
- After incubation, gently wash the cells with PBS to remove any uninternalized exosomes.
- Detach the cells using trypsin treatment to dissociate surface-bound but not internalized exosomes [85].
4. Retrieval of Internalized Exosomes
- Subject the harvested recipient cells to a freeze-thaw cycle (-20°C for 15 min).
- Lyse the cells using a hypotonic solution (e.g., 10% PBS + 0.1% Triton) on ice for 15 min to release intracellular contents.
- Centrifuge the lysate sequentially (300 g for 10 min, 2,000 g for 30 min, 10,000 g for 1 h) to remove large cellular debris and organelles.
- Perform ultracentrifugation on the supernatant at 100,000 g for 90 min (twice) to pellet the internalized exosomes [85].
5. Analysis of Retrieved Exosomes
- Resuspend the final pellet and use NTA to quantify the retrieved exosomes.
- Analyze the cargo (proteins, RNA) via immunoblotting or other molecular techniques to characterize the internalized subpopulations and their functional roles [85].
Quantification and Imaging Controls
- Distinguishing Internalized vs. Surface-Bound Signal: Treat cells with trypsin after the uptake incubation and washing steps. Trypsinization removes exosomes bound to the cell surface but not those internalized, allowing for specific quantification of internalized fluorescence via flow cytometry or a microplate reader [85].
- Inhibition of Active Uptake Pathways: Use pharmacological inhibitors to block major endocytic pathways (e.g., clathrin-mediated endocytosis, macropinocytosis, phagocytosis). A significant reduction in fluorescence signal upon inhibition confirms active cellular uptake versus passive attachment [85].
- Confocal Microscopy: Perform Z-stack imaging and orthogonal views to visually confirm the intracellular localization of the fluorescent signal, providing spatial validation of uptake [85].
The Scientist's Toolkit: Essential Reagents and Materials
Table 2: Key Research Reagent Solutions for Uptake Quantification
| Item | Function / Application | Example Specifics |
|---|---|---|
| PKH67 / PKH26 | Lipophilic fluorescent dyes for general exosome membrane labeling [85]. | Used for tracking and visualizing exosomes via fluorescence microscopy or flow cytometry. |
| Click Chemistry Reagents | Specific, covalent labeling of exosomes via metabolic engineering [85]. | Ac4ManNAz (azide donor), AZDye 488-DBCO (fluorescent label). Reduces dye transfer artifacts. |
| Tetraspanin Antibodies | Immunoaffinity capture and characterization of exosomes [74]. | Anti-CD63, Anti-CD81, Anti-CD9 for positive marker identification. |
| Pharmacologic Inhibitors | Blocking specific endocytic pathways to determine uptake mechanisms [85]. | e.g., Chlorpromazine (clathrin-mediated), EIPA (macropinocytosis). |
| Trypsin-EDTA | Enzymatic dissociation of cells to remove surface-bound exosomes after uptake [85]. | 0.5% trypsin-EDTA solution. Critical control to confirm internalization. |
| Density Gradient Medium | High-purity exosome isolation via density gradient centrifugation [62]. | Sucrose or iodixanol gradients. |
| Size-Exclusion Columns | High-purity, size-based exosome isolation maintaining biological activity [62] [74]. | e.g., qEV columns. |
Signaling Pathways in Recipient Cells
Understanding the downstream signaling cascades activated upon exosome uptake is crucial for elucidating the functional outcomes in recipient keratinocytes and endothelial cells. Research indicates that exosome cargo can modulate key cellular pathways.
The diagram illustrates two well-documented pathways in keratinocytes that could be modulated by MSC exosome cargo. The PI3K-Akt-Rac1 pathway is critically involved in insulin-stimulated keratinocyte migration, which is dependent on the insulin receptor and independent of EGF/EGF-R signaling [86]. Simultaneously, the KDR/GEF-H1/RhoA pathway is essential for VEGF-A-induced migration, where VEGF-A activates the kinase insert domain receptor (KDR/VEGFR2), leading to ERK1/2-mediated phosphorylation and activation of the guanine nucleotide exchange factor GEF-H1, which in turn activates RhoA to drive cytoskeletal rearrangements necessary for migration [87]. MSC exosomes, loaded with proteins and miRNAs, may deliver components that activate or modulate these and other pathways in recipient keratinocytes and endothelial cells, promoting processes critical for wound healing, such as proliferation, migration, and angiogenesis [62].
Proof of Concept: Validating Uptake and Comparing Therapeutic Platforms
Preclinical In Vivo Evidence of Uptake and Efficacy in Wound Healing and Ischemia Models
This technical review synthesizes current preclinical evidence on the uptake mechanisms and therapeutic efficacy of advanced biologics, particularly mesenchymal stem cell-derived exosomes (MSCs-Exo) and chemically modified mRNA (cmRNA) technologies, in wound healing and ischemia models. The analysis reveals robust proof-of-concept across multiple animal models, with exosomes demonstrating enhanced angiogenesis through specific miRNA cargo delivery and cmRNA platforms showing sustained localized protein expression. Quantitative data from controlled studies indicate significant improvement in wound closure rates (up to 90% within 10-14 days), enhanced neovascularization, and accelerated reperfusion in ischemic tissues. These therapeutic effects are mediated through defined molecular pathways including MEK/ERK, PI3K/Akt, and Wnt/β-catenin signaling. Despite promising efficacy and favorable safety profiles, challenges remain in standardization of production, quantification of biodistribution, and translation to clinical applications. This comprehensive analysis provides researchers with validated experimental protocols, critical methodological considerations, and strategic directions for advancing this promising therapeutic modality toward clinical development.
The therapeutic potential of mesenchymal stem cell (MSC) derivatives, particularly exosomes and emerging nucleic acid technologies, represents a paradigm shift in regenerative medicine approach to wound healing and ischemic diseases. MSC-derived exosomes (MSCs-Exo) are nanoscale, lipid bilayer-enclosed extracellular vesicles containing bioactive molecules that mediate intercellular communication by delivering protein, lipid, and nucleic acid cargos to target cells [30]. These vesicles exhibit multifaceted biological functions, including immunomodulation, tissue repair, and pro-angiogenic activity, while offering advantages over cell-based therapies through their non-immunogenicity, absence of tumorigenic risk, high accessibility, and ability to cross biological barriers [4]. Similarly, chemically modified mRNA (cmRNA) technologies have emerged as a promising alternative to recombinant protein therapy, offering transient but sustained protein expression without nuclear entry or genomic integration risks [88].
Understanding the uptake mechanisms of MSC exosomes by target cells such as keratinocytes and endothelial cells is fundamental to optimizing their therapeutic application. The molecular cargo of these exosomes—including specific miRNAs, proteins, and lipids—orchestrates complex therapeutic responses through modulation of key signaling pathways in recipient cells [30] [4]. This in-depth technical review critically evaluates the current preclinical evidence regarding the uptake, biodistribution, and mechanistic efficacy of these advanced therapeutic platforms in established models of wound healing and ischemia, providing researchers with validated experimental frameworks and analytical approaches for future investigations.
Uptake Mechanisms and Biodistribution
Cellular Uptake Pathways
Exosomes utilize multiple sophisticated mechanisms for cellular entry and cargo delivery, which have been characterized through extensive in vitro and in vivo tracking studies. The primary uptake pathways include direct interaction, membrane fusion, and internalization, with the specific mechanism varying based on exosome source, surface composition, and target cell type [30].
- Direct Interaction: Exosomes can directly interact with recipient cells via ligand-receptor binding, triggering downstream signaling cascades without full internalization. This mechanism facilitates rapid response to exosomal surface molecules.
- Membrane Fusion: The lipid bilayers of exosomes and target cells merge, facilitating direct luminal component release into the cytoplasm. This process initiates with a hemi-fusion stalk formation via hydrophobic exosomes-plasma membrane interactions, which then expands to create a consistent fusion pore [30].
- Internalization: Exosomes are engulfed by recipient cells through multiple endocytic pathways, including clathrin-dependent endocytosis, lipid raft-mediated uptake, caveolin-assisted endocytosis, phagocytosis, and macropinocytosis. Following internalization, exosomes merge with intracellular compartments or endosomal routes to discharge their contents [30].
These uptake pathways are not mutually exclusive, with the same population of exosomes potentially utilizing multiple routes for cellular entry depending on local microenvironmental conditions and specific exosome surface characteristics.
Biodistribution and Kinetics
Preclinical studies utilizing advanced imaging modalities have provided critical insights into the tissue distribution and persistence of exosomes and cmRNA therapeutics in vivo.
Table 1: Biodistribution Profiles of Therapeutic Platforms in Preclinical Models
| Therapeutic Platform | Model System | Administration Route | Tissue Distribution | Peak Expression | Duration | Detection Method |
|---|---|---|---|---|---|---|
| Luciferase cmRNA [88] | Murine skin | Local injection | Epidermal and dermal layers | 48 hours | Up to 11 days | IVIS imaging, immunohistochemistry |
| MSCs-Exo [4] | Various disease models | Intravenous, local | Liver, spleen, lungs, target organs | 2-6 hours | 24-72 hours | Fluorescent/radioactive tracking |
| MSCs-Exo [89] | Rat spinal cord ischemia-reperfusion | Local application | Neural tissue, crossing BBB | 6-12 hours | Up to 48 hours | Immunohistochemistry, Western blot |
| hpMSCs-Exos [89] | Rat spinal cord IR injury | Local delivery | Spinal cord tissue | Not specified | Detected at 48h | Western blot, functional assays |
In cutaneous applications, cmRNA formulated in citrate-saline buffer demonstrated highly efficient transfection in both keratinocytes and dermal fibroblasts, with efficiency exceeding 90% in vitro [88]. Following local administration in murine skin, cmRNA-encoded proteins showed localized and sustained expression, with immunohistochemistry revealing protein expression in both epidermal and dermal layers as early as 1 hour post-injection, peaking at 48 hours, and remaining detectable for up to 11 days via in vivo imaging systems [88].
Exosomes demonstrate more heterogeneous biodistribution patterns influenced by administration route and source characteristics. When administered systemically, exosomes predominantly accumulate in clearance organs (liver, spleen), while local application enhances target tissue retention [4]. Their natural ability to cross biological barriers, including the blood-brain barrier, enables therapeutic access to privileged sites [89].
Efficacy in Wound Healing Models
Exosome-Based Interventions
MSC-derived exosomes have demonstrated remarkable efficacy across multiple wound healing models, promoting healing through multifaceted mechanisms including enhanced angiogenesis, modulation of inflammation, and stimulation of cellular proliferation and migration.
Table 2: Efficacy of Exosome-Based Therapies in Wound Healing Models
| Exosome Source | Model | Key Findings | Mechanistic Insights | Reference |
|---|---|---|---|---|
| Human umbilical cord MSCs [30] | Murine full-thickness wound | Accelerated wound closure, enhanced angiogenesis | miR-27b activation of ITCH/JUNB/IRE1α pathway; promoted keratinocyte and fibroblast activation | [30] |
| BMSCs [30] | Murine diabetic wound | Improved healing in compromised models | miR-223 mediated promotion of M2 macrophage polarization | [30] |
| ADMSCs [30] | Natural aging and type-2 diabetic mouse models | Mitigated endothelial cell senescence, promoted angiogenesis | miR-146a/Src pathway modulation; promoted M2 macrophage polarization | [30] |
| Lactobacillus rhamnosus GG [30] | Murine wound model | Accelerated wound healing | Promoted angiogenesis and re-epithelialization | [30] |
The therapeutic effects of exosomes are largely mediated through their specific miRNA cargo, which regulates gene expression in recipient cells. For instance, exosomes from human umbilical cord MSCs (HUMSCs) containing miR-27b activate keratinocytes and fibroblasts through the ITCH/JUNB/IRE1α pathway, significantly accelerating wound closure in vivo [30]. Similarly, BMSC-derived exosomes containing miR-223 promote the M2 polarization of macrophages, facilitating the resolution of inflammation and transition to proliferative phase healing [30].
Additional studies demonstrate that adipose tissue-derived MSC (ADMSC) exosomes mitigate endothelial cell senescence and promote angiogenesis through miR-146a/Src regulation, showing particular efficacy in challenging healing environments such as natural aging and type-2 diabetic mouse models [30]. Beyond mammalian sources, bacterial EVs from beneficial species such as Lactobacillus rhamnosus GG have also demonstrated significant wound healing properties, promoting both angiogenesis and re-epithelialization in preclinical models [30].
cmRNA and Biomaterial Approaches
Emerging nucleic acid technologies and advanced biomaterials have shown complementary therapeutic potential in wound healing applications.
Chemically Modified mRNA (cmRNA): A recent innovative approach utilizing cmRNA encoding epidermal growth factor (EGF) demonstrated remarkable efficacy in full-thickness skin defect models. The cmRNA, formulated in biocompatible citrate-saline, achieved high transfection efficiency in human immortalized keratinocytes (HaCaT) and normal human dermal fibroblasts (NHDF) (93.97% ± 1.25% and 90.37% ± 0.97%, respectively), resulting in efficient production of biologically active EGF protein [88]. This platform significantly accelerated wound healing, with superior re-epithelialization observed compared to controls by day 6. By day 14, EGF cmRNA outperformed recombinant human EGF (rhEGF), as indicated by enhanced formation of hair follicles and cutaneous glands, better-organized collagen fibers, and a reduced collagen Type I/III ratio [88]. Mechanistic studies revealed marked increases in MEK/ERK signaling and Ki67 mRNA expression both in vitro and in vivo, indicating activation of proliferative pathways [88].
Keratin-Based Biomaterials: Biomaterials derived from natural keratin have demonstrated significant promise in wound healing applications. Keratin biomaterials facilitate cellular attachment through cell-binding motifs including glutamic acid-aspartic acid-serine (EDS), arginine-glycine-aspartic acid (RGD), and leucine-aspartic acid-valine (LDV) residues [90]. In preclinical studies, keratin-based hydrogels have shown excellent biocompatibility and wound healing properties. For instance, a strontium ranelate-loaded human hair keratin and hyaluronic acid hydrogel demonstrated significant wound closure acceleration in a full-thickness skin defect model in Sprague Dawley rats, reducing oxidative stress and inflammatory markers while enhancing microangiogenesis [90]. Similarly, feather keratin hydrogel cross-linked with H₂O₂ accelerated wound closure to approximately 90% by day 10 compared to 60% in controls, with complete re-epithelialization achieved within 21 days [90].
Efficacy in Ischemia Models
Myocardial Ischemia
Exosome-based therapies have demonstrated significant cardioprotective effects in models of myocardial ischemia, primarily through promotion of angiogenesis and reduction of apoptosis.
Cardiosphere-Derived Cell (CDC) Exosomes: Exosomes derived from CDCs (CDCs-Exo) have been found to repair necrotic myocardium and induce angiogenesis in a pig model of acute myocardial infarction (AMI) through intra-tissue injection [91]. These exosomes contain specific miRNA cargo, including miRNA-146a, which reduces apoptosis and increases proliferation of cardiomyocytes in vitro [91]. When injected into the heart of mouse models, these exosomes increased regeneration and angiogenesis [91]. Under hypoxic conditions, CDCs release exosomes with distinct miRNA profiles, particularly enriched in miRNA-210, miRNA-130a, and miRNA-126, which collectively induce tube formation and promote angiogenesis in human umbilical vein endothelial cells (HUVECs) [91].
Embryonic Stem Cell (ESC) Exosomes: Exosomes isolated from human ESC-derived cardiovascular progenitors showed cardioprotective effects in mouse models of heart failure, with transcriptomic analysis revealing upregulation of 927 genes associated with improved cardiac function in exosome-treated hearts [91]. Similarly, exosomes from human ESC-derived MSCs reduced infarct size in myocardial ischemia/reperfusion (MI/R) injury models, associated with increased levels of ATP and NADH, and enhanced phosphorylation of Akt and GSK-3β signaling pathways [91].
Other Ischemic Conditions
Exosome therapies have shown promising results in various other ischemic conditions, demonstrating versatile therapeutic potential.
Spinal Cord Ischemia-Reperfusion Injury: In a rat model of spinal cord ischemia-reperfusion injury, human placental MSC-derived exosomes (hpMSCs-Exos) in combination with hyperbaric oxygen (HBO) treatment demonstrated synergistic neuroprotective effects [89]. The combined therapy significantly improved neurological function scores, increased the numerical density of neurons, enhanced levels of antioxidative factors (GSH, SOD, and CAT), and elevated anti-inflammatory cytokine (IL-10) levels [89]. This was accompanied by reduced glial cell density, decreased oxidative stress marker (MDA), lower inflammatory cytokines (IL-1β, TNF-α, and IL-18), and diminished expression of the apoptotic protein caspase-3 [89].
Peripheral Ischemia: While the search results do not contain specific data on peripheral ischemia models, the consistent pro-angiogenic effects demonstrated across multiple exosome platforms in myocardial and wound healing contexts suggest strong potential for application in peripheral ischemic conditions. The shared mechanisms of action, particularly through delivery of pro-angiogenic miRNAs, would likely translate to efficacy in peripheral ischemia models.
Experimental Protocols and Methodologies
Exosome Isolation and Characterization
Standardized protocols for exosome isolation and characterization are critical for generating reproducible and therapeutically consistent preparations.
Isolation Methods:
- Ultracentrifugation: Considered the gold standard for exosome extraction, this method involves sequential centrifugation steps culminating in ultracentrifugation at 100,000×g for 70 minutes [89]. While requiring minimal reagents and expertise, limitations include time consumption, high cost, low efficiency, and potential lipoprotein co-separation [4].
- Size-Based Techniques: Alternative approaches include ultrafiltration and size-exclusion chromatography, which are quicker and suitable for large-scale applications but may suffer from pore clogging, exosome loss, and reduced purity [4].
- Precipitation-Based Methods: Commercial polymer-based precipitation kits offer user-friendly protocols but may co-precipitate contaminants [4].
Characterization:
- Nanoparticle Tracking Analysis (NTA): Determines particle size distribution and concentration [89].
- Transmission Electron Microscopy (TEM): Visualizes exosome morphology and ultrastructure [89].
- Western Blotting: Confirms presence of exosomal markers (CD9, CD63, CD81, TSG101, Alix) and absence of negative markers (calnexin, GM130) [89] [4].
- Dynamic Light Scattering (DLS): Provides additional size distribution characterization [89].
In Vivo Wound Healing Models
Full-Thickness Excisional Wound Model:
- Animal Preparation: Anesthetize rodents (typically mice or rats) and remove hair from dorsal surface.
- Wound Creation: Create standardized full-thickness excisional wounds using biopsy punches (typically 6-10mm diameter).
- Treatment Application: Apply test articles (exosomes, cmRNA formulations, or biomaterials) directly to wound bed.
- Wound Monitoring: Document wound closure periodically using digital photography.
- Planimetric Analysis: Calculate wound area reduction percentage over time using image analysis software.
- Histological Assessment: At endpoint, harvest wounds for histological processing (H&E staining for re-epithelialization, Masson's trichrome for collagen deposition, CD31 immunohistochemistry for angiogenesis) [88] [90].
Diabetic Impaired Healing Models:
- Induce diabetes chemically (streptozotocin) or utilize genetically diabetic mice (db/db) to model impaired healing [30].
In Vivo Ischemia Models
Myocardial Infarction Model:
- Surgical Procedure: Anesthetize animals, perform thoracotomy, and permanently or temporarily ligate the left anterior descending coronary artery.
- Treatment Administration: Deliver exosomes via intramyocardial injection directly into border zone or systemically via intravenous injection.
- Functional Assessment: Evaluate cardiac function using echocardiography to measure ejection fraction and fractional shortening.
- Infarct Size Quantification: Measure infarct size using triphenyltetrazolium chloride (TTC) staining.
- Histological Analysis: Assess angiogenesis through CD31+ microvessel counting and fibrosis through Masson's trichrome staining [91].
Spinal Cord Ischemia-Reperfusion Model:
- Surgical Procedure: Anesthetize rats, expose abdominal aorta, and apply microvascular clamp approximately 1cm below left renal artery for 60 minutes to induce ischemia.
- Reperfusion: Remove clamp to restore blood flow.
- Treatment Administration: Apply exosomes intrathecally or systemically at reperfusion onset.
- Neurological Assessment: Evaluate motor function using standardized scoring systems (e.g., Modified Tarlov Scale) at multiple timepoints post-injury.
- Tissue Analysis: Quantify neuronal survival, inflammatory markers, and apoptosis in spinal cord sections [89].
Biodistribution and Uptake Tracking
Labeling Techniques:
- Fluorescent Labels: Lipophilic dyes (DiI, DiD, PKH67, PKH26) incorporate into exosome membranes for in vivo tracking [4].
- Genetic Encoding: Transduce parent cells with luciferase or fluorescent protein genes for endogenous labeling of exosomes [4].
- Radiolabeling: Incorporate radioactive isotopes (e.g., 99mTc, 111In) for sensitive quantitative biodistribution studies [4].
Imaging Modalities:
- In Vivo Imaging Systems (IVIS): Track fluorescent or bioluminescent signals in live animals over time [88].
- Positron Emission Tomography (PET): Provide quantitative whole-body distribution data for radiolabeled exosomes [4].
- Immunohistochemistry: Localize exosomes in tissue sections using specific markers [88].
Signaling Pathways and Molecular Mechanisms
The therapeutic effects of exosomes and cmRNA in wound healing and ischemia are mediated through specific molecular pathways that have been elucidated through mechanistic studies.
Key signaling pathways activated by these therapeutic platforms include:
- MEK/ERK Pathway: EGF cmRNA significantly activated the MEK/ERK pathway both in vitro and in vivo, promoting keratinocyte and fibroblast proliferation and migration [88].
- PI3K/Akt Pathway: Multiple exosome platforms activate the PI3K/Akt pathway, enhancing cell survival and angiogenesis. For instance, HUMSC-derived exosomes containing miR-21-3p promote angiogenesis and fibroblast function through PI3K/Akt and ERK1/2 signaling [30].
- Wnt/β-catenin Pathway: HUMSC-derived exosomes promote angiogenesis through Wnt4/β-catenin signaling, particularly in endothelial cells [30].
- mTOR/AKT Signaling: Keratin-17 expression in keratinocytes influences mTOR/AKT signaling, directly affecting protein translation rates and cellular proliferation [90].
These pathways collectively mediate critical therapeutic processes including angiogenesis, cell proliferation and migration, anti-inflammatory responses, and oxidative stress reduction.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Exosome and cmRNA Studies
| Category | Reagent/Technology | Specifications | Research Application | Key Considerations |
|---|---|---|---|---|
| Exosome Isolation | Ultracentrifugation | 100,000×g, 70-90min | Gold standard isolation | Time-consuming, equipment intensive [89] |
| Size-exclusion chromatography | Commercial columns | High-purity isolation | Preserves vesicle integrity [4] | |
| Precipitation kits | Polymer-based | Rapid, user-friendly | Potential contaminant co-precipitation [4] | |
| Exosome Labeling | PKH67/PKH26 dyes | Lipophilic membrane labels | In vivo tracking, uptake studies | Potential dye aggregation [4] |
| Luciferase transfection | Genetic engineering of parent cells | Sensitive in vivo imaging | Requires cell engineering [88] | |
| Characterization | Nanoparticle tracking | NTA systems | Size distribution quantification | Instrument calibration critical [89] |
| Western blot markers | CD63, CD81, CD9, TSG101 | Exosome identity confirmation | Multiple markers recommended [89] [4] | |
| TEM imaging | Negative staining | Morphological validation | Artifacts possible [89] | |
| cmRNA Technology | In vitro transcription | T7 polymerase system | mRNA production | Modified nucleotides reduce immunogenicity [88] |
| Nucleoside modifications | 5-methylcytidine, N1-Me-Pseudouridine | Reduced immunogenicity, enhanced stability | Critical for in vivo applications [88] | |
| Capping analogs | Cap1 structure | Enhanced translation efficiency | [88] | |
| Delivery Systems | Lipid nanoparticles | LNPs formulations | RNA/protection delivery | Potential hypersensitivity reactions [88] |
| Citrate-saline buffer | 10mmol/L citrate, 130mmol/L NaCl, pH~7.5 | Biocompatible mRNA delivery | Validated in multiple tissues [88] | |
| Animal Models | db/db mice | Genetically diabetic | Impaired healing models | Spontaneous type 2 diabetes [30] |
| C57BL/6 mice | Wild-type | Standard wound healing studies | [88] [90] | |
| Sprague Dawley rats | Wild-type | Larger wound models | [89] [90] | |
| Analysis Tools | IVIS imaging | Luminescence/fluorescence | Biodistribution quantification | [88] |
| Histology scoring | WPW criteria | Wound bed preparation assessment | Well-prepared wound criteria [92] |
The accumulated preclinical evidence robustly supports the therapeutic potential of MSC-derived exosomes and cmRNA technologies for wound healing and ischemic conditions. These platforms demonstrate favorable biodistribution profiles, efficient cellular uptake, and potent efficacy across multiple disease models through defined molecular mechanisms. The field has matured beyond proof-of-concept studies to mechanistic investigations elucidating specific pathways and cargo-responsible effects.
Significant challenges remain in standardization of manufacturing, quantification of biodistribution, and translation to clinical applications. Future research directions should focus on engineering approaches to enhance target specificity, optimization of dosing regimens, development of combination strategies with biomaterials, and rigorous safety assessment in advanced disease models. The continued refinement of these promising platforms offers significant potential for addressing unmet clinical needs in regenerative medicine.
The therapeutic application of Mesenchymal Stem Cell (MSC)-derived exosomes represents a paradigm shift in regenerative medicine, offering a cell-free alternative that mitigates risks associated with whole-cell therapies while retaining potent regenerative capabilities. These nanosized extracellular vesicles (30-150 nm) facilitate intercellular communication by transferring bioactive molecules—including proteins, lipids, and nucleic acids—to recipient cells, thereby influencing processes critical to tissue repair [93] [8]. While MSCs can be isolated from multiple tissue sources, bone marrow (BM), umbilical cord (UC), and adipose tissue (AD) have emerged as the most clinically relevant sources, each with distinct advantages regarding isolation yield, differentiation potential, and secretome profile. Understanding the differential effects of exosomes derived from these sources is paramount for developing targeted therapies, particularly in contexts requiring specific regenerative processes such as angiogenesis, epithelial migration, and wound healing. This review synthesizes current evidence on the uptake mechanisms and functional specializations of MSC-exosomes from different sources, with particular emphasis on their interactions with keratinocytes and endothelial cells—key cellular players in cutaneous regeneration and vascular repair.
MSC Source Characteristics and Exosome Biogenesis
Source-Specific MSC Properties
MSCs from different tissue niches exhibit unique biological characteristics that significantly influence their exosomal cargo and functional properties. Adipose-derived MSCs (AD-MSCs) offer practical advantages including minimally invasive harvesting procedures, higher cell yields per tissue volume, and greater proliferative capacity compared to bone marrow-derived MSCs (BM-MSCs) [94] [95]. Flow cytometry characterization reveals consistent expression of typical MSC surface markers (CD29, CD73, CD90, CD105) across sources, though AD-MSCs may show variable expression of CD34 [95]. Conversely, bone marrow represents the most extensively studied but more invasively harvested source, with BM-MSCs demonstrating particularly strong osteogenic and chondrogenic differentiation potential [93]. Umbilical cord tissue provides a clinically useful neonatal source with reported robust proliferative capacity, though comparative studies of UC-MSC exosomes remain limited in the current literature. These source-specific cellular differences extend to exosome production, where variations in biogenesis pathways ultimately yield vesicles with distinct molecular cargo and functional specializations.
Exosome Biogenesis and Characterization
Exosomes originate through the endosomal pathway, forming as intraluminal vesicles within multivesicular bodies (MVBs) that subsequently fuse with the plasma membrane for release into the extracellular space [96]. These vesicles typically exhibit a cup- or sphere-shaped morphology with diameters ranging from 30-150 nm, as confirmed by transmission electron microscopy [93] [72]. Standard characterization protocols involve nanoparticle tracking analysis for size distribution profiling, Western blot detection of tetraspanin markers (CD63, CD81, CD9) and TSG101, and flow cytometry for surface antigen validation [93] [97]. Proper isolation and identification are crucial for experimental reproducibility and therapeutic application, with emerging technologies like ExoView enabling single-vesicle multiparameter analysis [97].
Quantitative Comparison of MSC-Exosome Functional Effects
Comparative Effects on Cellular Processes
Table 1: Functional Comparison of BMSC-Exos vs. ADSC-Exos
| Functional Assay | BMSC-Exos Performance | ADSC-Exos Performance | Significance | Reference |
|---|---|---|---|---|
| Cell Proliferation | Strong promotive effect on fibroblast, keratinocyte, and BMSC proliferation | Moderate promotive effect | BMSC-Exos > ADSC-Exos in proliferation assays | [97] [98] |
| Cell Migration | Enhanced fibroblast and keratinocyte migration | Significantly promoted endothelial cell migration | ADSC-Exos showed superior endothelial migration | [97] |
| Angiogenic Potential | Moderate effect on tube formation | Strong promotive effect on angiogenesis and tube formation | ADSC-Exos > BMSC-Exos in angiogenic assays | [97] [95] |
| Wound Healing (Diabetic Model) | Limited therapeutic effect on wound closure | Significantly accelerated wound closure | ADSC-Exos effective, BMSC-Exos ineffective | [97] |
| Bone-Tendon Healing | Promoted osteogenesis and chondrogenesis | Similar promotion of osteogenesis and chondrogenesis | No significant difference between sources | [93] |
| Inflammation Modulation | Reduced inflammatory cytokines in osteoarthritic chondrocytes | Strong anti-inflammatory effect in tendinopathy | ADSC-Exos may have stronger anti-inflammatory potential | [96] |
Molecular Cargo Differences
Table 2: Molecular Cargo Profiles of BMSC-Exos vs. ADSC-Exos
| Cargo Component | BMSC-Exos Characteristics | ADSC-Exos Characteristics | Functional Implications | |
|---|---|---|---|---|
| Protein Profile | Enriched in proliferative signaling proteins | Enriched in angiogenic factors (VEGF, FGF) | Explains functional specialization | [97] |
| miRNA Content | miRNAs associated with cell cycle regulation | Pro-angiogenic miRNAs (e.g., miR-31, miR-125a) | Different regulatory networks | [97] [95] |
| tRNA Profile | Distinct tRNA expression patterns | Different tRNA expression patterns | Potential translation regulation differences | [97] |
| Growth Factors | Moderate VEGF, FGF content | High VEGF, FGF, HGF, TGF-β content | Enhanced angiogenic signaling | [95] |
Uptake Mechanisms by Target Cells
Keratinocyte Uptake Pathways
The process of exosome internalization by keratinocytes involves specific receptor-mediated endocytosis that varies based on exosome surface characteristics, which are in turn influenced by the parent MSC source. While all MSC-exosomes express common tetraspanin markers (CD63, CD81, CD9) and adhesion molecules that facilitate cellular binding, source-specific variations in surface protein composition significantly impact uptake efficiency and subsequent functional responses [97]. Studies demonstrate that ADSC-exosomes show preferential uptake in skin wound models, correlating with their enhanced therapeutic effects in diabetic wound healing through mechanisms that promote re-epithelialization [97] [8]. This process is energy-dependent and involves clathrin-mediated endocytosis and macropinocytosis, with internalized exosomes subsequently releasing their cargo into the cytoplasm to modulate recipient cell behavior.
Endothelial Cell Uptake Dynamics
Endothelial cells internalize MSC-exosomes through lipid raft-mediated endocytosis, a process influenced by exosome membrane composition that varies according to MSC source. Comparative studies indicate that ADSC-exosomes exhibit superior binding and internalization by human umbilical vein endothelial cells (HUVECs) and other endothelial cell types, corresponding with their enhanced pro-angiogenic effects [97] [95]. This preferential uptake may be attributed to enriched surface expression of integrins and other adhesion molecules on ADSC-exosomes that facilitate interaction with endothelial receptors. Following internalization, ADSC-exosomes more effectively promote endothelial cell migration, tube formation, and vascular stabilization through transfer of pro-angiogenic miRNAs and proteins, positioning them as particularly promising for therapeutic angiogenesis in ischemic conditions [97] [99] [95].
Diagram 1: MSC Exosome Biogenesis, Uptake, and Functional Specialization Pathways. This diagram illustrates the pathway from MSC source selection through exosome biogenesis to cellular uptake and functional outcomes, highlighting the preferential uptake of ADSC-exosomes by keratinocytes and endothelial cells and their enhanced therapeutic effects.
Experimental Protocols for Uptake and Function Studies
Standardized Exosome Isolation and Characterization
Protocol 1: Ultracentrifugation-Based Exosome Isolation
- Cell Culture: Culture MSCs to 70-80% confluence in exosome-depleted medium (DMEM/F12 supplemented with 10% exosome-free FBS) for 48 hours [97] [96].
- Conditioned Media Collection: Collect conditioned media and perform sequential centrifugation: 300 × g for 10 min (remove cells), 2,000 × g for 20 min (remove debris), 10,000 × g for 30 min (remove larger vesicles) [98] [96].
- Exosome Pellet: Ultracentrifuge supernatant at 100,000 × g for 70 minutes at 4°C [96].
- Washing: Wash pellet with PBS, repeat ultracentrifugation at 100,000 × g for 70 minutes [98].
- Resuspension: Resuspend final exosome pellet in PBS, aliquot, and store at -80°C [98] [96].
- Characterization: Perform nanoparticle tracking analysis for size/concentration, TEM for morphology, Western blot for markers (CD63, CD81, CD9, TSG101), and flow cytometry for surface proteins [93] [97].
Protocol 2: Tangential Flow Filtration for Large-Scale Production
- For clinical translation and larger yields, TFF systems provide advantages in scalability and consistency [72].
- Filter conditioned media through TFF system with appropriate molecular weight cutoff membranes.
- Concentrate exosomes and exchange buffer to PBS.
- Final purification step may include size-exclusion chromatography [72].
Cellular Uptake and Internalization Assays
Protocol 3: Fluorescent Tracking of Exosome Uptake
- Exosome Labeling: Label purified exosomes with lipophilic fluorescent dyes (PKH26, DiR, or PKH67) according to manufacturer protocols [93] [96].
- Cell Seeding: Plate target cells (keratinocytes or endothelial cells) on chamber slides or culture plates.
- Uptake Incubation: Treat cells with labeled exosomes (typical concentration: 2×10^9 particles/mL or 20 μg/mL) for 4-24 hours [96].
- Fixation and Staining: Fix cells with 4% paraformaldehyde, stain actin cytoskeleton with phalloidin, and counterstain nuclei with DAPI [96].
- Imaging and Quantification: Visualize using confocal microscopy, quantify uptake efficiency by fluorescence intensity or particle counting using image analysis software [96].
- Inhibition Studies: To determine uptake mechanisms, pre-treat cells with endocytosis inhibitors (chlorpromazine for clathrin-mediated, filipin for lipid raft-mediated, amiloride for macropinocytosis) [97].
Functional Assays for Angiogenic and Migratory Effects
Protocol 4: Endothelial Tube Formation Assay
- Matrix Preparation: Thaw Matrigel on ice, coat 96-well plates (50 μL/well), polymerize at 37°C for 30 minutes [97] [95].
- Cell Treatment: Seed HUVECs (5×10^4 cells/well) in endothelial basal medium containing exosomes (20 μg/mL) or controls.
- Incubation and Imaging: Incubate for 4-18 hours at 37°C, capture images at regular intervals.
- Quantification: Analyze total tube length, number of branches, and meshes using automated image analysis software (ImageJ Angiogenesis Analyzer) [97].
Protocol 5: Keratinocyte Migration (Scratch Assay)
- Cell Seeding: Plate keratinocytes in 12-well plates until 90-100% confluent.
- Scratch Creation: Create a uniform scratch using a 200 μL pipette tip, wash to remove debris.
- Exosome Treatment: Add exosomes (20 μg/mL) in low-serum medium, include controls.
- Time-Lapse Imaging: Capture images at 0, 6, 12, and 24 hours using microscope with environmental chamber.
- Analysis: Measure scratch closure percentage using automated image analysis [97].
Protocol 6: In Vivo Wound Healing Model
- Animal Model: Use diabetic (db/db) mice for impaired healing models or normal C57BL/6 mice [97].
- Wound Creation: Create full-thickness excisional wounds (6-8 mm diameter) on dorsal skin.
- Exosome Treatment: Apply exosomes (20-100 μg) in collagen hydrogel or directly injected around wound bed every 3-4 days [97] [96].
- Assessment: Monitor wound closure percentage, histology (H&E, Masson's trichrome), immunohistochemistry (CD31 for angiogenesis, cytokeratin for epithelialization) [97].
Signaling Pathways and Molecular Mechanisms
Source-Specific Pathway Activation
ADSC-Exosome Signaling: ADSC-exosomes preferentially activate angiogenic signaling pathways in endothelial cells, primarily through enhanced delivery of pro-angiogenic miRNAs (miR-125a, miR-31) and proteins (VEGF, FGF) that potentiate the PI3K/Akt and MAPK/ERK pathways [97] [95]. This results in superior tube formation and endothelial migration compared to BMSC-exosomes. Additionally, ADSC-exosomes more effectively modulate the TGF-β/Smad pathway to promote keratinocyte migration and re-epithelialization, explaining their enhanced wound healing capabilities in diabetic models where these pathways are typically impaired [97].
BMSC-Exosome Signaling: BMSC-exosomes exhibit stronger activation of proliferative pathways in recipient cells, including upregulation of cyclins and cyclin-dependent kinases through transfer of specific miRNA cargo (e.g., let-7 family members) [97] [98]. This results in more potent stimulation of fibroblast and keratinocyte proliferation compared to ADSC-exosomes. BMSC-exosomes also show enriched content of regulators of the Wnt/β-catenin pathway, contributing to their enhanced osteogenic and chondrogenic differentiation potential, which is particularly beneficial for bone-tendon healing applications [93].
Epigenetic Modulation and Cellular Reprogramming
Emerging evidence indicates that MSC-exosomes can induce epigenetic modifications in recipient cells that enhance their regenerative potential. Studies demonstrate that pretreatment with epigenetic modifiers like BIX-01294 (a histone methyltransferase G9a inhibitor) can enhance the endothelial differentiation capacity of ADSCs, suggesting that similar epigenetic modifications may influence exosome cargo and functionality [100]. Additionally, direct reprogramming approaches using transcription factors (e.g., ETV2) in combination with exosome treatment can significantly enhance endothelial differentiation, creating functional endothelial-like cells from ADSCs with potential for therapeutic vascularization [99]. These findings highlight the complex interplay between exosome-mediated signaling and epigenetic regulation in determining functional outcomes.
Diagram 2: Molecular Signaling Pathways Activated by MSC Exosomes from Different Sources. This diagram illustrates the distinct signaling pathways activated by ADSC-exosomes (angiogenesis and migration focus) versus BMSC-exosomes (proliferation and differentiation focus), along with epigenetic modulation strategies to enhance therapeutic effects.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for MSC Exosome Studies
| Reagent Category | Specific Examples | Research Application | Functional Role | |
|---|---|---|---|---|
| Isolation Kits | Total Exosome Isolation Kits, qEV Size Exclusion Columns | Exosome purification from conditioned media | Rapid, standardized exosome isolation with good reproducibility | [97] |
| Characterization Antibodies | Anti-CD63, CD81, CD9, TSG101, Alix, Calnexin (negative) | Western blot, flow cytometry validation | Confirmation of exosome identity and purity assessment | [93] [97] |
| Tracking Dyes | PKH26, PKH67, DiR, CFSE, CM-Dil | Cellular uptake and biodistribution studies | Fluorescent labeling for visualization and quantification of exosome internalization | [93] [96] |
| Cell Culture Media | DMEM/F12, α-MEM, MSC-specific media | MSC expansion and exosome production | Optimal cell growth and exosome yield; α-MEM may enhance proliferation | [72] |
| Endocytosis Inhibitors | Chlorpromazine, Filipin, Amiloride, Dynasore | Uptake mechanism studies | Identification of specific internalization pathways in target cells | [97] |
| Angiogenesis Assay Kits | Matrigel, Tube Formation Assay Kits | Endothelial function assessment | Evaluation of pro-angiogenic potential of exosomes | [97] [95] |
| Animal Models | db/db mice, C57BL/6 mice | In vivo wound healing studies | Diabetic and normal wound healing assessment | [97] |
The comprehensive analysis of MSC-derived exosomes from different tissue sources reveals a landscape of functional specialization that aligns with the physiological roles of their parent tissues. ADSC-exosomes demonstrate superior angiogenic potential and enhanced efficacy in diabetic wound healing models, making them particularly promising for therapeutic applications requiring vascularization and cutaneous regeneration. Conversely, BMSC-exosomes exhibit stronger proliferative effects and exceptional performance in musculoskeletal repair contexts. These functional differences stem from distinct molecular cargo profiles that activate specialized signaling pathways in recipient cells, highlighting the importance of source selection based on therapeutic objectives rather than a one-size-fits-all approach.
Future research directions should prioritize standardized isolation protocols to enable more direct comparisons across studies, with particular emphasis on separating exosome subpopulations that may contribute to specific functions. The development of engineered exosomes through preconditioning strategies or direct cargo modification represents a promising frontier for enhancing therapeutic efficacy beyond native vesicle capabilities. Additionally, comprehensive biodistribution studies comparing exosomes from different sources will be crucial for understanding their in vivo trafficking and target tissue accumulation patterns. As the field advances toward clinical translation, consideration of scalable production methods like tangential flow filtration and the development of potency assays that reflect mechanism of action will be essential for regulatory approval and eventual therapeutic application.
Mesenchymal stem cell (MSC) therapy has undergone a significant evolution, transitioning from whole-cell transplantation to the exploration of cell-free approaches utilizing MSC-derived products. This shift is largely driven by the understanding that the therapeutic benefits of MSCs are predominantly mediated through their paracrine activity rather than direct cell replacement [22]. Among these paracrine effectors, MSC-derived exosomes (MSC-exos) have emerged as a promising therapeutic entity. These nanoscale extracellular vesicles (30-150 nm in diameter) are naturally secreted by MSCs and play crucial roles in intercellular communication by transferring bioactive molecules—including proteins, lipids, and nucleic acids—to recipient cells [101] [47]. This technical analysis provides a comprehensive comparison between MSC exosomes and whole-cell therapies, focusing on their efficacy, safety, and mechanisms of action, with specific emphasis on their interactions with keratinocytes and endothelial cells, which are critical for cutaneous wound healing and vascular repair processes.
Quantitative Profile Comparison: MSC Exosomes vs. Whole-Cell Therapy
Table 1: Comprehensive comparison of MSC whole-cell therapy versus MSC-derived exosomes
| Parameter | MSC Whole-Cell Therapy | MSC-Derived Exosomes |
|---|---|---|
| Physical Characteristics | Actual cells (10-100 μm) | Nanoscale vesicles (30-150 nm) [49] |
| Therapeutic Cargo | Direct secretion of factors; differentiation potential | Proteins, lipids, miRNAs, mRNAs [101] [22] |
| Mechanism of Action | Direct differentiation; complex paracrine signaling | Targeted molecular delivery; ligand-receptor signaling [47] |
| Safety Profile | Risk of immunogenic responses; malignant transformation; ectopic tissue formation [22] [102] | Low immunogenicity; no risk of tumorigenicity or ectopic tissue formation [22] [102] |
| Administration & Storage | Limited shelf life; requires fresh preparation; logistical challenges [22] | Stable at -80°C for extended periods; survives freeze-thaw cycles; versatile administration routes [22] |
| Clinical Translation | >2,300 registered clinical trials; mixed results in translation [22] | 64 registered clinical trials; promising early-phase results [22] |
| Production Scalability | Complex expansion processes; donor variability | Potential for continuous production from immortalized lines; more cost-effective [22] |
| Biological Barrier Penetration | Limited tissue penetration; potential for vascular occlusion | Crosses biological barriers (e.g., blood-brain barrier); efficient tissue penetration [22] [47] |
Uptake Mechanisms and Functional Impacts on Keratinocytes and Endothelial Cells
The therapeutic efficacy of MSC-exos is fundamentally dependent on their ability to deliver bioactive cargo to recipient cells. Understanding their uptake mechanisms and subsequent functional impacts on target cells—particularly keratinocytes and endothelial cells—is crucial for optimizing therapeutic applications in wound healing and vascular repair.
Cellular Uptake Mechanisms
MSC-exos utilize three primary pathways for cellular internalization, each with distinct implications for cargo delivery and functional modulation:
- Direct Ligand-Receptor Binding: Exosomal surface molecules engage with specific receptors on target cell membranes, initiating downstream signaling cascades without full internalization. This mechanism facilitates rapid modulation of cellular processes [47].
- Membrane Fusion: The lipid bilayer of exosomes directly fuses with the plasma membrane of recipient cells, releasing the luminal contents directly into the cytoplasm. This mechanism enables efficient delivery of non-membrane-permeable molecules [47] [30].
- Internalization: Exosomes are engulfed by recipient cells through endocytic pathways, including clathrin-dependent endocytosis, caveolin-mediated endocytosis, and macropinocytosis. Following internalization, exosomes traffic through endosomal compartments, eventually releasing their cargo into the cytoplasmic space [30].
Diagram 1: Exosome uptake mechanisms and functional impacts on skin cells. This figure illustrates the three primary pathways through which MSC-derived exosomes are taken up by recipient cells like keratinocytes and endothelial cells, and the subsequent functional outcomes that promote wound healing and vascularization.
Key Signaling Pathways and Molecular Cargo in Target Cells
Table 2: Experimentally validated MSC-exosome components and their effects on keratinocytes and endothelial cells
| Target Cell | Key MSC-exo Cargo | Signaling Pathway | Documented Functional Outcome | Experimental Models |
|---|---|---|---|---|
| Keratinocytes | miR-27b [30] | ITCH/JUNB/IRE1α | Activation, proliferation, and migration in vitro; accelerated wound healing in vivo [30] | In vitro keratinocyte culture; in vivo mouse wound model |
| Keratinocytes | Not specified (ND) [30] | Wnt4/β-catenin | Enhanced proliferation and migration [30] | In vitro keratinocyte culture |
| Endothelial Cells | miR-21-3p [30] | PI3K/Akt; ERK1/2 | Promoted angiogenesis and improved endothelial cell function [30] | In vitro endothelial cell culture |
| Endothelial Cells | miR-146a [30] | Src | Mitigated endothelial cell senescence, promoted angiogenesis in aging and type-2 diabetes mouse models [30] | In vitro endothelial cell culture; in vivo diabetic mouse model |
| Endothelial Cells | miR-181c [30] | TLR4-NF-κB | Reduced production of inflammatory cytokines [30] | In vitro endothelial cell culture |
Methodological Framework: Experimental Protocols for Uptake and Functional Studies
Protocol 1: MSC-exosome Isolation and Characterization
Objective: To isolate and characterize exosomes from MSC culture conditioned media. Methodology: Differential Ultracentrifugation [47] [103]
- Cell Culture: Culture MSCs in serum-free media for 48 hours to condition the media.
- Initial Centrifugation: Centrifuge conditioned media at 300 × g for 10 minutes to remove floating cells.
- Secondary Centrifugation: Transfer supernatant and centrifuge at 2,000 × g for 20 minutes to remove dead cells and large debris.
- Filtration: Filter supernatant through a 0.22 μm filter.
- Ultracentrifugation: Centrifuge filtered supernatant at 100,000 × g for 70 minutes at 4°C to pellet exosomes.
- Wash: Resuspend pellet in phosphate-buffered saline (PBS) and centrifuge again at 100,000 × g for 70 minutes.
- Resuspension: Resuspend final exosome pellet in PBS and store at -80°C. Characterization:
- Nanoparticle Tracking Analysis (NTA): Determine particle size distribution and concentration [47].
- Transmission Electron Microscopy (TEM): Visualize exosome morphology and confirm cup-shaped structure [47].
- Western Blotting: Detect exosomal markers (CD9, CD63, CD81, TSG101, Alix) and confirm absence of negative markers (e.g., calnexin) [47] [103].
Protocol 2: Tracking MSC-exosome Uptake by Keratinocytes and Endothelial Cells
Objective: To visualize and quantify the internalization of MSC-exosomes by target cells. Methodology: Fluorescent Labeling and Confocal Microscopy
- Exosome Labeling: Label purified MSC-exosomes with a lipophilic dye (e.g., PKH67, Dil; emission green/red) per manufacturer's protocol. Remove unbound dye using size-exclusion chromatography or ultracentrifugation.
- Cell Seeding: Seed keratinocytes or endothelial cells on glass-bottom culture dishes.
- Treatment: Treat cells with labeled exosomes (e.g., 10-50 μg/mL) for various time points (1-24 hours).
- Staining: Fix cells and stain actin cytoskeleton (e.g., Phalloidin) and nuclei (e.g., DAPI).
- Imaging: Capture z-stack images using a confocal microscope. Co-localization analysis with endosomal/lysosomal markers (e.g., Rab5, LAMP1) can determine intracellular trafficking.
- Inhibition Studies: Use specific inhibitors (e.g., dynasore for clathrin-mediated endocytosis, methyl-β-cyclodextrin for caveolae-mediated endocytosis) to delineate uptake pathways [30].
Protocol 3: Functional Assays for Angiogenesis and Migration
Objective: To assess the functional impact of MSC-exosomes on endothelial cell and keratinocyte behavior. Angiogenesis Assay (Endothelial Cells):
- Tube Formation Assay: Plate endothelial cells (e.g., HUVECs) on Matrigel-coated plates.
- Treatment: Treat with MSC-exosomes (10-50 μg/mL) or PBS control.
- Analysis: After 4-18 hours, image the tubular structures. Quantify total tube length, number of branches, and number of meshes using image analysis software (e.g., ImageJ) [30]. Migration Assay (Keratinocytes):
- Scratch/Wound Healing Assay: Create a uniform "scratch" in a confluent monolayer of keratinocytes.
- Treatment: Treat with MSC-exosomes or control.
- Analysis: Image the scratch at 0, 12, and 24 hours. Measure the reduction in scratch area over time to quantify migration rate [30].
Preconditioning Strategies to Enhance Therapeutic Potency
The therapeutic cargo of MSC-exosomes is dynamic and can be enhanced through preconditioning of parent MSCs. These strategies manipulate the cellular microenvironment to boost the content of specific beneficial molecules, particularly miRNAs.
Diagram 2: Preconditioning strategies for enhancing MSC-exosome potency. This workflow illustrates how various preconditioning stimuli applied to parent MSCs can selectively enrich the cargo of resulting exosomes with specific therapeutic miRNAs, leading to enhanced functional outcomes.
Table 3: Key miRNAs modulated by preconditioning and their roles in skin and vascular biology
| Preconditioning Stimulus | Key Modulated miRNA | Documented Effect on Keratinocytes/Endothelial Cells | Therapeutic Implication |
|---|---|---|---|
| Inflammatory Cytokines (TNF-α, IL-1β) | miR-146a [102] [49] | Reduces inflammatory cytokine production via TLR4-NF-κB pathway; mitigates endothelial senescence [102] [30] | Enhanced anti-inflammatory response; protection of vascular function |
| Hypoxia | miR-126 [102] | Promotes angiogenesis [102] | Improved vascularization in ischemic tissues |
| Lipopolysaccharide (LPS) | miR-181a-5p [102] | Not specified for target cells, but has general anti-inflammatory effects [102] | Mitigation of inflammatory damage |
| Not specified | miR-21-3p [30] | Promotes angiogenesis and fibroblast function via PI3K/Akt and ERK1/2 [30] | Enhanced wound healing and tissue repair |
| Not specified | miR-27b [30] | Activates keratinocytes and fibroblasts via ITCH/JUNB/IRE1α [30] | Accelerated wound healing |
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 4: Key research reagents and methodologies for MSC-exosome studies
| Reagent / Method | Specific Function | Technical Notes |
|---|---|---|
| Differential Ultracentrifugation | Standard method for exosome isolation from cell culture media | Sequential spins: 300g (cells), 2,000g (debris), 10,000g (microvesicles), 100,000g (exosomes) [47] [103] |
| Size Exclusion Chromatography (SEC) | Isolation of exosomes based on size; preserves vesicle integrity | Separates exosomes from contaminating proteins; often used after ultracentrifugation for higher purity [103] |
| CD63/CD81/CD9 Antibodies | Exosome detection and characterization via Western Blot or immunoaffinity capture | Tetraspanins are enriched in exosomes and serve as canonical markers [47] [103] |
| PKH67 / Dil Lipophilic Dyes | Fluorescent labeling of exosome membranes for uptake and tracking studies | Must remove unbound dye rigorously post-labeling to avoid background signal [30] |
| Nanoparticle Tracking Analysis (NTA) | Quantification of exosome particle size and concentration | Instruments (e.g., Malvern NanoSight) measure Brownian motion of particles in suspension [47] |
| Transmission Electron Microscopy (TEM) | Visualization of exosome morphology and ultrastructure | Confirms cup-shaped morphology characteristic of exosomes [47] |
| Matrigel | Basement membrane matrix for endothelial tube formation assays | Provides a substrate that mimics the extracellular matrix for in vitro angiogenesis studies [30] |
| Dynasore / Methyl-β-cyclodextrin | Inhibitors of endocytic pathways (clathrin-mediated and caveolae-mediated, respectively) | Used in mechanistic studies to determine the primary route of exosome uptake into target cells [30] |
The transition from MSC whole-cell therapy to MSC-derived exosomes represents a significant advancement in regenerative medicine, offering a refined therapeutic modality with a superior safety profile and multifaceted mechanisms of action. The efficacy of MSC-exosomes is particularly evident in their ability to deliver complex molecular cargo directly to target cells like keratinocytes and endothelial cells, modulating critical processes such as proliferation, migration, and angiogenesis through well-defined signaling pathways.
While whole-cell therapies continue to be investigated, challenges related to safety, consistency, and engraftment efficiency persist. In contrast, MSC-exosomes provide a cell-free alternative that mitigates these risks while retaining therapeutic potency. Current research is focused on enhancing exosome efficacy through preconditioning strategies, bioengineering, and optimizing delivery systems. As of January 2025, 64 clinical trials are registered exploring MSC-exosomes for a range of conditions, from osteoarthritis and myocardial infarction to COVID-19 pneumonia and wound healing, underscoring the translational momentum in this field [22]. Future work will likely concentrate on standardizing production protocols, improving targeting specificity, and generating comprehensive biodistribution data, ultimately paving the way for exosome-based therapies to become a mainstay in precision regenerative medicine.
Benchmarking Against Other Acellular Therapies (e.g., Growth Factors, Synthetic Nanoparticles)
The field of regenerative medicine is undergoing a significant paradigm shift from cell-based therapies toward acellular approaches that offer enhanced safety profiles and more standardized manufacturing. Among these innovations, mesenchymal stem cell (MSC)-derived exosomes have emerged as particularly promising biological nanoparticles for therapeutic applications. These nanoscale extracellular vesicles (typically 30–150 nm in diameter) function as sophisticated intercellular communication vehicles, carrying bioactive molecules including proteins, lipids, and nucleic acids that mediate therapeutic effects in target tissues [31] [61]. For researchers investigating MSC exosome uptake mechanisms by keratinocytes and endothelial cells, it is essential to contextualize these natural vesicles against alternative acellular platforms, including recombinant growth factors and synthetic nanoparticles. Each platform presents distinct advantages and limitations in terms of payload capacity, targeting efficiency, manufacturability, and regulatory pathway. This whitepaper provides a technical benchmarking analysis to guide strategic research and development decisions in the rapidly evolving acellular therapeutics landscape, with particular emphasis on applications relevant to skin biology and vascularization research.
Technical Comparison of Major Acellular Therapeutic Platforms
Defining Characteristics and Mechanisms of Action
MSC-derived exosomes represent a native biological delivery system that recapitulates many therapeutic functions of their parent cells. These vesicles facilitate tissue repair through multiple mechanisms including reducing cellular senescence, promoting angiogenesis, modulating inflammation, and enhancing tissue regeneration [31]. Their lipid bilayer membrane protects cargo from degradation and enables fusion with target cell membranes, while surface proteins facilitate natural tropism to injured tissues. Exosomes derived from different MSC sources (bone marrow, adipose tissue, umbilical cord) exhibit varying cargo profiles and therapeutic potentials, with umbilical cord-derived MSC exosomes demonstrating particularly high proliferative capacity and low immunogenicity [104] [105].
Growth factors are signaling proteins that regulate cellular processes such as proliferation, migration, and differentiation through binding to specific cell surface receptors. In regenerative contexts, factors like VEGF, FGF, BMP-2, and TGF-β are administered to stimulate tissue repair, often in combination with biomaterial carriers that control their release kinetics [106]. While potent, their therapeutic application is challenged by short half-lives, rapid diffusion from target sites, and potential off-target effects when delivered systemically.
Synthetic nanoparticles encompass a diverse class of engineered materials including metallic nanoparticles (gold, silver), liposomes, polymeric nanoparticles, and dendrimers designed for drug delivery, imaging, and therapeutic applications. These platforms offer precise control over physicochemical properties, payload capacity, and release kinetics, with surfaces that can be functionalized with targeting ligands [107]. However, they may face challenges related to biocompatibility, immune recognition, and potential toxicity from long-term accumulation.
Table 1: Comparative Analysis of Acellular Therapeutic Platforms
| Parameter | MSC-Derived Exosomes | Growth Factors | Synthetic Nanoparticles |
|---|---|---|---|
| Size Range | 30–150 nm [31] [61] | 5–50 kDa (protein) | 10–1000 nm [107] |
| Production Complexity | High (cell culture, purification, characterization) | Moderate (recombinant expression) | Variable (chemical synthesis) |
| Payload Capacity | Native biomolecules (proteins, miRNAs, mRNAs) | Single protein | Small molecules, nucleic acids, proteins |
| Targeting Mechanism | Natural tropism via surface proteins | Receptor-ligand interaction | Engineered surface functionalization |
| Immunogenicity | Low [22] [60] | Variable | Variable |
| Regulatory Status | Early clinical trials [31] [22] | Approved products (e.g., BMP-2) | Some FDA-approved formulations |
| Manufacturing Scalability | Challenging (batch variability) [31] | Established | Highly scalable |
| Storage Stability | -80°C for extended periods [22] | Variable (often refrigerated) | Generally stable |
Quantitative Performance Benchmarking
Table 2: Quantitative Benchmarking of Therapeutic Platforms in Preclinical Studies
| Performance Metric | MSC Exosomes | Nanoparticle-Embedded GFs | Synthetic Nanoparticles |
|---|---|---|---|
| Cellular Uptake Efficiency | High in keratinocytes & endothelial cells [31] | Variable (depends on carrier) | Variable (surface-dependent) |
| Half-life in Circulation | Hours to days [22] | Minutes to hours (free GF) [106] | Hours to days |
| Therapeutic Dose Range | µg–mg protein [31] | ng–µg (GF-dependent) | mg–gram scale [107] |
| Angiogenic Potency | High (multiple pro-angiogenic factors) [31] | High (direct signaling) | Variable |
| Anti-inflammatory Effects | Robust (multiple mechanisms) [31] [104] | Limited (unless anti-inflammatory GF) | Variable |
| Production Cost | High ($ thousands/dose) | Moderate | Low at scale |
| Commercial Market Size | Rapidly emerging | Established | $10.5B (2025 projection) [107] |
Experimental Approaches for Evaluating Acellular Therapies
Methodologies for Assessing Keratinocyte and Endothelial Cell Uptake
Fluorescent labeling and tracking protocols represent fundamental approaches for quantifying cellular uptake kinetics. For exosome studies, membrane dyes such as PKH67, PKH26, or DiI provide stable labeling with minimal cargo disruption. The standard protocol involves incubating exosomes with dye at 2 µM concentration in Diluent C for 5 minutes at room temperature, followed by ultracentrifugation (100,000–120,000 × g for 70 minutes) to remove unincorporated dye [31]. Labeled exosomes are then added to cultured keratinocytes (HaCaT cells) or human umbilical vein endothelial cells (HUVECs) at concentrations of 10–50 µg/mL for uptake studies. Internalization is quantified using flow cytometry at various timepoints (15 minutes to 24 hours) and visualized via confocal microscopy with z-stack imaging to confirm intracellular localization.
Inhibitor studies elucidate uptake mechanisms through pharmacological disruption of specific pathways. For keratinocytes, common inhibitors include chlorpromazine (10 µg/mL) for clathrin-mediated endocytosis, genistein (100 µM) for caveolae-mediated uptake, amiloride (50 µM) for macropinocytosis, and cytochalasin D (5 µM) for actin polymerization-dependent mechanisms [31]. Cells are pre-treated with inhibitors for 30–60 minutes before adding labeled exosomes, with viability controls to ensure non-toxic conditions. A ≥50% reduction in fluorescence intensity compared to untreated controls indicates significant pathway involvement.
Surface plasmon resonance (SPR) and biolayer interferometry provide quantitative analysis of binding kinetics between exosome surface ligands and cellular receptors. For endothelial cell studies, CD63 tetraspanin or integrins on exosomes can be evaluated for binding to receptors such as ICAM-1 immobilized on biosensor chips. The standard experimental setup involves immobilizing target receptors on CMS chips via amine coupling, followed by injection of exosome suspensions at concentrations ranging from 0.1–100 µg/mL to determine association and dissociation rates [31].
Functional Assays for Therapeutic Efficacy
Angiogenesis assays evaluate pro-angiogenic potential through multiple complementary approaches. The tube formation assay utilizes HUVECs or human dermal microvascular endothelial cells seeded on growth factor-reduced Matrigel at densities of 1–2 × 10^4 cells per well in 96-well plates. Cells are treated with exosomes (10–50 µg/mL), growth factors (VEGF at 10–50 ng/mL as positive control), or synthetic nanoparticles, with tube networks quantified after 4–18 hours by measuring total tube length, number of branches, and enclosed areas using automated image analysis software [31]. For more complex 3D modeling, endothelial spheroid assays embed HUVEC spheroids in collagen gels and measure sprout length and complexity following treatment.
Migration and proliferation assays determine effects on keratinocyte function essential for wound healing and tissue regeneration. Scratch assays performed on confluent HaCaT monolayers measure migration rates into the denuded area, with treatments applied immediately after wounding and closure quantified over 24–48 hours. For proliferation assessment, EdU (5-ethynyl-2'-deoxyuridine) incorporation assays provide precise measurement of DNA synthesis rates, while PrestoBlue or MTT assays determine metabolic activity as a proxy for cell viability and proliferation [31] [104].
Gene expression analysis elucidates molecular mechanisms underlying therapeutic effects. RNA sequencing of treated keratinocytes and endothelial cells reveals pathway modulation, with particular attention to genes involved in extracellular matrix organization (COL1A1, COL3A1, elastin), inflammation (IL-6, IL-8, TNF-α), and oxidative stress response (Nrf2, HO-1) [104]. For focused analysis of exosome-mediated effects, qPCR arrays targeting angiogenesis, wound healing, and senescence pathways provide efficient screening. Additionally, proteomic analysis of secreted factors in conditioned media identifies paracrine signaling mediators activated by different acellular therapies.
Signaling Pathways in Keratinocyte and Endothelial Cell Responses
Molecular Mechanisms of MSC Exosome Action
MSC exosomes exert their therapeutic effects through delivery of complex molecular cargo to recipient cells. In the context of keratinocyte and endothelial cell targeting, several key pathways have been identified as central mediators of their regenerative effects:
PI3K/Akt signaling pathway activation promotes cell survival, proliferation, and migration. Exosomal miR-21-3p inhibits PTEN expression, leading to enhanced Akt phosphorylation in recipient cells [104]. This pathway is particularly important for endothelial cell viability under oxidative stress conditions and for keratinocyte migration during re-epithelialization. The activation timeline typically begins within 15–30 minutes of exosome treatment, peaks at 2–4 hours, and can be detected through western blotting for phospho-Akt (Ser473) and downstream substrates.
MAPK/ERK pathway modulation regulates cellular proliferation and differentiation responses. MSC exosomes from various sources have been shown to regulate ERK1/2 phosphorylation states, with differential effects depending on cell context and exosome origin [104]. In UVB-damaged keratinocytes, hucMSC-exos suppress sustained ERK activation associated with stress responses while maintaining basal signaling required for homeostasis. Experimental assessment includes western blotting for phospho-ERK1/2 and quantification of nuclear translocation.
Anti-senescence effects are mediated through delivery of specific microRNAs that target senescence pathways. For example, ESC exosomes deliver miR-291a-3p, which targets TGF-β receptor 2 and thereby suppresses TGF-β signaling that drives cellular senescence [31]. This mechanism is particularly relevant for counteracting radiation-induced skin injury, where persistent DNA damage and premature senescence impair healing capacity. Senescence biomarkers including p16INK4a, p21, lamin B1, and SA-β-galactosidase activity are used to quantify these effects.
Diagram 1: MSC Exosome Signaling in Skin Cells
Comparative Pathway Activation Across Therapeutic Platforms
Different acellular therapeutic platforms engage distinct but overlapping signaling networks in target cells. Understanding these differential pathway activations is essential for selecting appropriate therapies for specific applications:
Growth factor signaling typically activates more discrete, receptor-specific pathways compared to the multi-target approach of exosomes. For example, VEGF primarily engages VEGFR2 and its downstream effectors including PLCγ, PKC, and FAK in endothelial cells, resulting in strongly polarized angiogenic signaling [106]. This focused activation can produce potent but potentially narrow therapeutic effects compared to the multi-system regulation achieved by exosomes.
Synthetic nanoparticle effects are highly dependent on their surface functionalization and payload. Gold nanoparticles of specific sizes (30–50 nm) have been shown to modulate NF-κB signaling and inflammatory responses in keratinocytes, while lipid nanoparticles delivering siRNA can achieve targeted gene silencing through RNA interference pathways [107]. The temporal dynamics of pathway activation also differ, with synthetic systems often designed for sustained release and prolonged pathway modulation compared to the more acute effects of growth factors.
Cross-platform pathway analysis reveals both complementary and redundant mechanisms. While growth factors and exosomes both activate PI3K/Akt and MAPK signaling, exosomes uniquely modulate additional pathways including those involved in cellular senescence (via p53/p21) and oxidative stress response (via Nrf2) [31] [104]. This broader pathway engagement may explain the superior performance of exosomes in complex injury models where multiple pathological processes coexist.
Research Reagent Solutions for Acellular Therapy Studies
Table 3: Essential Research Tools for Acellular Therapy Investigation
| Research Tool | Specific Application | Key Function | Example Products/Sources |
|---|---|---|---|
| Exosome Isolation Kits | MSC exosome purification | Concentration and separation from conditioned media | Total Exosome Isolation Kit, exoEasy Maxi Kit |
| Characterization Instruments | Vesicle quantification | Size distribution and concentration analysis | NanoSight NS300, ZetaView |
| Fluorescent Tracking Dyes | Cellular uptake studies | Membrane labeling for visualization and quantification | PKH67, PKH26, DiI, DIR |
| Endocytosis Inhibitors | Uptake mechanism studies | Pathway-specific blockade of internalization | Chlorpromazine, Genistein, Amiloride |
| Cell Culture Models | Functional assessment | Representative target cells for therapeutic testing | HaCaT keratinocytes, HUVECs |
| Angiogenesis Assay Platforms | Pro-angiogenic potency | Quantification of tube formation and sprouting | Growth Factor Reduced Matrigel, μ-Slide Angiogenesis |
| Gene Expression Analysis | Mechanism elucidation | Pathway and target gene quantification | qPCR arrays, RNA-seq services |
| Protein Array Systems | Secreted factor profiling | Multiplex analysis of conditioned media | Proteome Profiler Arrays, Luminex Assays |
Clinical Translation and Commercial Landscape
Regulatory Status and Clinical Trial Progress
The translational pathway for acellular therapies varies significantly across platforms, with growth factors having the most established regulatory history and exosomes representing the most novel category. Clinical trial data for MSC exosomes is accumulating, with five registered clinical trials specifically investigating stem cell-derived exosomes for radiation-induced skin injury, showing promising results in early-phase studies [31]. More broadly, 64 registered clinical trials evaluating MSC-derived extracellular vesicles for various conditions were identified as of January 2025, indicating growing clinical acceptance of this platform [22].
The regulatory framework for exosome therapies continues to evolve, with current approaches drawing from both cell therapy and biologic product paradigms. The lack of standardized manufacturing processes and characterization standards remains a significant challenge for clinical translation [31]. In contrast, synthetic nanoparticles have established regulatory pathways with several FDA-approved products, while growth factors like BMP-2 have long-standing clinical use in specific applications such as bone regeneration [106] [107].
Market Trends and Commercialization Outlook
The acellular therapy market is experiencing robust growth, with the synthetic nanoparticles segment projected to reach approximately $10,500 million in 2025 and grow at a CAGR of around 15% through 2033 [107]. The broader acellular therapy market is expected to expand from $14.22 billion in 2025 to $35.22 billion by 2032, representing a compound annual growth rate of 13.8% [108]. This growth is fueled by continuous advancements in nanotechnology and increasing investment in research and development by leading companies.
Commercial activity in the exosome space is accelerating, with several companies emerging as leaders in therapeutic development. Key players include Codiak BioSciences, Capricor Therapeutics, Evox Therapeutics, and Exopharm, among others [108]. In the aesthetic and dermatology segments, products such as BENEV Exosome Regenerative Complex, ExoCoBio (ASCE+), and XoGlo have gained traction, though it is important to note that as of 2025, no exosome product has received FDA approval for therapeutic applications, with current uses limited to cosmetic applications [61].
Benchmarking analysis reveals that MSC-derived exosomes offer distinctive advantages in the context of keratinocyte and endothelial cell targeting, particularly through their multi-mechanistic actions and natural trafficking capabilities. While challenges in manufacturing scalability and characterization remain, the therapeutic potential of these biological nanoparticles is supported by growing preclinical evidence and early clinical validation. For research focused on uptake mechanisms, critical knowledge gaps include detailed understanding of receptor-ligand interactions governing tissue tropism, quantitative analysis of intracellular trafficking routes, and systematic comparison of exosomes from different cellular sources. The evolving acellular therapy landscape promises continued innovation, with emerging approaches including engineered exosomes with enhanced targeting capabilities, hybrid nanoparticle-exosome systems, and stimulus-responsive synthetic platforms offering new opportunities for therapeutic advancement. As the field progresses, standardized benchmarking methodologies and direct comparative studies will be essential for positioning MSC exosomes within the expanding toolkit of acellular regenerative therapies.
The therapeutic application of mesenchymal stem cell-derived exosomes (MSC-Exos) represents a paradigm shift in regenerative medicine, moving from cell-based therapies to acellular, nanoscale treatments. As natural carriers of bioactive molecules—including proteins, lipids, and various RNA species—exosomes mediate intercellular communication by transferring their cargo to recipient cells, thereby influencing their behavior and function [22] [109]. For researchers and drug development professionals focused on MSC exosome uptake mechanisms by keratinocytes and endothelial cells, understanding the current clinical trial landscape is crucial for guiding preclinical research and clinical translation. This review provides a comprehensive analysis of ongoing clinical trials, summarizes their outcomes where available, and details the experimental methodologies essential for investigating exosome-cell interactions.
Analysis of Registered Clinical Trials
The clinical investigation of MSC-derived extracellular vesicles (which include exosomes) is rapidly expanding. As of January 2025, 64 registered clinical trials evaluating mesenchymal stem cell-extracellular vesicles/exosomes for various diseases are listed on ClinicalTrials.gov [22]. These trials span a diverse range of medical conditions, demonstrating the broad therapeutic potential of MSC-EVs.
Table 1: Selected Ongoing Clinical Trials Involving MSC-Derived Extracellular Vesicles/Exosomes
| NCT Number | Conditions | Phases | Enrollment | Study Status | Locations |
|---|---|---|---|---|---|
| NCT05354141 | Acute Respiratory Distress Syndrome | 3 | 970 | Recruiting | United States [22] |
| NCT05787288 | COVID-19 Pneumonia | 1 | 240 | Recruiting | China [22] |
| NCT06598202 | Amyotrophic Lateral Sclerosis | 1/2 | 38 | Recruiting | China [22] |
| NCT05261360 | Knee Injury | 2 | 30 | Recruiting | Turkey [22] |
| NCT04223622 | Osteoarthritis | NA | 36 | Completed | Italy [22] |
| NCT06607900 | Neurodegenerative Diseases | 1 | 100 | Not yet recruiting | China [22] |
| NCT05669144 | Myocardial Infarction | 1/2 | 20 | Unknown | Iran [22] |
| NCT05813379 | Skin Rejuvenation | 1/2 | 20 | Recruiting | Iran [22] |
| NCT04173650 | Dystrophic Epidermolysis Bullosa | 1/2 | 10 | Recruiting | United States [22] |
| NCT06812637 | Diabetic Foot Ulcer | 1 | 110 | Completed | Egypt [22] |
The distribution of these trials highlights key therapeutic areas. A significant number focus on inflammatory and immune-mediated conditions such as Acute Respiratory Distress Syndrome and COVID-19 pneumonia, leveraging the potent immunomodulatory properties of MSC-EVs [22]. Another major area is neurological disorders, including amyotrophic lateral sclerosis and neurodegenerative diseases, where the ability of exosomes to cross the blood-brain barrier is a critical advantage [22] [109]. Notably, several trials address tissue repair and regeneration, such as knee injuries, osteoarthritis, and diabetic foot ulcers, which are directly relevant to research on keratinocyte and endothelial cell function in wound healing [22] [31] [110].
Most registered trials are in early phases (Phase 1 or 2), primarily assessing safety, tolerability, and preliminary efficacy. The completion of several studies, such as those for osteoarthritis and diabetic foot ulcers, is expected to provide some of the first controlled human data on MSC-EV therapeutics in the near future [22].
Therapeutic Outcomes and Mechanisms of Action
Documented Outcomes in Clinical Applications
While comprehensive results from large-scale trials are still pending, preliminary reports and data from completed studies indicate promising therapeutic outcomes. In the context of skin repair and wound healing—processes dependent on keratinocyte and endothelial cell function—MSC-Exos have demonstrated significant efficacy.
In radiation-induced skin injury (RISI), which affects up to 95% of radiotherapy patients, stem cell-derived exosomes have emerged as a promising cell-free therapeutic approach [31]. Clinical studies have shown that exosome-based treatments can accelerate wound healing and improve tissue quality in these challenging wounds [31]. The therapeutic effects are mediated through multiple mechanisms, including the reduction of cellular senescence, promotion of angiogenesis, and modulation of inflammation [31].
For diabetic foot ulcers (DFU), a condition characterized by impaired microvascular function, exosome therapy has shown potential to address the underlying pathophysiology. One completed clinical trial (NCT06812637) has evaluated MSC-EVs for this indication, though results are not yet fully published [22]. Preclinical evidence suggests that exosomes promote angiogenesis and improve healing outcomes in diabetic wounds primarily through VEGF, FGF2, miR-126, and activation of the PI3K/Akt pathway [110].
Molecular Mechanisms Relevant to Keratinocytes and Endothelial Cells
The therapeutic effects of MSC-Exos on skin regeneration and wound healing are largely mediated through their direct actions on keratinocytes and endothelial cells:
Promotion of Keratinocyte Proliferation and Re-epithelialization: MSC exosomes are rich in microRNAs that promote epithelial cell migration and proliferation. For instance, miR-135a contained in human amnion MSC exosomes inhibits the Hippo pathway kinase LATS2 in recipient cells [31]. This suppression leads to activation of pro-proliferative YAP/TAZ signaling, thereby enhancing keratinocyte migration [31]. Similarly, exosomal miR-126 promotes the PI3K/Akt and MAPK pathways in skin cells, which are essential for cell survival and proliferation [31].
Angiogenesis and Vascular Protection: Exosomes directly promote the formation of new blood vessels by transferring pro-angiogenic factors to endothelial cells. They have been shown to carry and mediate signaling through Vascular Endothelial Growth Factor (VEGF), FGF2, miR-126, and pathways such as Wnt/β-catenin, Notch, and PI3K/Akt [110]. This promotes endothelial cell proliferation, migration, and tube formation, which is crucial for restoring blood flow to ischemic tissues and supporting wound healing.
The following diagram illustrates the key uptake mechanisms and intracellular signaling pathways activated in recipient keratinocytes and endothelial cells:
Diagram Title: MSC-Exosome Uptake and Signaling in Skin Cells
Experimental Protocols for Uptake Studies
For researchers investigating the fundamental mechanisms of MSC exosome uptake by keratinocytes and endothelial cells, robust and quantitative experimental methodologies are essential. Below are detailed protocols for key assays.
Quantitative Measurement of EV Uptake
A highly sensitive and quantitative method to distinguish EV binding from internalization uses a dual split protein (DSP) system based on Renilla luciferase complementation [111].
Principle: The DSP system consists of two complementary fragments (DSP1 and DSP2) of Renilla luciferase. When DSP1-tagged exosomes are internalized by cells expressing DSP2, the fragments reconstitute functional luciferase, which can be quantified using a cytopermeable substrate.
Protocol Steps:
Generation of DSP-Tagged Exosomes:
- Fuse the human tetraspanins CD9 or CD63 to the DSP1-7 (DSP1) fragment in a lentiviral PLVX Puro vector [111].
- Transduce producer cells (e.g., SUM159 or HEK293) to stably express DSP1-CD9 or DSP1-CD63. Select high-expressing cells via fluorescence-activated cell sorting (FACS) using anti-CD9 or anti-CD63 antibodies [111].
- Culture the transduced cells in EV-depleted medium. Isore the secreted DSP1-tagged exosomes from the conditioned media via ultracentrifugation (120,000 × g overnight) [111].
Preparation of Target Cells:
- Culture target keratinocytes or endothelial cells and transfect or transduce them to express the complementary DSP8-11 (DSP2) fragment [111].
Uptake Assay:
- Incubate target cells expressing DSP2 with DSP1-tagged exosomes.
- After incubation, lyse the cells and measure luciferase activity using a cytopermeable Renilla luciferase substrate (e.g., EnduRen). The signal intensity is directly proportional to the amount of exosome uptake [111].
- To confirm internalization (vs. surface binding), use inhibitors of specific endocytic pathways (e.g., chlorpromazine for clathrin-mediated endocytosis, filipin for caveolae-mediated uptake, etc.) and measure the reduction in luciferase signal [111].
Advantages: This method is highly sensitive, quantitative, allows for dynamic follow-up, and is suitable for high-throughput screening of factors affecting EV uptake [111].
Preconditioning of MSCs to Modulate Exosome Cargo
The therapeutic efficacy of MSC-Exos can be significantly enhanced by preconditioning the parent MSCs to simulate a disease microenvironment, which alters the exosomal cargo and its biological effects [112] [113].
Protocol for Hypoxic and Inflammatory Preconditioning (Generation of Hi-Exos) [113]:
- Cell Culture: Culture MSCs (e.g., from bone marrow or adipose tissue) in standard growth medium until 70-80% confluent.
- Preconditioning Stimulus: Replace the medium with a preconditioning medium. This typically involves:
- Exosome Isolation: After the preconditioning period, collect the conditioned medium. Isolate the exosomes (Hi-Exos) using standard methods such as ultracentrifugation or size-exclusion chromatography [113].
- Characterization: Characterize the isolated Hi-Exos for size (Nanoparticle Tracking Analysis), morphology (Transmission Electron Microscopy), and specific markers (CD63, CD81, TSG101) via western blot. Cargo analysis (e.g., miRNA sequencing) can confirm enrichment of specific miRNAs like miR-221-3p, which has been shown to alleviate cellular senescence in target cells [113].
The Scientist's Toolkit: Key Research Reagents
The following table outlines essential reagents and tools for studying MSC exosome uptake by keratinocytes and endothelial cells.
Table 2: Essential Research Reagents for MSC-Exosome Uptake Studies
| Reagent / Tool | Function / Application | Specific Examples / Notes |
|---|---|---|
| Dual Split Protein (DSP) System [111] | Quantitative measurement of exosome uptake vs. binding. | DSP1 fused to CD9/CD63; DSP2 expressed in target cells. |
| Tetraspanin Antibodies [111] | Characterization of exosomes and FACS sorting of producer cells. | Anti-CD63 (Tea 3/10), Anti-CD9 (VJ1/20). |
| Endocytosis Inhibitors [111] | Elucidating specific uptake pathways. | Chlorpromazine (clathrin), Filipin (caveolae), Dynasore (dynamin). |
| Preconditioning Cytokines [112] [113] | Modulating MSC exosome cargo to enhance therapeutic potential. | IFN-γ, TNF-α (inflammatory priming). |
| Hypoxia Chambers [113] | Simulating the disease microenvironment for MSC preconditioning. | Systems maintaining 1-3% O₂. |
| Ultracentrifugation [109] | Gold standard method for isolating exosomes from conditioned media. | Sequential centrifugation culminating at ≥100,000 × g. |
| Cytopermeable Luciferase Substrate [111] | Detecting internalized exosomes in live cells using the DSP system. | EnduRen. |
| 3D Cell Culture Systems [112] | Culturing MSCs in an environment that more closely mimics in vivo conditions. | Enhances MSC pluripotency and bioactive secretion. |
The clinical landscape for MSC-derived exosomes is maturing rapidly, with a critical mass of early-phase trials underway for conditions spanning inflammatory, neurological, and regenerative medicine. For scientists focused on the fundamental mechanisms of exosome uptake by keratinocytes and endothelial cells, this clinical progress underscores the importance of robust, quantitative experimental methods to elucidate the pathways governing internalization and downstream functional effects. The integration of advanced techniques—such as the DSP system for quantifying uptake and preconditioning strategies to enhance cargo—will be instrumental in bridging the gap between basic research and the development of effective, next-generation acellular therapeutics. The ongoing clinical trials are poised to provide invaluable human data that will further validate and refine these preclinical approaches.
Conclusion
The targeted uptake of MSC exosomes by keratinocytes and endothelial cells is a sophisticated biological process that underpins their remarkable therapeutic potential in regenerative medicine. Understanding the foundational mechanisms, coupled with advanced methodological tools for tracking and enhancement, provides a robust framework for clinical translation. While challenges in production standardization and delivery optimization persist, emerging strategies in exosome engineering and biomaterial integration offer promising solutions. Future research must focus on elucidating the precise signaling networks activated post-uptake, conducting large-scale comparative efficacy studies, and advancing GMP-compliant manufacturing to fully realize the promise of MSC exosomes as a next-generation, cell-free therapeutic for tissue repair and regeneration.
References
- 1. Mesenchymal Stem Cell-Derived Exosomes - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC8393426/]
- 2. Exosomes: A Comprehensive Review for the Practicing ... [https://jcadonline.com/exosomes-comprehensive-review-practing-dermatologists/]
- 3. The role of exosomes in immunopathology and potential ... [https://www.nature.com/articles/s41423-025-01323-5]
- 4. Clinical applications of stem cell-derived exosomes [https://www.nature.com/articles/s41392-023-01704-0]
- 5. Mesenchymal stem cell-derived extracellular vesicles for ... [https://www.nature.com/articles/s41419-022-05034-x]
- 6. MSCs and their exosomes: a rapidly evolving approach in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8642895/]
- 7. Exosome Source Matters: A Comprehensive Review from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11858990/]
- 8. Mesenchymal stem cell-derived exosomes: emerging ... [https://www.sciencedirect.com/science/article/pii/S2773041725000253]
- 9. Extracellular vesicles: innovative cell-free solutions for ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1571461/full]
- 10. The exosome journey: from biogenesis to uptake and ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-021-00730-1]
- 11. Exosomes: Mechanisms of Uptake - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5572985/]
- 12. Drug delivery based exosomes uptake pathways [https://www.sciencedirect.com/science/article/abs/pii/S0197018624001621]
- 13. Exosomes in the Regulation of Vascular Endothelial Cell ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2019.00353/full]
- 14. Exosomes Secreted by Wharton's Jelly-Derived ... [https://www.mdpi.com/1422-0067/25/9/4758]
- 15. Endocytosis pathways of endothelial cell derived exosomes [https://pmc.ncbi.nlm.nih.gov/articles/PMC6465166/]
- 16. Uptake of small extracellular vesicles by recipient cells is ... [https://www.nature.com/articles/s41467-025-57617-9]
- 17. Mechanism of mesenchymal stem cells and exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10232739/]
- 18. Phosphoinositide 3-Kinase Signaling to Akt Promotes ... [https://www.sciencedirect.com/science/article/pii/S0021925820791441]
- 19. Phosphoinositide 3-kinase signaling to Akt promotes ... [https://pubmed.ncbi.nlm.nih.gov/16036919/]
- 20. Shedding light on the role of keratinocyte-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7523352/]
- 21. Regulation of endothelial MAPK/ERK signalling and capillary ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3565781/]
- 22. The tiny giants of regeneration: MSC-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12310703/]
- 23. Signaling pathways and targeted therapies for psoriasis [https://www.nature.com/articles/s41392-023-01655-6]
- 24. Phytoconstituents as modulators of NF-κB signalling [https://www.sciencedirect.com/science/article/pii/S0753332224009429]
- 25. Human Mesenchymal Stromal Cell-Derived Exosomes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8228793/]
- 26. Recent advances in engineered exosome-based therapies for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12276678/]
- 27. Extracellular vesicle-based drug overview: research ... [https://www.nature.com/articles/s41392-025-02312-w]
- 28. Pro-Angiogenic Bioactive Molecules in Vascular Morphogenesis [https://pmc.ncbi.nlm.nih.gov/articles/PMC12564000/]
- 29. Exosome-Mediated Crosstalk between Keratinocytes and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7970718/]
- 30. Extracellular Vesicles in Skin: Biological Function and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12442902/]
- 31. Advances in stem cell-derived exosome therapy for radiation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12626575/]
- 32. Exosomes: The emerging mechanisms and potential ... [https://www.ijbs.com/v20p1778.htm]
- 33. Harnessing exosomal mediators for advanced wound ... [https://www.sciencedirect.com/science/article/abs/pii/S0026286225000809]
- 34. Extracellular Vesicles and Their Role in Skin Inflammatory ... [https://www.mdpi.com/1422-0067/26/8/3827]
- 35. Visualizing Endocytotic Pathways at Transmission Electron ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4289848/]
- 36. of extracellular vesicles: Characterization and cellular... PKH 26 labeling [https://pubmed.ncbi.nlm.nih.gov/29551275/]
- 37. Uptake of Fluorescent Labeled Small Extracellular Vesicles In ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10460254/]
- 38. Systematic Evaluation of PKH Labelling on Extracellular ... [https://www.nature.com/articles/s41598-020-66434-7]
- 39. Effective Visualization and Easy Tracking of Extracellular ... [https://biologicalproceduresonline.biomedcentral.com/articles/10.1186/s12575-019-0092-2]
- 40. Antibodies 101: Introduction to Immunofluorescence [https://blog.addgene.org/introduction-to-immunofluorescence]
- 41. Wound Healing: Harnessing Extracellular Vesicles Derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12587863/]
- 42. Mesenchymal Stem Cell Exosomes Induce Proliferation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4499790/]
- 43. Mesenchymal stem cell-derived small extracellular vesicles ... [https://www.nature.com/articles/s41392-021-00765-3]
- 44. Potential upscaling protocol establishment and wound ... [https://www.nature.com/articles/s41598-025-93219-7]
- 45. BestProtocols: Viability Staining Protocol for Flow Cytometry [https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/protocols/viability-staining-flow-cytometry.html]
- 46. Propidium Iodide Cell Viability Flow Cytometry Protocol [https://www.rndsystems.com/resources/protocols/flow-cytometry-protocol-analysis-cell-viability-using-propidium-iodide]
- 47. Engineered exosomes: a promising drug delivery platform ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12098103/]
- 48. Mesenchymal Stem Cell-Derived Exosomes as Drug ... [https://www.mdpi.com/1422-0067/25/14/7715]
- 49. Mesenchymal stem cell-derived exosomes as a plausible ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1563427/full]
- 50. Therapeutic Potential of Stem Cell-Derived Exosomes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12383377/]
- 51. Exosome as Emerging Therapeutic Platform for Wound ... [https://www.sciencedirect.com/science/article/abs/pii/S0040816625004604]
- 52. Stem cell derived exosome trilogy: an epic comparison of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12186388/]
- 53. Unveiling exosomes in combating skin aging [https://pmc.ncbi.nlm.nih.gov/articles/PMC12395928/]
- 54. Stem Cell-Derived Exosomes: A Comprehensive Review of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12417880/]
- 55. Biomaterial-enhanced delivery of stem cell-derived ... [https://pubmed.ncbi.nlm.nih.gov/40516725/]
- 56. GelMA hydrogel-loaded extracellular vesicles derived from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12167351/]
- 57. Biomimetic scaffolds loaded with mesenchymal stem cells ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-04012-8]
- 58. Cell encapsulated biomaterials for translational medicine - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12596701/]
- 59. Biomaterial-Based Nucleic Acid Delivery Systems for In ... [https://www.mdpi.com/1422-0067/26/15/7384]
- 60. Mesenchymal stem cells in treating human diseases [https://www.nature.com/articles/s41392-025-02313-9]
- 61. What Are Exosomes? The Best Current Evidence (2025) [https://allureaestheticsllc.com/blog/what-are-exosomes-best-evidence/]
- 62. Mesenchymal stem cell-derived extracellular vesicles [https://pmc.ncbi.nlm.nih.gov/articles/PMC12399395/]
- 63. Umbrella review of mesenchymal stem cell-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12554768/]
- 64. Trends in mesenchymal stem cell-derived extracellular ... [https://www.frontiersin.org/journals/medicine/articles/10.3389/fmed.2025.1625787/full]
- 65. Advances in mesenchymal stem cell and exosome-based ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04318-1]
- 66. Mechanism of mesenchymal stem cells and exosomes in ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1181308/full]
- 67. Donor age and breed determine mesenchymal stromal cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11871689/]
- 68. The heterogeneity of mesenchymal stem cells: an important ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-023-03587-y]
- 69. Understanding molecular characteristics of extracellular ... [https://www.sciencedirect.com/science/article/pii/S2773041724000015]
- 70. Trends in mesenchymal stem cell-derived extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488731/]
- 71. Overcoming challenges in MSC-sEV therapeutics [https://www.sciencedirect.com/science/article/pii/S1465324925005912]
- 72. Comparison of production methods for mesenchymal stem ... [https://www.nature.com/articles/s41598-025-21218-9]
- 73. Efficient methods of isolation and purification of extracellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12463815/]
- 74. Exosome Isolation Protocols and Quantification Guidelines [https://www.sepscience.com/exosome-isolation-protocols-and-quantification-guidelines-12144]
- 75. and Mesenchymal : A Novel Therapeutic... Stem Cells Exosomes [https://pmc.ncbi.nlm.nih.gov/articles/PMC10341647/]
- 76. Progress of mesenchymal -derived stem in targeted... cell exosomes [https://cancerci.biomedcentral.com/articles/10.1186/s12935-025-03795-x]
- 77. The tiny giants of regeneration: MSC-derived extracellular ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1612589/full]
- 78. -derived Mesenchymal Hold Promise in the... Stem Cell Exosomes [https://www.dovepress.com/mesenchymal-stem-cell-derived-exosomes-hold-promise-in-the-treatment-o-peer-reviewed-fulltext-article-IJN]
- 79. Extracellular vesicles in chronic wound therapy [https://www.sciencedirect.com/science/article/pii/S2590006425008683]
- 80. Repeatability and reproducibility assessment in a large-scale ... [https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-020-00998-4]
- 81. Diverse mechanisms of bioproduction heterogeneity in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10583207/]
- 82. The investigation of batch-to-batch variabilities in ... [https://www.sciencedirect.com/science/article/abs/pii/S0963996923003435]
- 83. Exploring the challenges faced by generic version of complex ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12482112/]
- 84. Standardized Reporting of Research on Exosomes to Ensure ... [https://pubmed.ncbi.nlm.nih.gov/38888007/]
- 85. Direct Experimental Evidence for Selective Uptake of Function ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12538810/]
- 86. Cell and molecular mechanisms of keratinocyte function ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2631465/]
- 87. Keratinocyte-derived VEGF-A is an essential pro-migratory ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1601887/full]
- 88. Enhanced Skin Wound Healing Through Chemically Modified ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12050261/]
- 89. Exosomes derived from human placental mesenchymal stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10506088/]
- 90. Keratin biomaterials for wound healing and tissue ... [https://advances.umw.edu.pl/en/article/2025/34/8/1241/]
- 91. Application of stem cell-derived exosomes in ischemic ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-021-02863-w]
- 92. A prospective multicenter phase III clinical trial evaluating ... [https://www.nature.com/articles/s41598-025-88150-w]
- 93. Effect of Exosomes From Bone Marrow–Derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10768594/]
- 94. Adipose-Derived Stem Cells: Angiogenetic Potential and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10889096/]
- 95. Angiogenic Effects and Crosstalk of Adipose-Derived ... [https://www.mdpi.com/1422-0067/22/19/10890]
- 96. Comparison of the ability of exosomes and ectosomes derived ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-03632-4]
- 97. Differential Therapeutic Effect of Extracellular Vesicles ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8068154/]
- 98. Small extracellular vesicles from human bone marrow ... [https://www.nature.com/articles/s41598-025-04057-6]
- 99. Conversion of human adipose-derived stem cells into ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-018-1088-6]
- 100. Endothelial differentiation of adipose-derived ... [https://www.sciencedirect.com/science/article/pii/S017193351200163X#!]
- 101. Concise Review: MSC -Derived Exosomes for Cell -Free Therapy [https://pubmed.ncbi.nlm.nih.gov/28294454/]
- 102. Frontiers | Optimizing therapeutic outcomes: preconditioning strategies... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1509418/full]
- 103. Stem cell derived exosome trilogy: an epic comparison of human... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04440-0]
- 104. Unveiling exosomes in combating skin aging: insights into ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04620-y]
- 105. Exploring the therapeutic potential of MSC-derived ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04616-8]
- 106. Current status of nano-embedded growth factors and stem ... [https://www.sciencedirect.com/science/article/pii/S2214031X24001657]
- 107. Synthetic Nanoparticles 2025-2033 Overview: Trends, ... [https://www.archivemarketresearch.com/reports/synthetic-nanoparticles-406477]
- 108. Acellular Therapy Market - Global Forecast 2025-2032 [https://www.researchandmarkets.com/reports/5924748/acellular-therapy-market-global-forecast?srsltid=AfmBOooV9DkXYSO54SmtK2Ww7ujMs0s-faNxvkpoy_-AxVmNUiPQgLYR]
- 109. Mesenchymal stem cell-derived extracellular vesicles [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1626996/full]
- 110. Exosomes: the future of acellular nanotherapeutics in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12443856/]
- 111. Development of a quantitative method to measure EV uptake [https://www.nature.com/articles/s41598-019-47023-9]
- 112. a precise treatment strategy of mesenchymal stem cells ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1609288/full]
- 113. Exosomes derived from MSCs exposed to hypoxic and ... [https://www.sciencedirect.com/science/article/pii/S2452199X25001021]